Abstract
Sinorhizobium meliloti is a soil bacterium that fixes nitrogen after being established inside nodules that can form on the roots of several legumes, including Medicago truncatula. A mutation in an S. meliloti gene (lpsB) required for lipopolysaccharide synthesis has been reported to result in defective nodulation and an increase in the synthesis of a xylose-containing glycan. Glycans containing xylose as well as arabinose are also formed by other rhizobial species, but little is known about their structures and the biosynthetic pathways leading to their formation. To gain insight into the biosynthesis of these glycans and their biological roles, we report the identification of an operon in S. meliloti 1021 that contains two genes encoding activities not previously described in bacteria. One gene encodes a UDP-xylose synthase (Uxs) that converts UDP-glucuronic acid to UDP-xylose, and the second encodes a UDP-xylose 4-epimerase (Uxe) that interconverts UDP-xylose and UDP-arabinose. Similar genes were also identified in other rhizobial species, including Rhizobium leguminosarum, suggesting that they have important roles in the life cycle of this agronomically important class of bacteria. Functional studies established that recombinant SmUxs1 is likely to be active as a dimer and is inhibited by NADH and UDP-arabinose. SmUxe is inhibited by UDP-galactose, even though this nucleotide sugar is not a substrate for the 4-epimerase. Unambiguous evidence for the conversions of UDP-glucuronic acid to UDP-α-d-xylose and then to UDP-β-l-arabinose (UDP-arabinopyranose) was obtained using real-time 1H-NMR spectroscopy. Our results provide new information about the ability of rhizobia to form UDP-xylose and UDP-arabinose, which are then used for the synthesis of xylose- and arabinose-containing glycans.
INTRODUCTION
The plant symbiont Sinorhizobium meliloti 1021 is a Gram-negative soil bacterium that induces the formation of nitrogen-fixing nodules on the roots of leguminous plants, including alfalfa, yellow sweet clover and fenugreek (Galibert et al., 2001; van Rhijn & Vanderleyden, 1995). Nodule formation is a multi-step process that is initiated when the bacteria attach to a root hair and cause it to curl. A tubular infection thread is then formed by invagination of the host root cell membrane and allows the bacteria to pass through the root cortex and reach newly formed meristematic cells. These cells then develop into the nodule that contains the nitrogen-fixing bacteroids, which convert atmospheric nitrogen to ammonia and allow the plants to grow in the absence of added nitrogen (Gage, 2004; Jones et al., 2007). There is increasing evidence that changes in the structures and types of the rhizobial cell surface polysaccharides during infection and penetration are required for successful nodulation.
The rhizobial cell surface contains several structurally complex carbohydrates, including extracellular polysaccharides (EPSs), lipopolysaccharides (LPSs), K antigens (capsular polysaccharides; CPSs) and cyclic β-glucans (Carlson et al., 1999). Mutations that impair the production and structure of LPS, CPS and EPS typically affect infection and nodulation (Hirsch, 1999). For example, Campbell et al. (2002) have reported that a loss of function mutation in the S. meliloti lpsB gene impairs host plant nodulation and leads to the synthesis of an altered LPS containing 40-fold more xylose (Xyl) residues than the wild-type. However, the biochemical basis for the increased amounts of Xyl residues and the biological function of the altered LPS are unknown.
Several studies have shown that rhizobia synthesize glycans that contain Xyl and arabinose (Ara). For example, polymers containing these pentoses as well as rhamnose (Rha), galactose (Gal) and glucose (Glc) were identified in four rhizobia species (De Leizaola & Dedonder, 1955; Humphrey & Vincent, 1959). Wild-type and mutant strains of Bradyrhizobium japonicum grown on gluconate or mannitol produce an EPS comprised of Glc, Gal, Xyl and glucuronic acid (GlcA) (Karr et al., 2000). The LPS isolated from R. leguminosarum RBL5523 contains Xyl, 3-deoxy-d-manno-2-octulosonic acid (Kdo), galacturonic acid (GalA), mannose (Man), Glc, N-acetyl quinovoamine (QuiNAc) and N-acetylglucosamine (GlcNAc). The LPS of wild-type B. japonicum (Puvanesarajah et al., 1987) contains fucose (Fuc), Xyl, Ara, Man, Glc, N-acetylfucosamine, QuiNAc, GlcNAc and Kdo. However, a non-nodulating B. japonicum mutant, HS123, forms LPS that lacks Xyl and Ara. In addition, partially purified LPS isolated from R. leguminosarum 3841 has been reported to contain a glycan named xylomannan, which consists of Man, Xyl and Glc (Forsberg & Carlson, 2008). Moreover, the ratio between Xyl and Man is affected by growth conditions (Kannenberg & Brewin, 1989; Kannenberg et al., 1994; Kannenberg & Carlson, 2001). Taken together, these observations suggest that rhizobia change their cell surface glycans to adapt to changes in their environment. Such changes may be critical for the survival of a rhizobium as a ‘free-living’ soil bacterium or during its interactions with the host plant root. However, little is known about the biochemical pathways and the corresponding genes involved in these processes. Biosynthesis of glycans requires glycosyltransferases, a class of enzymes that transfer sugar from an activated donor, usually a nucleotide sugar, to an appropriate glycan acceptor, leading to extension of the glycan. To identify genes involved in the synthesis of Ara- and Xyl-containing glycans we decided to investigate nucleotide–sugar biosynthetic genes in rhizobia species.
UDP-Xyl is the sugar donor for the synthesis of diverse Xyl-containing glycans in animals (Götting et al., 2000; Kuhn et al., 2001), fungi (Klutts & Doering, 2008) and plants (Egelund et al., 2006; Peña et al., 2007). UDP-Xyl is formed from UDP-GlcA by a decarboxylation reaction catalysed by UDP-xylose synthase (Uxs) (Bar-Peled et al., 2001; Harper & Bar-Peled, 2002; Hwang & Horvitz, 2002; Pattathil et al., 2005; Zhang et al., 2005). To date, UDP-Ara, which is formed from UDP-Xyl in a reversible reaction catalysed by UDP-xylose 4-epimerase (Uxe) (Burget & Reiter, 1999), has only been identified in green plants, where it is a donor for the synthesis of pectic and hemicellulosic polysaccharides, proteoglycans and glycoproteins (Porchia et al., 2002; Konishi et al., 2007). In contrast, little is known about the formation of activated Xyl and Ara in bacteria and their subsequent use in the synthesis of Xyl- or Ara-containing glycans.
Here we describe the functional characterization of two S. meliloti 1021 genes that encode Uxs and Uxe (Fig. 1a). Such enzyme activities have not, to our knowledge, previously been described in bacteria, and thus our data provide the basis for understanding the formation of Xyl- and Ara-containing glycans by rhizobia and their roles in the nodulation of legumes.
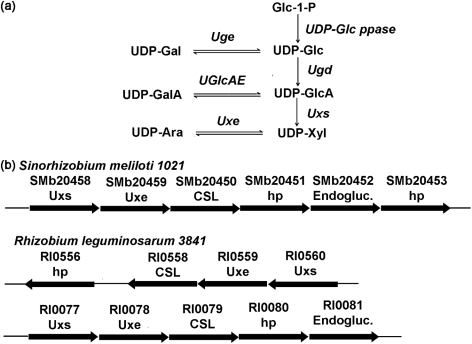
Fig. 1.
Biosynthesis of UDP-Xyl and UDP-Ara in rhizobia. (a) In rhizobia, UDP-glucose pyrophosphorylase (UDP-Glc ppase) converts glucose 1-phosphate (Glc-1-P) and UTP to UDP-Glc. UDP-Glc and UDP-Gal are interconverted by UDP-glucose 4-epimerase (Uge) [exoB (Sánchez-Andújar et al., 1997)]. UDP-glucose 6-dehydrogenase (Ugd) [exo5 (Laus et al., 2004)] converts UDP-Glc and NAD+ to UDP-GlcA and NADH. UDP-GlcA and UDP-GalA are interconverted by UDP-glucuronic acid 4-epimerase (UGlcAE) [lpsL (Keating et al., 2002)]. UDP-GlcA is decarboxylated to give UDP-Xyl by UDP-xylose synthase (Uxs; this study). UDP-Xyl and UDP-Ara are interconverted by UDP-xylose 4-epimerase (Uxe; this study). (b) Organization of some of the operons containing Uxs and Uxe genes in S. meliloti 1021 and R. leguminosarum 3841. The locus number for each gene within the operon is shown and the annotated function is indicated below. The biochemical functions of SMb20458 and SMb20459 based on this report are UDP-xylose synthase (SmUxs1) and UDP-xylose 4-epimerase (SmUxe), respectively. The R. leguminosarum 3841 genes Rl0560 and Rl0077 encode active Uxs, and Rl0559 and Rl0078 encode active Uxe (S. Roy, X. Gu & M. Bar-Peled, unpublished data). hp, hypothetical protein; Endogluc., endoglucanase.
METHODS
Cloning of SmUxs1 and SmUxe from S. meliloti 1021.
Total DNA was isolated from a two-day-old culture of S. meliloti 1021 grown in LB liquid medium (2 ml) using phenol/chloroform (Syn & Swarup, 2000). The coding sequences of SMb20458 (herein named UDP-xylose synthase, SmUxs1) and SMb20459 (herein named UDP-xylose 4-epimerase, SmUxe) were PCR-amplified using 1 unit of proof-reading Platinum Taq DNA polymerase high-fidelity (Invitrogen), 0.2 μM of each forward and reverse primer (for Uxs1, 5′-TCATGAATTATTTTAGAAATGACTTCAG-3′ and 5′-AAGCTTGACCAGCTCCGCCTTTCC-3′; for Uxe, 5′-CCATGGTTGCGCCACGTATCCTCGTC-3′ and 5′-GGATCCAAGCTTTGACCGGACCTCCAGC-3′), 200 μM dNTPs and S. meliloti 1021 DNA as template. Each PCR product was separated by agarose gel electrophoresis, extracted and then cloned to generate plasmids pGEM-T : SmUxs1#2 and pCR4 : SmEPI#2, respectively. Following DNA sequence analyses and subsequent biochemical characterization (see below) of the cloned genes, they were annotated as SmUxs1 and SmUxe, and their sequences were deposited in GenBank. The BspHI–HindIII fragment (1053 bp) containing the full-length SmUxs1 gene and the NcoI–HindIII fragment (986 bp) containing the full-length SmUxe, both without stop codons, were cloned into an Escherichia coli expression vector (pET28b : SmUxs1#1 and pET28b : SmUxe#1) for the production of recombinant proteins with a six-histidine extension at the C terminus.
Expression and purification of recombinant SmUxs1 and SmUxe.
E. coli cells containing pET28b : SmUxe#1 or an empty vector (pET28b) were cultured for 16 h at 37 °C in LB medium (10 ml) supplemented with kanamycin (50 μg ml−1) and chloramphenicol (34 μg ml−1). A portion (8 ml) of the cultured cells was transferred into fresh LB liquid medium (250 ml) supplemented with the same antibiotics, and the cells were then grown at 37 °C at 250 r.p.m. until the cell density reached OD600 0.8. Gene expression was then induced by the addition of IPTG (to 0.5 mM), and the cells were then grown for 4 h at 30 °C at 250 r.p.m. Overnight cultures of cells harbouring pET28b : SmUxs1#1 or an empty vector (pET28) were transferred into 190 ml fresh E. coli Expression Medium (Invitrogen) supplemented with kanamycin (50 μg ml−1) and chloramphenicol (34 μg ml−1), and grown for 24 h at 30 °C at 250 r.p.m. The induced cells were collected by centrifugation (6000 g for 10 min at 4 °C), suspended in 20 ml lysis buffer [50 mM sodium phosphate, pH 7.5, containing 10 % (v/v) glycerol, 1 mM EDTA, 1 mM DTT and 0.5 mM PMSF] and lysed in an ice bath by sonication (24 cycles of 10 s on, 20 s off) using a Misonix S-4000 sonicator (Misonix) equipped with a microtip probe. The lysed cells were centrifuged at 6000 g for 10 min at 4 °C. The supernatant was supplemented with 1 mM DTT and centrifuged again (30 min at 20 000 g). The resulting supernatant (termed S20) was recovered and kept at −20 °C. His-tagged proteins were purified using a Ni-Sepharose fast-flow column [2 ml resin (GE Healthcare Life Sciences) that had been packed in a 10 mm internal diameter×150 mm column]. The column was pre-equilibrated with loading buffer [50 mM sodium phosphate (pH 7.5), 0.3 M NaCl]. The bound His-tagged protein was eluted with the same buffer containing an increasing amount of imidazole (10–250 mM). Fractions containing the desired enzymic activity were pooled, supplemented with 10 % (v/v) glycerol and 5 mM DTT, and then dialysed (6000–8000 molecular mass cut-off, Spectrum Laboratories) three times for 30 min at 4 °C against 50 mM sodium phosphate, pH 7.5, containing 0.1 M NaCl, 10 % (v/v) glycerol and 5 mM DTT. The dialysate was flash-frozen in liquid nitrogen and stored at −80 °C. Proteins extracted from E. coli cells expressing the empty vector were obtained using the same column purification protocol and were used as controls in enzyme assays and SDS-PAGE analyses. Protein concentrations were determined using the Bradford reagent with BSA as standard. The native molecular mass of each recombinant protein was estimated by size-exclusion chromatography, as described previously (Gu et al., 2010).
Uxs, Uxe enzyme assays, HPLC and NMR product analyses.
Standard Uxs reactions (50 μl final volume) contained 50 mM sodium phosphate (pH 7.6), 1 mM NAD+, 1 mM UDP-GlcA and 0.3 μg purified recombinant SmUxs1. Uxe reactions (50 μl final volume) contained 50 mM sodium phosphate (pH 7.6), 0.5 mM NAD+, 0.5 mM UDP-Xyl and 0.04 μg purified recombinant SmUxe. Reactions were kept for up to 30 min (Uxs) or 10 min (Uxe) at 37 °C and then terminated by heating for 45 s at 100 °C. Chloroform (50 μl) was then added and the mixture was vortexed for 30 s. The suspension was centrifuged at room temperature for 5 min at 14 000 g and the upper aqueous phase was collected. Reaction products for SmUxs1 were chromatographed on a Q15 anion-exchange column (2 mm×250 mm, Amersham) using an Agilent 1100 Series HPLC system equipped with an autosampler, diode-array detector and ChemStation software. Reaction products for SmUxe were chromatographed on a Prep C18 Scalar column (4.6 mm internal diameter×250 mm, Agilent) at a flow rate of 1 ml min−1 using a gradient from solution A (20 mM tert-butylamine-H3PO4, pH 6.6, 2 %, v/v, acetonitrile) to solution B (20 mM tert-butylamine-H3PO4, 20 %, v/v, acetonitrile) over 35 min. Nucleotides were detected by their UV absorbance, and the maximum absorbances for UDP-sugars and NAD+ were at 261 and 259 nm, respectively. The amount of product formed was determined using calibration curves of standard UDP-Xyl and UDP-GlcA. The products formed by the Uxs reaction (eluted at 16.8 min from Q-column) and the Uxe reaction (eluted at 8.2 min from the C18 column) were collected, lyophilized, dissolved in 99.9 % D2O and analysed by 1H-NMR spectroscopy. UDP-Xyl and UDP-Ara were purchased from CarboSource Services (CCRC); UDP-GlcA, UDP-Glc and NAD+ were obtained from Sigma.
Real-time 1H-NMR enzymic reaction assays (200 μl) were performed at 37 °C in a final mixture of D2O : H2O (1 : 9, v/v), and contained 50 mM sodium phosphate, pH/pD 7.6, 1 mM UDP-GlcA, 1 mM NAD+ and 1.2 μg recombinant SmUxs1. The NMR assay for SmUxe (0.16 μg) used the same buffer, with NAD+ and 1 mM UDP-Xyl. Real-time 1H-NMR spectra were obtained using a Varian DirectDrive spectrometer operating at 900 or 600 MHz. Data acquisition was started between 5 and 10 min after the addition of enzyme to allow spectrometer acquisition conditions to be optimized. Sequential 1D proton spectra with pre-saturation of the water resonance were acquired over the course of the enzymic reaction. All chemical shifts (p.p.m.) are referenced to 2,2-dimethyl-2-silapentane-5-sulphonate.
Characterization and kinetic analyses of the recombinant enzymes.
SmUxs1 and SmUxe activities were determined using different buffers, different temperatures, and in the presence of selected cations or potential inhibitors. For optimal pH studies, solutions of recombinant SmUxs1 (0.3 μg) were mixed in each individual buffer (50 mM). Subsequently, NAD+ (1 mM) and UDP-GlcA (1 mM) were added and reactions were incubated for 30 min at 37 °C. Inhibition assays were performed by first mixing the enzyme and buffer with various additives (e.g. nucleotides) on ice for 10 min. UDP-GlcA and NAD+ (1 mM each) were then added. After 30 min at 37 °C, the amount of UDP-Xyl formed was determined by Q15 anion-exchange chromatography. Optimal temperature studies were performed using standard conditions for 30 min at different temperatures, and the amount of UDP-Xyl was determined by HPLC. SmUxe (0.04 μg protein) was characterized in a similar manner using 10 min reactions with NAD+ (0.5 mM) and UDP-Xyl (0.5 mM), and the products were analysed by reverse-phase HPLC.
The catalytic activity of SmUxs1 (0.3 μg) or SmUxe (0.04 μg) was determined at 37 °C for 8 min (Uxs) or 5 min (Uxe) in 50 mM sodium phosphate, pH 7.6, containing 1 mM NAD+, with variable concentrations (0.08–1 mM) of UDP-GlcA (Uxs) or UDP-Xyl (Uxe). Km values were calculated from a plot of the reciprocal initial velocity against the reciprocal UDP-sugar concentration.
RESULTS
Identification and cloning of S. meliloti 1021 UDP-xylose synthase 1 (SmUxs1) and UDP-xylose 4-epimerase (SmUxe)
blast searches using the amino acid sequence of Arabidopsis UDP-xylose synthase (AtUxs3, GenBank accession number AF387789) identified two S. meliloti 1021 Uxs homologues (SmUxs1 and SmUxs2; see Supplementary Fig. S1). Both homologues reside on the 1.68 Mb pSymb megaplasmid of S. meliloti 1021. SmUxs1 has 59, 55 and 57 % amino acid sequence identity to known plant, fungal and human Uxs, respectively. The operon containing SmUxs1 (SMb20458) has an additional gene (SMb20459, herein named SmUxe). This gene encodes a protein with 41, 42 and 44 % amino acid sequence identity to human, yeast and Burkholderia UDP-galactose 4-epimerases, respectively, and 42 % amino acid sequence identity to the Arabidopsis UDP-xylose 4-epimerase (MUR4, At1g30620; Burget et al., 2003) (Supplementary Fig. S2). Such sequence identity, however, is insufficient to predict the function of SMb20459. Nonetheless, a comparison of the SmUxs1 operon with other sequenced rhizobial genomes identified similar genes and operons (see Fig. 1b), suggesting the widespread occurrence of these genes.
To determine whether the S. meliloti homologues (SmUxs1 and SmUxe) function as a UDP-xylose synthase and as a UDP-sugar epimerase, we cloned and expressed both genes in E. coli and then determined the biochemical properties of the recombinant proteins.
SmUxs1 encodes active UDP-xylose synthase; SmUxe encodes active UDP-xylose 4-epimerase
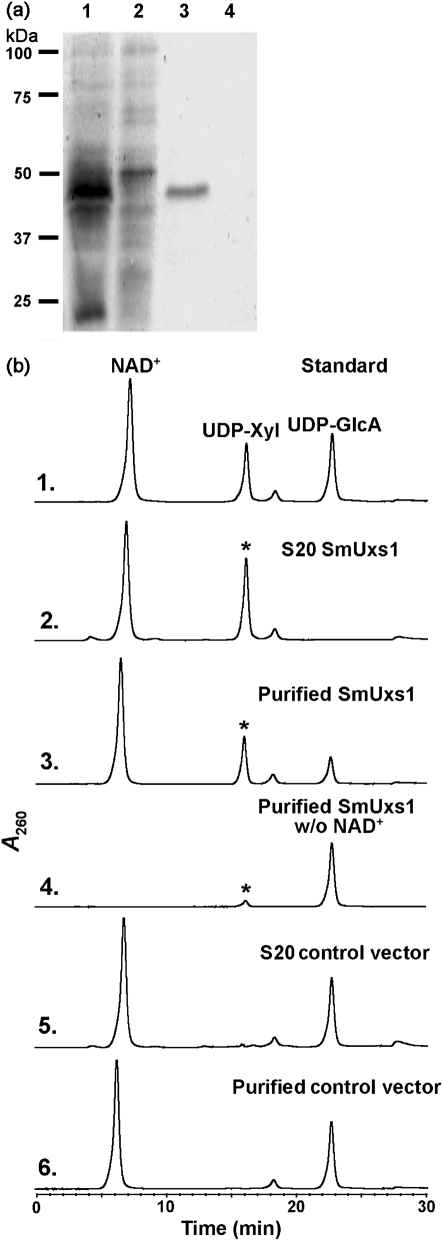
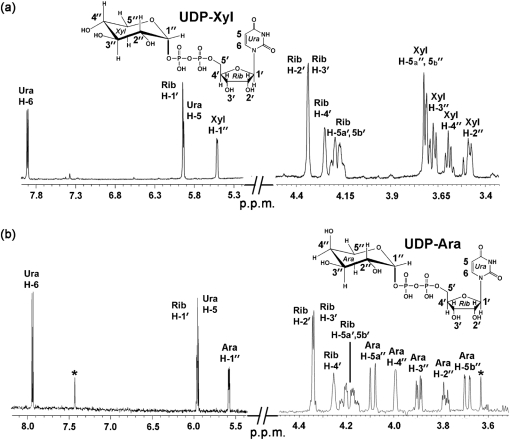
A clearly visible protein band (∼40 kDa) was detected by SDS-PAGE analysis of the extracts from E. coli cells expressing recombinant SmUxs1 (Fig. 2a, lane 1) but was absent in E. coli cells expressing the empty vector. The expressed protein was purified (Fig. 2a, lane 3) and shown, using an HPLC-based assay, to convert UDP-GlcA to a new UDP-sugar in the presence of NAD+. The newly formed UDP-sugar had the same retention time (16.8 min) as the UDP-Xyl standard (Fig. 2b, panel 3, indicated by an asterisk), and its 1H-NMR spectrum (Fig. 3a, Supplementary Table S1) was consistent with UDP-Xyl. The distinct chemical shift (Supplementary Table S1) of H″1 at 5.51 p.p.m. and the coupling constant value of 7.0 Hz for J1′,P show the linkage of the anomeric Xyl residue with the phosphate of UDP. The coupling constant value of 3.4 Hz for J1′,2′ and the J2′,3′, J3′,4′ values of 9.5 and 9.5 Hz, respectively, indicate an α-xylopyranose configuration. The collective analyses thus confirm that the recombinant SmUxs1 is a UDP-xylose synthase, as illustrated in Fig. 1(a).
Fig. 2.
Expression and characterization of recombinant SmUxs1. (a) SDS-PAGE of total soluble protein isolated from E. coli cells expressing SmUxs1 (lane 1), control empty vector (lane 2), and column-purified SmUxs1 or control (lanes 3 and 4, respectively). (b) High-performance anion-exchange chromatography of the products formed by SmUxs1. Purified recombinant SmUxs1 was reacted with UDP-GlcA for 45 min in the presence (panel 3) or absence (panel 4) of added NAD+. The corresponding column-purified protein isolated from cells expressing the empty vector was reacted with UDP-GlcA and NAD+ for 45 min (panel 6) as a control. The reaction products were separated on a Q15 anion-exchange column. The distinct UDP-sugar peak indicated by an asterisk (panels 2 and 3 with the same retention time as UDP-Xyl) was collected and characterized by 1H-NMR spectroscopy. The activity of total soluble protein (S20) isolated from cells expressing recombinant SmUxs1 (panel 2) or vector control (panel 5) is shown.
Fig. 3.
1H-NMR spectroscopic analyses of the UDP-α-d-xylose and UDP-β-l-arabinose formed by SmUxs1 and SmUxe, respectively. (a) The peak (Fig. 2b, panel 2, marked by an asterisk) corresponding to the product formed by SmUxs1 was collected and analysed by 1H-NMR spectroscopy at 600 MHz. Portions of the NMR spectrum between 3.4–4.5 and 5.2–8.0 p.p.m. are shown. The assigned signals have the symbol H for the uracil protons (Ura), the symbol H′ for the ribose (Rib) protons, and the symbol H″ for the xylose (Xyl) protons. (b) The Uxe enzymic product (Fig. 4b, panel 2, marked with an asterisk) was collected from the column, lyophilized and analysed by 1H-NMR at 25 °C. The 600 MHz NMR spectrum of the product is shown between 3.5–4.5 and 5.4–8.2 p.p.m. The assigned signals have the symbol H for the uracil protons (Ura), the symbol H′ for the ribose (Rib) protons, and the symbol H″ for the arabinose (Ara) protons.
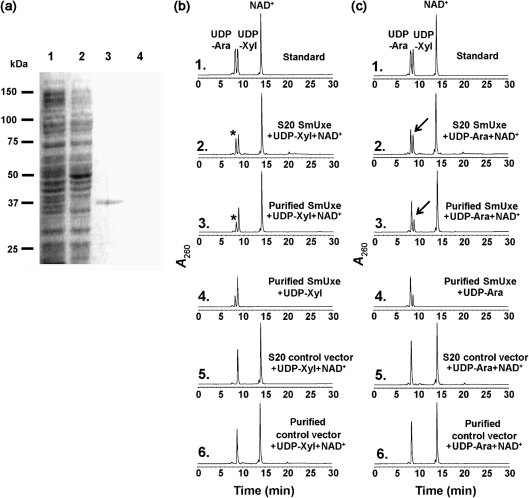
We next analysed the enzymic function of recombinant SmUxe. The protein was expressed and purified from E. coli (Fig. 4a, lanes 1 and 3). Initial HPLC-based assays determined that SmUxe was unable to epimerize UDP-Glc or UDP-Gal, even when the reactions were performed for extended times (data not shown). However, SmUxe readily converted UDP-Xyl to a new UDP-sugar that had the same retention time (8.2 min) as UDP-Ara (see Fig. 4b, panels 2 and 3). Proteins isolated from E. coli cells expressing empty vector had no activity (Fig. 4b, panels 5 and 6). The 1H-NMR spectrum of the product eluting at 8.2 min (Fig. 3b, Supplementary Table S1) contained a signal for an anomeric proton at 5.59 p.p.m. (J1′,2′ 3.4 Hz; J1′,P 6.8 Hz), and had J2′,3′ (10 Hz), J3′,4′ (3.4 Hz), J4,5a′ (1.8 Hz), J4,5b′ (<1 Hz), and J5a′,5b′ (12.9 Hz) values (Fig. 4b, panel 3) that were consistent with UDP-β-l-arabinopyranose (UDP-Ara). SmUxe also converted UDP-Ara to a peak that eluted at 8.9 min (Fig. 4c, panels 2 and 3). The 1H-NMR spectrum of this product (Fig. 4c, panel 3) was identical to that of UDP-Xyl. Together, these data establish that recombinant SmUxe is a 4-epimerase that interconverts UDP-d-xylopyranose and UDP-β-l-arabinopyranose (Fig. 1a, Supplementary Table S1). Thus, we conclude that S. meliloti has genes that encode proteins forming UDP-Xyl and UDP-Ara from UDP-GlcA.
Fig. 4.
Expression and characterization of recombinant SmUxe. (a) SDS-PAGE of total soluble protein isolated from E. coli cells expressing SmUxe (lane 1), control empty vector (lane 2), and column-purified SmUxe or control (lanes 3 and 4, respectively). (b) High-performance reverse-phase chromatography of the products formed by SmUxe reacting with UDP-Xyl. Purified recombinant SmUxe was reacted with UDP-Xyl for 45 min in the presence (panel 3) or absence (panel 4) of added NAD+. The purified protein isolated from cells expressing control empty vector was incubated with UDP-Xyl and NAD+ for 45 min (panel 6) and is a control. The distinct UDP-sugar peak marked by the asterisk (panels 2 and 3 with the same retention time as UDP-Ara) was collected and analysed by 1H-NMR spectroscopy. The activity of total soluble protein (S20) isolated from cells expressing recombinant SmUxe (panel 2) or vector control (panel 5) is shown. (c) High-performance reverse-phase chromatography of the products formed by SmUxe reacting with UDP-Ara. Purified recombinant SmUxe was reacted with UDP-Ara for 45 min in the presence (panel 3) or absence (panel 4) of exogenous NAD+. The purified protein isolated from cells expressing control empty vector was incubated with UDP-Xyl and NAD+ for 45 min (panel 6) and is a control. The peak marked by the arrows (panels 2 and 3 with the same retention time as UDP-Ara) was collected and analysed by 1H-NMR spectroscopy. The activity of total soluble protein (S20) isolated from cells expressing recombinant SmUxe (panel 2) or vector control (panel 5) is shown.
Real-time NMR assays of SmUxs1 and SmUxe activity
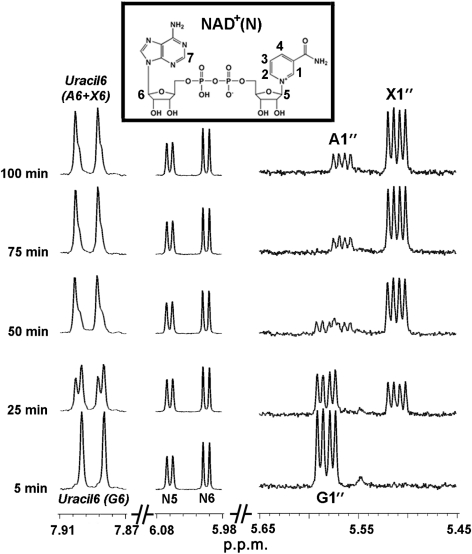
Time-resolved NMR-based assays were performed with SmUxs1 or SmUxe and their corresponding substrates to monitor product formation and the presence of intermediates. The enzymic progression of SmUxs1, as monitored by analysing selected regions of the NMR spectra, is shown in Fig. 5. The characteristic NMR signal having a quadruplet peak that corresponds to the C1-anomeric proton of UDP-GlcA (G) (labelled G1′ at 5.58 p.p.m. in Fig. 5b; and in an expansion of this region in Fig. 5c) was decreased, and simultaneously the quadruplet peak signal corresponding to the UDP-Xyl anomeric proton (X1′, 5.51 p.p.m.) was increased during the time-course of the Uxs reaction. No variation was observed for the peaks corresponding to NAD+ (N) protons, as expected. In addition, performing the assay at a higher field strength (900 MHz) led to a better resolution of signals and product identification, showing clear differences in the chemical shifts for the proton attached to the C-6 of uracil during the formation of UDP-Xyl from UDP-GlcA (Fig. 5d, marked by X6 and G6, respectively).
Fig. 5.
Real-time 1H-NMR analyses of dual-enzyme (SmUxe and SmUxs1) activities. SmUxe and SmUxs1 were mixed with UDP-GlcA and NAD+. Approximately 5 min after enzyme addition and NMR shimming, NMR data were collected at 37 °C using the 600 MHz spectrometer (bottom trace). Selected regions of the spectra acquired between 5 and 100 min are shown. 1H-NMR signals for the H-1 proton of UDP-GlcA, UDP-Xyl and UDP-Ara (from 5.45 to 5.65 p.p.m., quadruplet peak) are labelled as G1″, X1″ and A1″, respectively. The N5, N6 protons of NAD+ (5.98–6.08 p.p.m.) are labelled arbitrarily as shown in the insert. Peaks marked by Uracil6 (A6+X6) (7.87–7.91 p.p.m.) are the mixture of signals from the H-6 proton of the uracil of UDP-Xyl and UDP-Ara.
The real-time 1H-NMR spectra the of SmUxe assay (see Supplementary Fig. S3) revealed that the Xyl H-1 signal of UDP-Xyl (X1′) decreased with a concomitant increase in the Ara H-1 signal of UDP-Ara (A1′). The appearance of signals with chemical shifts corresponding to H-5 (A5a′, 3.71 p.p.m.) and H-4 (A4′, 4.01 p.p.m.) of the arabinosyl moiety provides additional evidence for the formation of UDP-Ara. In addition, NMR established that NAD+ is not released from the epimerase during the C-4 oxidation (NAD+→NADH)/reduction (NADH→NAD+) cycle.
We then used real-time 1H-NMR to monitor the product formation when SmUxs1 and SmUxe were reacted together with UDP-GlcA. The conversion of UDP-GlcA to UDP-Xyl was immediately apparent (Supplementary Fig. S4; 5.6–5.5 p.p.m.). As the reaction progressed, the UDP-Xyl was inter-converted to UDP-Ara, until UDP-GlcA was completely consumed and an equilibrium between UDP-Xyl and UDP-Ara was achieved. At equilibrium, the ratio of UDP-Ara to UDP-Xyl was 0.8 (Table 1). Overall, the real-time NMR-based enzyme assay provides a powerful analytical tool to observe the transformation of nucleotide sugars.
Table 1.
Enzymic properties of recombinant SmUxs1 and SmUxe
| Property | SmUxs1 | SmUxe |
|---|---|---|
| Optimal pH | 7.6–8.4 | 7.6–8.0 |
| Optimal temperature (°C)* | 37 | 37 |
| Km (mM)† | 0.095±0.003 | 0.31±0.05 |
| Vmax (μM s−1) | 0.25±0.01 | 0.25±0.04 |
| kcat (s−1) | 1.7±0.1 | 13.0±1.9 |
| kcat/Km (mM−1 s−1) | 18.3±0.2 | 42.1±1.7 |
| Mass of active protein/monomer (kDa)‡ | 75/40.5 | 66/36.7 |
| Equilibrium constant§ | – | 0.80±0.01 |
*Optimal temperature assays were conducted in phosphate buffer.
†The reciprocal initial velocity was plotted against the reciprocal UDP-sugar concentration according to Lineweaver and Burk to calculate the corresponding Km values. The data presented are the mean Km values from three experiments.
‡The mass of active SmUxs1 or SmUxe eluted from a Superdex-200 gel filtration column (28.7 or 29.3 min, respectively) was estimated based on extrapolation (R2=0.981) from the relative time for a standard protein marker.
§The ratio between UDP-Ara and UDP-Xyl for SmUxe activity was determined after a 4 h assay.
Characterization and properties of recombinant SmUxs1 and SmUxe
The recombinant SmUxs1 gene encodes a polypeptide with a molecular mass of ∼40 kDa. However, the functional SmUxs1 protein elutes from a size-exclusion column in the region for a ∼75 kDa protein (Table 1), suggesting that it exists as a dimer. The SmUxe polypeptide (∼37 kDa) also eluted from the column as a dimer (Table 1). Kinetic data for SmUxs1 and SmUxe are summarized in Table 1. SmUxs1 had an apparent Km of 94.8 μM (UDP-GlcA) and a kcat/Km value (s−1 mM−1) of 18.3. SmUxe had an apparent Km value of 0.31 mM for UDP-Xyl and a kcat/Km value (s−1 mM−1) of 42.1 (Table 1).
Recombinant SmUxs1 and SmUxe had optimal pH values of 8.0 and 7.6, respectively, in phosphate buffer, and their optimal temperature was 37 °C (Table 1). Both enzymes were fully active in the presence of EDTA (data not shown), suggesting that metal ions are not required for SmUxs1 and SmUxe activity. Similar to the Uxs1p from the fungus Cryptococcus neoformans (Bar-Peled et al., 2001) but distinct from the plant Uxs homologues (Harper & Bar-Peled, 2002; Pattathil et al., 2005), the activity of S. meliloti Uxs1 was inhibited by NADH (Supplementary Table S2). UDP-Ara also inhibited SmUxs1 although it does not inhibit known plant and fungal Uxs. Thus, in S. meliloti 1021, UDP-Ara may control the GlcA→Xyl→Ara pathway by feedback inhibition. Both SmUxe and SmUxs1 require the co-factor NAD+. However, SmUxe, in contrast to SmUxs, is not inhibited by NADH (Supplementary Table S3). This suggests that SmUxe has a stronger interaction with the co-factor than SmUxs1. Moreover, SmUxs1 does not utilize NADP+ as a co-factor, suggesting that the enzyme requires the ribosyl C2-OH for activity.
No decarboxylation activity was observed when SmUxs1 reacted for up to 60 min with UDP-GalA, which is an isomer of UDP-GlcA, suggesting that the recombinant enzyme is substrate specific. In addition, SmUxe had no detectable activity when reacted with different nucleotide sugars, including UDP-Glc, UDP-GlcA, UDP-GalA, UDP-GlcNAc and GDP-Man (data not shown).
DISCUSSION
Our study describes the functional characterization of recombinant S. meliloti 1021 Uxs1 and Uxe, and thus provides biochemical evidence for genes that are involved in producing UDP-Ara and UDP-Xyl in bacteria.
Two Uxs genes located in different operons were identified in S. meliloti 1021. blast analyses revealed that the R. leguminosarum 3841 genome contains three Uxs homologues (RL0560, RL0077 and PRL90147) located in different operons. Preliminary studies suggest that RL0560 and RL0077 encode active Uxs (S. Roy, X. Gu & M. Bar-Peled, unpublished data). One possibility for the existence of two Uxs isoforms in S. meliloti 1021 is that each SmUxs functions at different stages of the Sinorhizobium life cycle. No data are currently available to evaluate the transcription level of specific SmUxs at different developmental stages. However, such a possibility could be supported by examining the data from large microarray analyses (Karunakaran et al., 2009) of many rhizobial genes, including the Uxs homologues in R. leguminosarum 3841. Transcriptomic data for the three R. leguminosarum Uxs genes (RL0560, RL0077 and PRL90147) indicate that the expression of RL0560, but not RL0077 and PRL90147, is upregulated approximately fivefold in a 15-day pea-bacteroid when compared with free-living bacteria (Karunakaran et al., 2009).
Both S. meliloti 1021 and R. leguminosarum 3841 operons carrying Uxs and Uxe genes also contain a gene encoding a protein with homology to a cellulose synthase-like protein (CSL) (Fig. 1b), suggesting that they provide UDP-Xyl and UDP-Ara for the formation of specific glycans. These glycans, for example, could be the Ara- and Xyl-containing glycans found in the viscous EPSs, the nodule-specific polysaccharide (NPS), the CPS or the altered LPS structure that is displayed under acidic conditions. In addition, it is possible that upregulation of Uxs gene(s) accounts for the 40-fold increase of xylosyl residues and lack of GalA and GlcA residues in the ‘modified LPS glycan’ prepared from the S. meliloti 1021 lpsB mutant (Campbell et al., 2002). The increased production of Uxs enzymes in the lpsB mutant may alter the UDP-GalA and UDP-GlcA pool by driving the formation of UDP-Xyl (Fig. 1a) and its incorporation into the modified LPS.
SmUxs1 requires NAD+ for the oxidative decarboxylation of UDP-GlcA. Many short-chain dehydrogenase/reductases (SDR proteins) also require NAD+ for activity. In some of these enzymes, the co-factor is tightly bound to the enzyme during the catalytic oxidation and reduction steps (NAD+→NADH→NAD+). However, other SDR enzymes do not bind NAD+ tightly and the co-factor is typically lost during purification. The extent of binding between NAD+ and Uxs from animals, plants and fungi is typically assessed by the inhibition effects of NADH. For example, Uxs1p from C. neoformans and SmUxs1 are both inhibited by NADH, whereas the plant and animal Uxs enzymes characterized to date are not (Harper & Bar-Peled, 2002; Hwang & Horvitz, 2002; Pattathil et al., 2005; Zhang et al., 2005). The ‘binding stringency’ of the enzyme to NAD+ has been reported to be determined by the number of hydrogen bonds formed between specific amino acid residues and the co-factor. For example, Thoden et al. (1996) have proposed that 17 hydrogen bonds are required for the tight binding between NAD+ and UDP-glucose 4-epimerase. In contrast, the six hydrogen bonds that have been proposed to mediate the interaction between NAD+ and ArnA (Gatzeva-Topalova et al., 2005) are not sufficient to retain the co-factor during enzyme purification.
Some bacteria utilize Ara as a carbon source (Ricke et al., 1996), whereas other bacteria, including Salmonella typhimurium and E. coli, produce UDP-β-4-deoxy-4-formamido-l-arabinose via decarboxylation of UDP-GlcA (Breazeale et al., 2002). As far as we are aware, there have been no previous reports of a bacterial 4-epimerase that interconverts UDP-Xyl and UDP-Ara. Preliminary studies suggest that the Uxe homologues from R. leguminosarum 3841 are also specific for these UDP-pentoses (S. Roy, X. Gu & M. Bar-Peled, unpublished data). Thus, it is likely that Uxs and Uxe genes involved in UDP-Xyl and UDP-Ara synthesis exist in rhizobial species that produce Xyl- and Ara-containing glycans.
In summary, the functional identification of bacterial enzymes involved in UDP-Xyl and UDP-Ara biosynthesis, along with the existence of putative glycosyltransferases in the same operons, provides the basis to elucidate the pathway for these glycans in rhizobia. Work is currently under way to determine whether the CSL gene in the operon encodes a functional glycosyltransferase.
Acknowledgments
We thank Dr John Glushka for his help with NMR spectroscopy, and Malcolm O'Neil for constructive comments on the manuscript. This research was supported in part by the National Science Foundation (NSF) IOB-0453664 (to M. B.-P.) and by the BioEnergy Science Center (grant DE-PS02-06ER64304), which is supported by the Office of Biological and Environmental Research in the Department of Energy (DOE) Office of Science. This research also benefited from activities at the South-East Collaboratory for High-Field Biomolecular NMR, a research resource at the University of Georgia, funded by the National Institute of General Medical Sciences (NIGMS grant number GM66340) and the Georgia Research Alliance. This research was also supported in part by the DOE-funded Center for Plant and Microbial Complex Carbohydrates (DE-FG05-93ER20097).
Abbreviations
CPS, capsular polysaccharide
CSL, cellulose synthase-like protein
EPS, extracellular polysaccharide
Footnotes
References
- Bar-Peled, M., Griffith, C. L. & Doering, T. L. (2001). Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci U S A 98, 12003–12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breazeale, S. D., Ribeiro, A. A. & Raetz, C. R. (2002). Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid A species modified with 4-amino-4-deoxy-l-arabinose. J Biol Chem 277, 2886–2896. [DOI] [PubMed] [Google Scholar]
- Burget, E. G. & Reiter, W. D. (1999). The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho l-arabinose. Plant Physiol 121, 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget, E. G., Verma, R., Molhoj, M. & Reiter, W. D. (2003). The biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell 15, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, G. R., Reuhs, B. L. & Walker, G. C. (2002). Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci U S A 99, 3938–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, R. W., Reuhs, B. L., Forsberg, L. S. & Kannenberg, E. L. (1999). Rhizobial cell surface carbohydrates: their structures, biosynthesis and functions. In Genetics of Bacterial Polysaccharides, pp. 53–90. Edited by Goldberg, J. B.. Boca Raton, FL. : CRC Press.
- De Leizaola, M. & Dedonder, R. (1955). Etude de quelques polyoisides produits par des souches de Rhizobium. C R Hebd Seances Acad Sci 240, 1825–1827. [PubMed] [Google Scholar]
- Egelund, J., Petersen, B. L., Motawia, M. S., Damager, I., Faik, A., Olsen, C. E., Ishii, T., Clausen, H., Ulvskov, P. & Geshi, N. (2006). Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-α-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18, 2593–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, L. S. & Carlson, R. W. (2008). Structural characterization of the primary O-antigenic polysaccharide of the Rhizobium leguminosarum 3841 lipopolysaccharide and identification of a new 3-acetimidoylamino-3-deoxyhexuronic acid glycosyl component: a unique O-methylated glycan of uniform size, containing 6-deoxy-3-O-methyl-d-talose, N-acetylquinovosamine, and rhizoaminuronic acid (3-acetimidoylamino-3-deoxy-d-gluco-hexuronic acid). J Biol Chem 283, 16037–16050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68, 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert, F., Finan, T. M., Long, S. R., Puhler, A., Abola, P., Ampe, F., Barloy-Hubler, F., Barnett, M. J., Becker, A. & other authors (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668–672. [DOI] [PubMed] [Google Scholar]
- Gatzeva-Topalova, P. Z., May, A. P. & Sousa, M. C. (2005). Structure and mechanism of ArnA: conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure 13, 929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götting, C., Kuhn, J., Zahn, R., Brinkmann, T. & Kleesiek, K. (2000). Molecular cloning and expression of human UDP-d-xylose : proteoglycan core protein β-d-xylosyltransferase and its first isoform XT-II. J Mol Biol 304, 517–528. [DOI] [PubMed] [Google Scholar]
- Gu, X., Glushka, J., Yin, Y., Xu, Y., Denny, T., Smith, J. A., Jiang, Y. & Bar-Peled, M. (2010). Identification of a bifunctional UDP-4-keto-pentose/UDP-xylose synthase in the plant pathogenic bacterium, Ralstonia solanacearum str. GMI1000: a distinct member of the 4,6-dehydratase and decarboxylase family. J Biol Chem 285, 9030–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, A. D. & Bar-Peled, M. (2002). Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene family, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130, 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A. M. (1999). Role of lectins (and rhizobial exopolysaccharides) in legume nodulation. Curr Opin Plant Biol 2, 320–326. [DOI] [PubMed] [Google Scholar]
- Humphrey, B. A. & Vincent, J. M. (1959). Extracellular polysaccharides of Rhizobium. J Gen Microbiol 21, 477–484. [DOI] [PubMed] [Google Scholar]
- Hwang, H. Y. & Horvitz, H. R. (2002). The SQV-1 UDP-glucuronic acid decarboxylase and the SQV-7 nucleotide-sugar transporter may act in the Golgi apparatus to affect Caenorhabditis elegans vulval morphogenesis and embryonic development. Proc Natl Acad Sci U S A 99, 14218–14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K. M., Kobayashi, H., Davies, B. W., Taga, M. E. & Walker, G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat Rev Microbiol 5, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg, E. L. & Brewin, N. J. (1989). Expression of a cell surface antigen from Rhizobium leguminosarum 3841 is regulated by oxygen and pH. J Bacteriol 171, 4543–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg, E. L. & Carlson, R. W. (2001). Lipid A and O-chain modifications cause Rhizobium lipopolysaccharides to become hydrophobic during bacteroid development. Mol Microbiol 39, 379–391. [DOI] [PubMed] [Google Scholar]
- Kannenberg, E. L., Perotto, S., Bianciotto, V., Rathbun, E. A. & Brewin, N. J. (1994). Lipopolysaccharide epitope expression of Rhizobium bacteroids as revealed by in situ immunolabelling of pea root nodule sections. J Bacteriol 176, 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr, D. B., Liang, R. T., Reuhs, B. L. & Emerich, D. W. (2000). Altered exopolysaccharides of Bradyrhizobium japonicum mutants correlate with impaired soybean lectin binding, but not with effective nodule formation. Planta 211, 218–226. [DOI] [PubMed] [Google Scholar]
- Karunakaran, R., Ramachandran, V. K., Seaman, J. C., East, A. K., Mouhsine, B., Mauchline, T. H., Prell, J., Skeffington, A. & Poole, P. S. (2009). Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J Bacteriol 191, 4002–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating, D. H., Willits, M. G. & Long, S. R. (2002). A Sinorhizobium meliloti lipopolysaccharide mutant altered in cell surface sulfation. J Bacteriol 184, 6681–6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts, J. S. & Doering, T. L. (2008). Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem 283, 14327–14334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, T., Takeda, T., Miyazaki, Y., Ohnishi-Kameyama, M., Hayashi, T., O'Neill, M. A. & Ishii, T. (2007). A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17, 345–354. [DOI] [PubMed] [Google Scholar]
- Kuhn, J., Götting, C., Schnolzer, M., Kempf, T., Brinkmann, T. & Kleesiek, K. (2001). First isolation of human UDP-d-xylose: proteoglycan core protein β-d-xylosyltransferase secreted from cultured JAR choriocarcinoma cells. J Biol Chem 276, 4940–4947. [DOI] [PubMed] [Google Scholar]
- Laus, M. C., Logman, T. J., Van Brussel, A. A., Carlson, R. W., Azadi, P., Gao, M. Y. & Kijne, J. W. (2004). Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 186, 6617–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil, S., Harper, A. D. & Bar-Peled, M. (2005). Biosynthesis of UDP-xylose: characterization of membrane-bound AtUxs2. Planta 221, 538–548. [DOI] [PubMed] [Google Scholar]
- Peña, M. J., Zhong, R., Zhou, G. K., Richardson, E. A., O'Neill, M. A., Darvill, A. G., York, W. S. & Ye, Z. H. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porchia, A. C., Sorensen, S. O. & Scheller, H. V. (2002). Arabinoxylan biosynthesis in wheat. Characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol 130, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvanesarajah, V., Schell, F. M., Gerhold, D. & Stacey, G. (1987). Cell surface polysaccharides from Bradyrhizobium japonicum and a nonnodulating mutant. J Bacteriol 169, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke, S. C., Martin, S. A. & Nisbet, D. J. (1996). Ecology, metabolism, and genetics of ruminal selenomonads. Crit Rev Microbiol 22, 27–56. [DOI] [PubMed] [Google Scholar]
- Sánchez-Andújar, B., Coronado, C., Philip-Hollingsworth, S., Dazzo, F. B. & Palomares, A. J. (1997). Structure and role in symbiosis of the exoB gene of Rhizobium leguminosarum bv trifolii. Mol Gen Genet 255, 131–140. [DOI] [PubMed] [Google Scholar]
- Syn, C. K. & Swarup, S. (2000). A scalable protocol for the isolation of large-sized genomic DNA within an hour from several bacteria. Anal Biochem 278, 86–90. [DOI] [PubMed] [Google Scholar]
- Thoden, J. B., Frey, P. A. & Holden, H. M. (1996). Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism. Biochemistry 35, 5137–5144. [DOI] [PubMed] [Google Scholar]
- van Rhijn, P. & Vanderleyden, J. (1995). The Rhizobium–plant symbiosis. Microbiol Rev 59, 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Shirley, N., Lahnstein, J. & Fincher, G. B. (2005). Characterization and expression patterns of UDP-d-glucuronate decarboxylase genes in barley. Plant Physiol 138, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]