Abstract
Sinorhizobium meliloti has multiple systems for iron acquisition, including the use of haem as an iron source. Haem internalization involves the ShmR haem outer membrane receptor and the hmuTUV locus, which participates in haem transport across the cytoplasmic membrane. Previous studies have demonstrated that expression of the shmR gene is negatively regulated by iron through RirA. Here, we identify hmuP in a genetic screen for mutants that displayed aberrant control of shmR. The normal induction of shmR in response to iron limitation was lost in the hmuP mutant, showing that this gene positively affects shmR expression. Moreover, the HmuP protein is not part of the haemin transporter system. Analysis of gene expression and siderophore production indicates that disruption of hmuP does not affect other genes related to the iron-restriction response. Our results strongly indicate that the main function of HmuP is the transcriptional regulation of shmR. Sequence alignment of HmuP homologues and comparison with the NMR structure of Rhodopseudomonas palustris CGA009 HmuP protein revealed that certain amino acids localized within predicted β-sheets are well conserved. Our data indicate that at least one of the β-sheets is important for HmuP activity.
INTRODUCTION
The alpha-proteobacterium Sinorhizobium meliloti has the ability to fix nitrogen in symbiotic association with certain legumes. It is also a free-living organism in the soil and rhizosphere, and adaptation to diverse ecological niches involves differential regulation of gene expression. Nitrogen-fixing bacteria have a high demand for iron during symbiosis, since nitrogenase and other iron-containing proteins are required for N2 fixation (Georgiadis et al., 1992; Hennecke, 1992; Rees & Howard, 2000; Sangwan & O'Brian, 1992). In soil, iron is mostly insoluble, and therefore scarce. Like other bacteria, S. meliloti possesses highly efficient iron-acquisition systems. These systems comprise the synthesis and transport of the di-hydroxamate siderophore rhizobactin 1021, the use of the xenosiderophores ferrichrome and ferrioxamine B, and iron acquisition from different iron-porphyrin compounds such as haemin, haemoglobin and leghaemoglobin (Cuiv et al., 2008; Lynch et al., 2001; Noya et al., 1997; Persmark et al., 1993). The haem transport mechanism involves ShmR, an outer membrane haem receptor, and the HmuTUV transport system (Amarelle et al., 2008; Cuiv et al., 2008). Yersinia enterocolitica HemTUV was among the first ABC transporters involved in haem uptake to be characterized, and is considered as a prototype for haem transport (Stojiljkovic & Hantke, 1994). Its homologue in Yersinia pestis is called HmuTUV (Hornung et al., 1996). According to the model of Hem/Hmu transport, once the haem moiety is translocated into the periplasm it is bound by a periplasmic binding protein, HmuT, which in turn presents it to the inner membrane permease–ATP hydrolase complex HmuU/HmuV. Intact haem is then delivered to the bacterial cytoplasm (Hornung et al., 1996; Stojiljkovic & Hantke, 1994).
Genes that encode rhizobactin 1021 biosynthesis and transport are located in a regulon that comprises the biosynthesis operon rhbABCDEF; rhtA, encoding the ferri-rhizobactin 1021 outer membrane receptor; rhtX, encoding a permease that belongs to a novel family of siderophore transporters; and rhrA, the AraC-like regulator of the receptor and biosynthetic genes (Cuiv et al., 2004; Lynch et al., 2001). Ferrichrome and ferrioxamine B transport systems consist of FhuA1 and FoxA, the respective outer membrane receptors; FhuP, a periplasmic binding protein for both siderophores; and the HmuUV complex, also involved in haem transport across the inner membrane (Cuiv et al., 2008).
Studies of the control of bacterial iron homeostasis have focused largely on the ferric uptake regulator Fur. This protein senses the intracellular ferrous ion concentration, through the formation of a Fur–Fe2+ complex, which in turn interacts with specific DNA targets in the promoters of iron-repressed genes (Andrews et al., 2003). Nonetheless, in S. meliloti and other alpha-proteobacteria, Fur homologues are manganese-responsive regulators (Chao et al., 2004; Diaz-Mireles et al., 2004; Hohle & O'Brian, 2009; Platero et al., 2004). In the alpha-proteobacteria S. meliloti, Rhizobium leguminosarum and Agrobacterium tumefaciens, a different protein, RirA, which belongs to the Rrf2 superfamily of regulators, is responsible for the regulation of most genes involved in iron uptake (Chao et al., 2005; Ngok-Ngam et al., 2009; Todd et al., 2002; Viguier et al., 2005). In particular, under iron-replete conditions, RirA represses the expression of the rhizobactin biosynthesis and transport regulon, shmR gene expression and the hmuPSTUV locus.
With the aim of finding novel transcriptional regulators involved in shmR expression, we performed a generalized mutagenesis approach in S. meliloti. In this work, we discover that the small protein HmuP is essential for shmR expression in iron-depleted conditions.
METHODS
Bacteria, plasmids and growth conditions.
Bacteria and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown aerobically at 37 °C in Luria–Bertani (LB) medium. S. meliloti strains were grown at 30 °C either in tryptone-yeast extract (TY) medium (Beringer, 1974) or in defined minimal medium M9 (Sambrook et al., 1989) supplemented with 6 mM glutamate, 200 μM methionine and 1 μM biotin. Low-iron conditions were obtained by supplementation with ethylenediamine-di-o-hydroxyphenylacetic acid (EDDHA). When required, 50 μg kanamycin ml−1, 100 μg neomycin ml−1, 100 μg streptomycin ml−1, 10 μg gentamicin ml−1, 50 μg ampicillin ml−1 or 1 μg tetracycline ml−1 was added to the media.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| S. meliloti strains | ||
| SM1021 | Streptomycin derivative of SU47 | Meade et al. (1982) |
| SHMR | SM1021 shmR : : lacZ-Gmr | Amarelle et al. (2008) |
| B20 | SHMR hmuP : : Tn5-1063a | This work |
| HMUP | SM1021 hmuP : : Tn5-1063a | This work |
| SM1021 : : pRG1SMc02726 | SM1021 with pRG1SMc02726 construct integrated into genome | This work |
| HMUP : : pRG1SMc02726 | HMUP with pRG1SMc02726 construct integrated into genome | This work |
| E. coli strains | ||
| DH5α | supE44ΔlacU169(φ80lacZΔM15)hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Hanahan (1983) |
| S17-1 | pro hsdR recA [RP4-2(Tc : : Mu)(Km : : Tn7)] mobilization strain for biparental matings | Simon et al. (1983) |
| S17-1 λpir | λpir lysogen of S17-1 | Simon et al. (1983) |
| Plasmids | ||
| pBluescript SK | Cloning vector, AmR | Stratagene |
| pOT2 | Promoter probe vector based on pBBR-1-MCS-5 replicon. Contains promoterless gfpuv and multiple cloning site (MCS) between two transcriptional terminators, GmR | Allaway et al. (2001) |
| pGEM-T | Cloning vector for PCR products, AmR | Promega |
| pRK2013 | ColE1 replicon with RK2 tra genes. Used for mobilizing incP and incQ plasmids, KmR | Ditta et al. (1980) |
| pRL1063a | Tn5 derivative; promoterless luxAB; oriV KmR | Wolk et al. (1991) |
| pRG1SMc02726 | pMK2030 with SMc02726 ORF inserted, TetR | Humann et al. (2008) |
| pOT-HmuP | hmuP under its native promoter and fused to gfpuv | This work |
| pOT-HmuPΔCt | hmuP lacking the region encoding the C-terminal GKLILNK residues, under its native promoter and fused to gfpuv | This work |
| pOT-HmuPΔK | hmuP lacking the codon encoding the C-terminal lysine residue, under its native promoter and fused to gfpuv | This work |
| pOT-HmuPY | hmuP with a codon encoding tyrosine inserted before the stop codon, under its native promoter and fused to gfpuv | This work |
| pOT-HmuPpr | hmuP presumptive promoter region fused to gfpuv | This work |
Tn5-1063a transposon mutagenesis and selection of transconjugants.
Plasmid pRL1063a (Wolk et al., 1991), containing the transposable element Tn5-1063a, was used to generate over 10 000 insertional mutants of S. meliloti strain SHMR (Amarelle et al., 2008). The transposon was delivered to strain SHMR by triparental mating with E. coli DH5α (pRL1063a) and E. coli DH5α (pRK2013) (Ditta et al., 1980) as donor and helper strains, respectively. Transconjugants were screened for changes in shmR expression using TY solid media supplemented with streptomycin, neomycin, 20 μg X-Gal ml−1 and either 37 μM FeCl3 or 50 μM EDDHA. Differences in shmR expression were confirmed by measuring β-galactosidase activity, using S. meliloti strain SHMR as control. Cells were grown for 48 h in TY medium and diluted 100-fold in TY medium supplemented with either 37 μM FeCl3 or 100 μM EDDHA. Cultures were grown to early stationary phase at 200 r.p.m. and 30 °C. The β-galactosidase assay was performed according to Miller (1972), with the modifications described by Poole et al. (1994).
Sequence analysis of S. meliloti SHMR Tn1063a-tagged locus and transposon transduction.
To identify the location of the transposon insertion in the B20 mutant, genomic DNA was isolated using the UltraClean Microbial DNA kit (MoBio). Arbitrary PCR (Knobloch et al., 2003) was used to amplify fragments containing transposon junctions using arbitrary primers ARB1-A or ARB2 (Griffitts & Long, 2008) and the transposon-specific primers Tn5-4 or Tn5-2 (Yurgel & Kahn, 2005). The DNA sequence was determined using the sequencing primer TZTn5 (Yurgel & Kahn, 2005) and the obtained nucleotide sequence was searched in the S. meliloti 1021 genome using the blast algorithm.
Lysates obtained from B20 mutant cells infected with the ΦM12 phage (Finan et al., 1984) were used to transduce the hmuP : : Tn5-1063a mutation to S. meliloti 1021, as described by Humann et al. (2009). Transductants were selected by three consecutive passages in LB medium supplemented with neomycin.
Integration of pRG1SMc02726 in the S. meliloti genome and β-glucuronidase activity assays.
The pRG1SMc02726 (Humann et al., 2008) construction was transferred to S. meliloti 1021 and HMUP (SM1021 hmuP : : Tn5-1063a) strains by biparental conjugation with strain E. coli S17-1 λ pir (pRG1SMc02726), and transconjugants were selected in Min-succinate-NH4 medium (Yurgel & Kahn, 2005) supplemented with tetracycline. Transconjugants with the plasmid integrated by homologous recombination were confirmed by colony PCR using primers shmRforward (Amarelle et al., 2008) and 2030F (Humann et al., 2008).
Differences in shmR : : gusA expression were assessed by measuring β-glucuronidase activity. Cells were grown for 48 h in M9 minimal medium and diluted 100-fold in M9 medium supplemented with 300 μM EDDHA. Cultures were grown to early stationary phase at 200 r.p.m. and 30 °C. β-Glucuronidase activity assays were performed as described by Jefferson et al. (1986), and β-glucuronidase arbitrary units were defined as 1000 A415 units min−1 ml−1 (OD620 unit)−1 (Jefferson et al., 1986).
Construction of plasmids containing the hmuP gene or the hmuP mutated versions, and in vivo complementation of the HMUP mutant strain.
PCRs were carried out with Tli Polymerase (Promega) and S. meliloti 1021 genomic DNA as template. Primers used are listed in Supplementary Table S1. The hmuP gene and its presumptive promoter region were amplified using primers HSF and HSR. The resulting 1952 bp fragment was cloned in the SmaI site of pBluescript SK (Promega), generating plasmid pBSK-HmuPS. Plasmid pBSK-HmuPS was subsequently digested with XhoI, and the 620 bp fragment obtained was subcloned in the XhoI site of pBluescript SK to obtain the pBSK-HmuP plasmid. This plasmid was digested with PstI, and a 650 bp fragment was cloned into the PstI site of pOT2 (Allaway et al., 2001), creating pOT-HmuP.
For the construction of pOT-HmuPY, pOT-HmuPΔK and pOT-HmuPΔCt, primer HPprF was used as a forward primer and HPY, HPK or HPCt as reverse primer, respectively. The amplicons were cloned into the EcoRV site of pBluescript SK, giving rise to plasmids pBSK-HmuPY, pBSK-HmuPΔK and pBSK-HmuPΔCt. The BamHI–HindIII fragments obtained from these plasmids were subcloned in pOT2, creating plasmids pOT-HmuPY, pOT-HmuPΔK and pOT-HmuPΔCt. These constructions were confirmed by sequencing.
For the construction of the pOT-HmuPpr plasmid (hmuP presumptive promoter region fused to gfpuv), a 464 bp fragment was amplified using the primers HPprF and HPprR. This amplicon was cloned into the EcoRV site of pBluescript SK, giving rise to plasmid pBSK-HmuPpr. Finally, pBSK-HmuPpr was digested with HindIII/XbaI and subcloned into pOT2, resulting in plasmid pOT-HmuPpr.
Plasmids were introduced into S. meliloti strains by triparental mating as described above.
Bioassays and growth assays.
Bioassay experiments were carried out as previously described (Noya et al., 1997). Briefly, 10 μl of the stock solutions to be tested were added to wells in solid TY medium (15 g agar l−1) supplemented with 300 μM EDDHA and containing about 106 c.f.u. ml−1. Stock solutions of the following compounds were used as iron sources: 37 mM FeCl3, 0.15 mM haemoglobin, 1 mM haemin, 0.3 mM ferrichrome and 46 mM Desferal.
For colony size experiments, appropriate dilutions of mid-exponential cultures grown in TY media were made. Dilutions were spotted with a replica plater in TY solid media or TY solid medium supplemented with 300 μM EDDHA and 10 μM haemin. Cells were grown for 5 days at 30 °C and colony sizes were recorded.
RNA purification.
Wild-type and hmuP mutant strains were grown in M9 minimal medium supplemented with 300 μM EDDHA. At mid-exponential phase (OD620 0.8–1.0), 20 ml of culture was treated with 4 ml RNAprotect (Qiagen) and harvested at 7000 r.p.m. at 4 °C for 10 min. Pellets were frozen in liquid nitrogen and stored at −80 °C. Total RNA was isolated using a hot-phenol procedure, as described elsewhere (Yang et al., 2006). RNA samples were treated with RQ1 RNase-free DNase I (Promega) and purified using the clean-up procedure of the RNeasy Bacterial RNA Purification kit (Qiagen).
Determination of the transcription start site and RT-PCR.
The transcription start site of the hmuP gene was determined by rapid amplification of 5′ complementary DNA ends (5′-RACE) using a kit from Invitrogen according to the manufacturer's instructions. We used total RNA purified from cells grown in iron-limited media. A 5′-RACE amplification product of 465 bp was obtained. Conventional cloning methods were used to clone the fragment in the pGEM-T vector (Promega). The plasmid was sequenced and the transcriptional start site was determined by comparison with the S. meliloti 1021 published genome using the blast algorithm.
Co-transcription of hmuPST was assessed by RT-PCR using the primer sets HmuPF/GSP1 and HmuSF/HmuTR shown in Supplementary Table S1. cDNA was synthesized using the iScript cDNA Synthesis kit (Bio-Rad) with total RNA obtained from iron-starved cells as a template. A negative control in which no reverse transcriptase was included in the reverse transcription reaction was used as a template in order to evaluate genomic DNA contamination.
Quantitative real-time PCR (qPCR).
Expression of the shmR, rhrA, rhbE, hmuS, hmuT and SMc01515 genes was assessed by qPCR. Reverse transcription was carried out with total RNA using the iScript cDNA Synthesis kit (Bio-Rad). For real-time PCR, 0.02 μg cDNA and 0.25 μM of each primer (IDT DNA Technologies) were used in a 20 μl reaction, using the iQ SYBR Green Supermix (Bio-Rad). Primers used are listed in Supplementary Table S1. PCRs were run on an iCycler thermal cycler (Bio-Rad) using a 3 min hot start at 95 °C, and then 40 cycles with steps of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s. The generation of specific PCR products was confirmed by melting curve analysis. gapA (SMc03979) was used as a housekeeping gene control. Samples in which the reverse transcriptase was omitted in reverse-transcriptase reactions were used as negative controls. For relative quantification, the standard curve method was used. S. meliloti 1021 genomic DNA was used as the PCR template for standard curves for each gene. Relative starting quantities of mRNAs for each gene were calculated from the corresponding standard curve and were normalized to gapA. Every reaction was done in triplicate.
Bioinformatics analysis.
Using the S. meliloti 1021 HmuP protein as query, the translated nucleotide database of NCBI was searched (tblastn). An expect value of 100 was used. Results were selected based on the following criteria: similarity along the HemP domain (from residues 10 to 46 in S. meliloti 1021); absence of gaps larger than two amino acids in region 20–46 (where the β-sheets are localized); GKLILTK motif conserved in at least four to five residues; Y22 and T27 conserved.
Plant assays.
Medicago sativa cv. Creola was used for screening the symbiotic phenotype of the hmuP mutant. The wild-type strain was used as a control. Plant assays were done in nitrogen-free Jensen medium, as previously described (Platero et al., 2004). Nitrogen-fixation efficiencies were estimated by determining plant dry weights 60 days after planting.
RESULTS
Identification of hmuP in a genetic screen for mutants deregulated in shmR gene expression
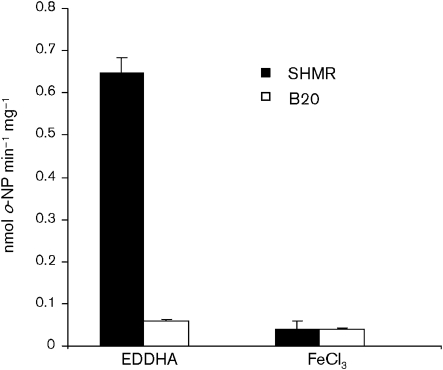
We previously demonstrated the iron responsiveness of the shmR promoter using a chromosomal transcriptional fusion shmR : : lacZ in the S. meliloti SHMR mutant strain (Amarelle et al., 2008). Colonies of S. meliloti SHMR are white when they are grown in TY solid medium supplemented with X-Gal and FeCl3, while the colonies are blue in TY supplemented with X-Gal and EDDHA, as a result of the repression or induction of shmR : : lacZ expression, respectively. Here, we used this construction as a tool to search for mutants that show deregulation of iron-dependent shmR expression. S. meliloti SHMR was mutagenized with Tn5-1063a and transconjugants were screened for loss of metal regulation in TY solid medium supplemented with FeCl3 or EDDHA. From over 10 000 transconjugants analysed, one mutant named B20 was selected for further analyses. Colonies of B20 were white in TY medium supplemented with EDDHA and X-Gal, and this mutant presented a reduced β-galactosidase activity in TY EDDHA broth when compared with the reporter strain, indicating a loss in shmR : : lacZ expression (Fig. 1).
Fig. 1.
β-Galactosidase activity in the SM1021 shmR : : lacZ mutant (SHMR) and in its derivative mutant (B20). Cells were grown to early stationary phase in TY broth. One hundred-fold dilutions were made in TY supplemented with either 150 μM EDDHA or 37 μM FeCl3 and grown for 48 h. β-Galactosidase activity is expressed as nmol o-nitrophenol min−1 (mg protein)−1. The data shown are the mean of two independent experiments done in triplicate. Error bars, 1 sd.
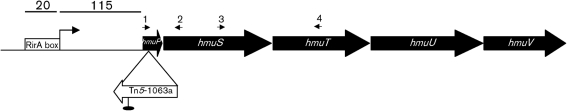
The DNA sequence of the transposon-flanking region in the B20 mutant was determined by arbitrary PCR. The PCR fragment was sequenced and the results obtained from blast searches in the S. meliloti 1021 genome indicated that the tagged locus was SMc01747. The deduced product of SMc01747 is a small protein (5.3 kDa) that belongs to the HemP superfamily. An hmuP/hemP homologue has been shown to be involved in the utilization of haem as an iron source in R. leguminosarum (Wexler et al., 2001), but the function of this protein is not known. Based on this homology, we designate SMc01747 as hmuP. In the S. meliloti genome, hmuP is localized together with the hmuS gene and upstream of the hmuTUV cluster (Fig. 2). The function of the putative hmuS gene has not yet been described in S. meliloti. The hmuTUV cluster has recently been described as encoding an ABC transport system involved in haem, ferrichrome and ferrioxamine B transport in S. meliloti 2011 (Cuiv et al., 2008).
Fig. 2.
Genetic organization of the hmuP gene in S. meliloti 1021. A black oval indicates a rho-independent transcriptional terminator. The Tn5-1063a insertion site and orientation are illustrated with a white arrow not to scale. The small numbered arrows indicate the positions of primers used for RT-PCR: HmuPF (1), GSP1 (2), HmuSF (3), HmuTR (4). The transcription start site of hmuP was determined by 5′-RACE. The bent arrow indicates the transcription start site and the numbers show the distance (bp) from it to the initiation codon (GTG) and to the putative RirA box.
The hmuP and hmuS genes are part of the hmuPSTUV transcriptional unit
The small intergenic space between the hmuP and hmuS genes suggested that both genes are part of the same transcriptional unit. On the other hand, a putative rho-independent transcriptional terminator downstream of the hmuS gene is indicated in the published S. meliloti 1021 genome, suggesting that hmuPS could be a transcriptional unit independent of the hmuTUV cluster. Here, we wanted to establish whether hmuPSTUV are independently transcribed. First, we performed a 5′-RACE, and we determined the transcription start site for hmuP as located 115 bp upstream of the initiation codon (Fig. 2). However, we were unable to determine the transcription start site for hmuT, suggesting that the hmuT gene could be cotranscribed together with the hmuP and hmuS genes. To assess this possibility, we employed an RT-PCR approach using different sets of primers. The locations of the primers used are shown in Fig. 2. The length of the products obtained indicates that the hmuP and hmuS genes as well as the hmuS and hmuT genes are transcribed in the same mRNA unit. No products could be detected when we used as a template a mock control in which no reverse transcriptase was included in the reverse transcription reaction (data not shown). The results obtained indicate that hmuP, hmuS and hmuTUV are part of the same transcriptional unit.
HmuP is essential for shmR expression and iron acquisition from haem compounds
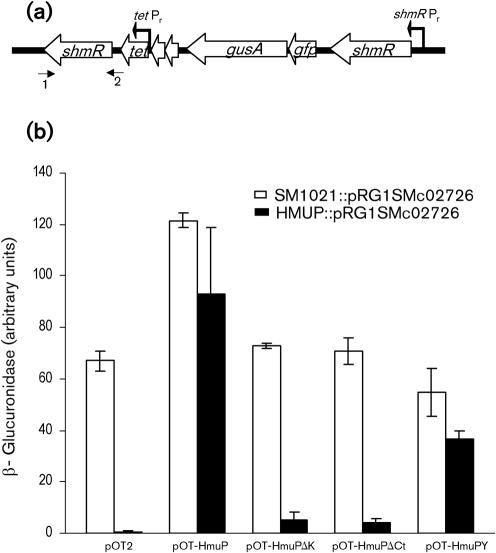
Since the B20 mutant was obtained from a generalized mutagenesis in an shmR mutant context, the hmuP : : Tn5-1063a mutation was transduced to the S. meliloti 1021 wild-type strain in order to obtain a mutant with a single transposon insertion in an shmR+ context. This mutant was named HMUP. To investigate shmR expression, we used the transcriptional gene fusion system reported by Humann et al. (2008) with the pRG1SMc02726 plasmid kindly provided by Michael L. Kahn, Washington State University, Pullman, WA, USA. With this approach, after integration of pRG1SMc02726 by homologous recombination, two functional copies of shmR are present; one is under the control of the native shmR promoter, which also drives the expression of the gusA and gfp reporter genes, and the other is expressed at a low and constitutive level from the tetracycline (tet) promoter located in pRG1SMc02726 (Fig. 3a). To evaluate the effect of hmuP disruption on shmR expression, this construction was integrated into S. meliloti 1021 wild-type and HMUP mutant strains.
Fig. 3.
In vivo effect of hmuP mutation on shmR : : gusA activity. (a) The pRG1SMc02726 construction integrated into the SM1021 or HMUP genome. Small numbered arrows indicate the position of primers shmRforward (1) and 2030F (2). (b) Strains SM1021 : : pRG1SMc02726 and HMUP : : pRG1SMc02726 containing the indicated pOT2 derivatives were grown until early stationary phase in M9 broth. One hundred-fold dilutions were made in M9 supplemented with 300 μM EDDHA and grown for 48 h. β-Glucuronidase activity is expressed as β-glucuronidase arbitrary units. The data shown are the mean of two independent experiments done in triplicate. Error bars, 1 sd.
As shown in Fig. 3(b), expression of the shmR : : gusA reporter in iron-restricted media was drastically reduced in the HMUP : : pRG1SMc02726 (pOT2) strain. In order to confirm that the lack of shmR expression was due to the lack of HmuP, we expressed hmuP in trans. Complementation of the HMUP : : pRG1SMc02726 strain with the pOT-HmuP plasmid completely restored the expression of the shmR : : gusA reporter. These results clearly demonstrate that HmuP is required for shmR gene expression.
Furthermore, the presence of multiple copies of hmuP in the SM1021 : : pRG1SMc02726 (pOT-HmuP) strain resulted in an 81 % increase of shmR : : gusA expression with respect to 1021 : : pRG1SMc02726 (pOT2) (Fig. 3b), indicating that HmuP may act as a positive regulator of shmR gene expression.
We previously demonstrated that the shmR gene is necessary for iron acquisition from haem compounds (Amarelle et al., 2008). Here, we wanted to evaluate whether hmuP disruption affects iron nutrition from haem and haem compounds. The hmuP mutant was unable to use either haem or haemoglobin as the sole iron source, but retained the ability to grown on FeCl3 (Table 2, Fig. 4a). Complementation with the hmuP gene in trans completely restored the ability to use haem iron sources (Table 2, Fig. 4a), indicating that the observed phenotype was due to hmuP disruption. These results clearly demonstrate that HmuP is necessary for haem-mediated iron acquisition in S. meliloti.
Table 2.
Assessment of the ability of S. meliloti strains to use different compounds as sole iron sources
Stock solutions were added to wells in TY medium supplemented with 300 μM EDDHA, containing about 106 c.f.u. ml−1.
| S. meliloti strain | Bacterial growth around wells containing different iron sources [mean diameter±sd (cm)]* | |||
|---|---|---|---|---|
| 370 nmol FeCl3 | 5 nmol Hb† | 20 nmol Hm† | 456 nmol Desferal | |
| SM1021 | 0.55±0.05 | 0.7±0.1 | 0.8±0.1 | 2±0.2 |
| SHMR | 0.5±0.1 | 0 | 0 | nd‡ |
| HMUP | 0.5±0.1 | 0 | 0 | 2.2±0.1 |
| HMUP (pOT-HmuP) | 0.5±0.1 | 0.7±0.1 | 0.8±0.1 | 2±0.1 |
| HMUP (pOT-HmuPΔCt) | 0.55±0.05 | 0 | 0 | 2±0.2 |
| HMUP (pOT-HmuPΔK) | 0.8±0.2 | 0.75±0.1§ | 0.8±0.1§ | 2±0.1 |
| HMUP (pOT-HmuPY) | 0.45±0.05 | 0.8±0.1 | 0.9±0.1 | 1.9±0.1 |
| SM1021 : : pRGSMc02726 | 0.45±0.05 | 0.7±0.1 | 0.7±0.1 | nd |
| HMUP : : pRGSMc02726 | 0.5±0.1 | 0.7±0.1 | 0.75±0.05 | nd |
*Results are the mean±sd of three independent experiments.
†Hb, haemoglobin; Hm, haemin.
‡nd, Not determined.
§Haloes were fainter than the rest.
Fig. 4.
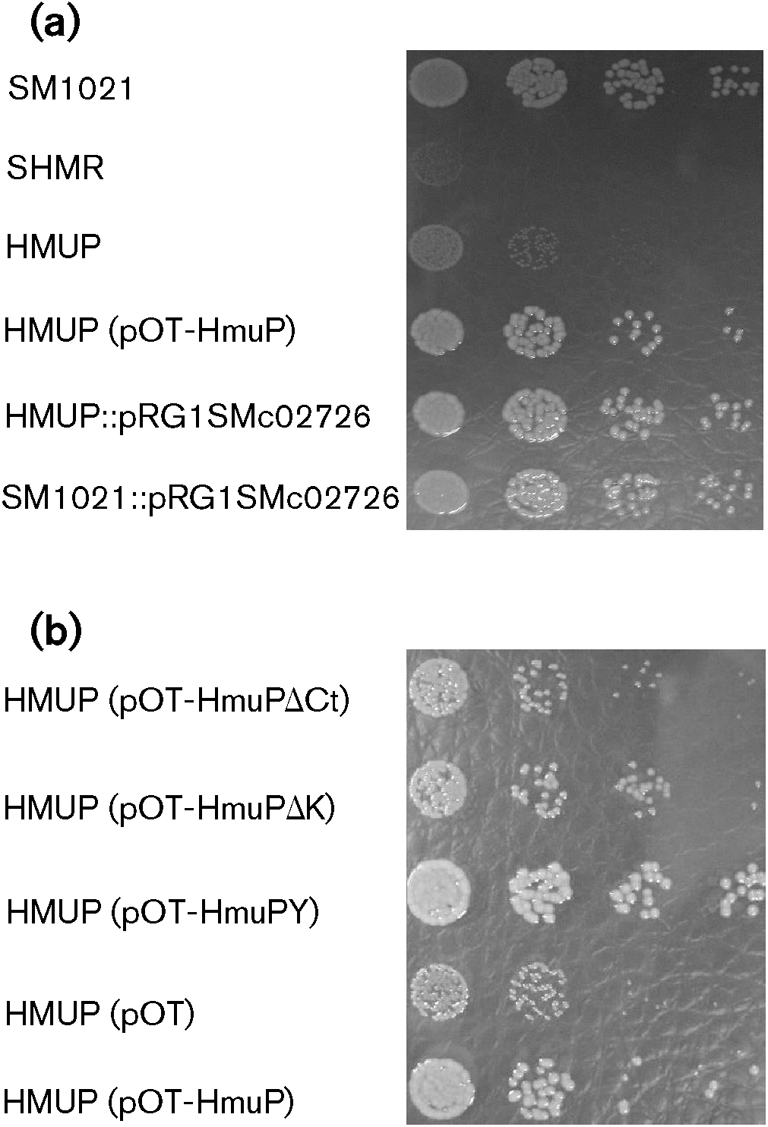
Growth assays. Cells were grown in TY broth until early stationary phase. Appropriate dilutions were made in TY broth and then plated in TY solid media supplemented with 300 μM EDDHA and 10 μM haemin. Dilutions were also spotted in TY media, and all the strains displayed a similar growth pattern (data not shown). Cells were grown for 5 days and colony sizes were determined. The experiments were done in triplicate with similar results. (a) Effect of HmuP on the ability of S. meliloti 1021 to use haemin as an iron source. (b) Effect of hmuP mutations on HmuP activity.
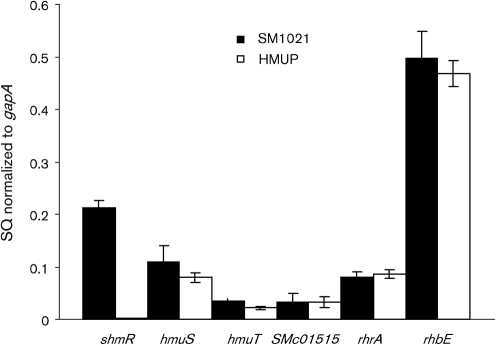
Since hmuP is part of the hmuPSTUV transcriptional unit, the inability to use haemin as an iron source could be due in part to polar effects. By using qPCR we observed that mRNA levels for hmuS and hmuT were similar in the wild-type and the hmuP mutant strains, indicating that the transposon insertion had no detectable effect on the expression of downstream genes (Fig. 5). The absence of polar effects on downstream genes can be explained by the direction in which the transposon was inserted. In the HMUP mutant strain, the transcriptional terminator present in the Tn5-1063a construction is localized in the opposite strand of the hmuP transcriptional promoter, as illustrated in Fig. 2.
Fig. 5.
Effect of HmuP on the expression of iron-regulated genes. mRNAs from wild-type (black bars) and hmuP mutant (white bars) cells grown in M9 minimal media supplemented with 300 μM EDDHA were analysed by qPCR. The genes assessed are indicated below the bars. The data are expressed as the relative starting quantity (SQ) of mRNA normalized to the housekeeping gene gapA. The data are expressed as the mean of three replicates; error bars, 1 sd.
The observed impairment of the hmuP mutant strain in growth with haemin as sole iron source could be due to the loss not only of shmR expression but also of other HmuP-dependent factors involved in haemin utilization. To test this possibility we assessed the ability to use haemin as an iron source of strain HMUP : : pRG1SMc02726, which expresses the shmR gene at a low constitutive level from the tet promoter (Table 2, Fig. 4a). The results obtained demonstrate that constitutive expression of shmR completely restores the ability to use iron from haemin in an hmuP mutant context, indicating that no other defects in haemin transport and utilization exist in the hmuP mutant except the lack of ShmR.
HmuP is not required for the global response to iron limitation
In order to evaluate a possible effect of the hmuP mutation on other genes involved in iron uptake and metabolism, we compared by qPCR the expression of shmR, rhrA, rhbE and SMc01515 genes in the wild-type and HMUP mutant strains grown in iron-restricted media. The gapA housekeeping gene was used as control. As shown in Fig. 5, expression of the rhrA and rhbE genes was not modified by the hmuP disruption. Siderophore quantification assays reinforced these results, as the siderophore production in iron-restricted media was similar in the wild-type and the hmuP mutant strain (data not shown). These observations suggest that HmuP is not involved in this iron-restriction response.
The expression of SMc01515, a putative gene encoding a TonB homologue, was similar in the wild-type and mutant strains (Fig. 5). SMc01515 is adjacent to and transcribed in the opposite direction from hmuP. These results indicate that HmuP is not involved in SMc01515 expression.
The hmuTUV gene cluster has recently been described as the ABC transport system involved in the utilization not only of haem but also of ferrichrome and ferrioxamine B in S. meliloti 2011 (Cuiv et al., 2008). The utilization of Desferal for iron nutrition was not affected in the hmuP mutant strain (Table 2). Similar results were obtained when ferrichrome was included as the sole iron source (data not shown). These results indicate that the transposon insertion does not affect xenosiderophore internalization and corroborate our previous data showing that the hmuP mutation has no polar effect on hmuUV expression.
Wild-type and HMUP strains containing the pOT-HmuPpr plasmid (hmuP presumptive promoter region fused to gfpuv) showed no GFP fluorescence difference when bacteria were grown in low-iron conditions (data not shown). This result agrees with our previous findings which show that the hmuS and hmuT genes, which are co-transcribed with hmuP, are expressed in the hmuP mutant (Fig. 5). These data demonstrate that HmuP is not involved in its own transcriptional regulation.
In sum, these results suggest that HmuP is not required for the global response to iron limitation.
HmuP is well conserved among proteobacteria
Using the blast algorithm with the deduced amino acid sequence of S. meliloti 1021 HmuP as a query, HmuP homologues were found mainly in the alpha-, beta- and gamma-proteobacteria, although some examples were also found in the delta- and epsilon-proteobacteria. As expected, HmuP from S. meliloti is more similar to other HmuP homologues from the alpha-proteobacteria, mainly among the Rhizobiales, having 81 % identity with R. leguminosarum bv. viciae strain 3841, 66 % with Mesorhizobium loti MAFF303099 and 44 % with Rhodopseudomonas palustris CGA009. The alignment of the last 36 amino acid residues of the C terminus indicates a high conservation in this region (Supplementary Fig. S1). The presence of a KLILXK motif at the end of the region is highly conserved. The structure of the HmuP homologue in Rhodopseudomonas palustris CGA009 has been determined by NMR (PDB ID: 2jra), showing that this protein is a dimer with four β-sheets and one small α-helix per monomer. According to the SWISS-MODEL SIB Service (Arnold et al., 2006; Guex & Peitsch, 1997; Schwede et al., 2003), the S. meliloti HmuP protein is structured in three β-sheets and one small α-helix. The C-terminal β-sheet has a well-conserved KLILXK motif, suggesting that this region is important for protein function (Supplementary Fig. S1).
The analysis of the genomic context of the hmuP homologues using GenCont (Hinrichs et al., 2006) revealed that the hmuP gene is usually located near other genes related to iron or haem uptake and metabolism. Usually, it is located adjacent to hmuTUV in the alpha-proteobacteria, and also to hmuS, tonB, exbB/exbD, haem outer membrane receptors and bacterioferritin (data not shown). This localization suggests that HmuP has a conserved role in haem and/or iron metabolism.
The C-terminal motif is essential for HmuP function
The high conservation of the KLILXK C-terminal motif in HmuP proteins suggests that it is important for HmuP function. To investigate this hypothesis, we mutagenized this motif by site-directed mutagenesis, and the mutated hmuP gene was used to complement the HMUP mutant in trans. When the S. meliloti HmuP GKLILNK motif or the C-terminal lysine was deleted (in pOT-HmuPΔCt and pOT-HmuPΔK, respectively), no significant shmR : : gusA expression was observed (Fig. 3b). Furthermore, when an extra tyrosine was present at the C terminus (pOT-HmuPY), a partial complementation was obtained (Fig. 3b). These results show that the integrity of the C-terminal GKLILNK motif of HmuP is essential for transcriptional activation of the shmR gene. It is worth mentioning that the presence of mutagenized versions of hmuP in trans in the wild-type context did not affect expression of shmR : : gusA, which means that these mutations are recessive with respect to the wild-type allele (Fig. 3b).
Subsequently, we evaluated by bioassays and growth assays the ability of the different mutagenized versions of HmuP to complement the hmuP mutant strain phenotype. The results obtained showed that the presence of pOT-HmuPY in trans was able to fully complement the hmuP mutant phenotype, restoring the ability to use haemin as iron source. The presence of pOT-HmuPΔK showed only a minor effect, while the presence of pOT-HmuPΔCt was unable to restore the use of haem-iron sources (Table 2, Fig. 4b). These observations were corroborated by using the toxic haemin analogue gallium protoporphyrin-IX (Ga-PPIX). Ga-PPIX was highly toxic for HMUP (pOT-HmuP) and for HMUP (pOT-HmuPY), but non-toxic for HMUP (pOT-HmuPΔCt). Toxicity was moderate for HMUP (pOT-HmuPΔK) (data not shown). A plausible explanation for the results obtained for the HMUP (pOT-HmuPΔK) strain could be that the small activation of shmR gene expression produced by the HmuP mutated protein results in a small but active haemin transport, although the expression is not enough to produce a detectable level of β-glucuronidase activity.
Altogether, these results support the conclusion that the GKLILNK C-terminal motif is required for HmuP-dependent expression of the haem receptor.
HmuP is not essential for nitrogen fixation
The hmuP mutant strain was able to develop nitrogen-fixing nodules in Medicago sativa cv. Creola plants, indicating that HmuP is not essential for symbiosis or nitrogen fixation in alfalfa under the conditions assayed (data not shown). These results agree with previously reported data which show that the shmR gene is not necessary for nitrogen fixation (Amarelle et al., 2008).
DISCUSSION
In this work, hmuP was found to be essential for transcriptional expression of the haemin receptor gene shmR in S. meliloti 1021. We found that an S. meliloti hmuP mutant was unable to express shmR by using transcriptional fusion analyses and qPCR (Figs 3 and 5). Not only does disruption of hmuP prevent transcriptional activation of shmR under iron-limited conditions, but multiple copies of hmuP result in increased transcriptional activation. In addition, we found that shmR expression was recovered by the presence of hmuP in trans (Fig. 3). These results were corroborated by growth analysis that showed that the hmuP mutant was unable to use haem and haem compounds as iron sources and that this phenotype was recovered by the presence of HmuP (Fig. 4, Table 2). Altogether, these results demonstrate that HmuP works under iron limitation to activate transcription of shmR. It has been demonstrated that the shmR gene is negatively regulated by iron via RirA (Chao et al., 2005; Viguier et al., 2005). RirA is the major iron-responsive regulator in S. meliloti, and the RirA regulon comprises numerous genes involved in iron transport. In particular, besides the shmR gene, different genes involved in rhizobactin synthesis and transport are also repressed by RirA when cells are grown in iron-sufficient conditions (Chao et al., 2005; Viguier et al., 2005). Interestingly, the rhizobactin synthesis and transport system responds to a second level of regulation, requiring the presence of the RhrA transcriptional activator under iron-limiting conditions. It has been established that RhrA belongs to the AraC-like family of transcriptional activators, although the induction signal and the mechanism of action of RhrA are still unknown (Lynch et al., 2001). In addition, it seems to be a general rule that xenosiderophore receptors require different types of positive regulators for their expression. These results, together with our observations, suggest that the expression of genes involved in high-affinity iron transport requires not only the relief of RirA iron repression but also the positive action of a transcriptional regulator.
Furthermore, analysis of gene expression and siderophore production indicates that disruption of hmuP does not affect other genes related to the iron-restriction response (Fig. 5, Table 2). It is possible, however, that another role for hmuP exists but is masked by redundancy with other components. The fact that constitutive expression of shmR restores the ability to use haemin of an hmuP mutant indicates that it is not required for transcriptional activation of other haemin-related genes, and that it has no direct role as an effector in haemin-transport mechanisms. Our results strongly indicate that the main function of HmuP in S. meliloti 1021 is the regulation of shmR transcription.
In numerous bacterial genomes, hmuP homologues are located next to genes which are predicted to be involved in haemin transport, e.g. in Y. pestis the hmuP homologue is present in the hmuPRSTUV gene cluster, while in Ralstonia solanacearum it is in the tonBexbB1exbD1hemP genetic organization. Co-localization of hmuP with genes related to the iron-restriction response, together with the results obtained in this work, suggests that HmuP has a conserved role in haem and/or iron metabolism in bacteria.
Sequence alignment of HmuP homologues, as well as comparison with the NMR solution structure of the Rhodopseudomonas palustris CGA009 HmuP protein, revealed that certain amino acids localized within the predicted β-sheets are well conserved. Moreover, the C-terminal β-sheet presents a KLILXK motif highly conserved among the C termini of the HmuP homologues (Supplementary Fig. S1). Here, we demonstrate that the deletion of the conserved GKLILNK motif produces a non-functional HmuP protein, indicating that this motif is important for HmuP activity.
It is not possible from the data obtained here to conclude that HmuP functions as a direct positive transcriptional regulator, or whether it is an indirect regulator. The wide distribution of HmuP homologues among bacteria makes this regulator an interesting subject for further studies.
Acknowledgments
This research was supported by a grant from the NIH Fogarty International Research Collaboration, award R03 TW007353, to M. R. O'B., with E. F. as the foreign collaborator; by NIH grant GM067966 to M. R. O'B.; and by a grant from Programa de Desarrollo de las Ciencias Básicas (PEDECIBA) and Comisión Sectorial de Investigación Cientıfica (CSIC), Uruguay, to E. F. and F. N.
Abbreviations
EDDHA, ethylenediamine-di-o-hydroxyphenylacetic acid
qPCR, quantitative real-time PCR
5′-RACE, rapid amplification of 5′ complementary DNA ends
Footnotes
A supplementary figure, showing an alignment of the last 36 amino acid residues (HemP domain) of some HmuP homologues from the alpha-, beta-, gamma-, delta- and epsilon-proteobacteria, and a supplementary table, showing primers used in the study, are available with the online version of this paper.
References
- Allaway, D., Schofield, N. A., Leonard, M. E., Gilardoni, L., Finan, T. M. & Poole, P. S. (2001). Use of differential fluorescence induction and optical trapping to isolate environmentally induced genes. Environ Microbiol 3, 397–406. [DOI] [PubMed] [Google Scholar]
- Amarelle, V., O'Brian, M. R. & Fabiano, E. (2008). ShmR is essential for utilization of heme as a nutritional iron source in Sinorhizobium meliloti. Appl Environ Microbiol 74, 6473–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. C., Robinson, A. K. & Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27, 215–237. [DOI] [PubMed] [Google Scholar]
- Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201. [DOI] [PubMed] [Google Scholar]
- Beringer, J. E. (1974). R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84, 188–198. [DOI] [PubMed] [Google Scholar]
- Chao, T. C., Becker, A., Buhrmester, J., Puhler, A. & Weidner, S. (2004). The Sinorhizobium meliloti fur gene regulates, with dependence on Mn(II), transcription of the sitABCD operon, encoding a metal-type transporter. J Bacteriol 186, 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, T. C., Buhrmester, J., Hansmeier, N., Puhler, A. & Weidner, S. (2005). Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl Environ Microbiol 71, 5969–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuiv, P. O., Clarke, P., Lynch, D. & O'Connell, M. (2004). Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J Bacteriol 186, 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuiv, P. O., Keogh, D., Clarke, P. & O'Connell, M. (2008). The hmuUV genes of Sinorhizobium meliloti 2011 encode the permease and ATPase components of an ABC transport system for the utilization of both haem and the hydroxamate siderophores, ferrichrome and ferrioxamine B. Mol Microbiol 70, 1261–1273. [DOI] [PubMed] [Google Scholar]
- Diaz-Mireles, E., Wexler, M., Sawers, G., Bellini, D., Todd, J. D. & Johnston, A. W. (2004). The Fur-like protein Mur of Rhizobium leguminosarum is a Mn2+-responsive transcriptional regulator. Microbiology 150, 1447–1456. [DOI] [PubMed] [Google Scholar]
- Ditta, G., Stanfield, S., Corbin, D. & Helinski, D. R. (1980). Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A 77, 7347–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan, T. M., Hartweig, E., LeMieux, K., Bergman, K., Walker, G. C. & Signer, E. R. (1984). General transduction in Rhizobium meliloti. J Bacteriol 159, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadis, M. M., Komiya, H., Chakrabarti, P., Woo, D., Kornuc, J. J. & Rees, D. C. (1992). Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257, 1653–1659. [DOI] [PubMed] [Google Scholar]
- Griffitts, J. S. & Long, S. R. (2008). A symbiotic mutant of Sinorhizobium meliloti reveals a novel genetic pathway involving succinoglycan biosynthetic functions. Mol Microbiol 67, 1292–1306. [DOI] [PubMed] [Google Scholar]
- Guex, N. & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hennecke, H. (1992). The Role of Respiration in Symbiotic Nitrogen Fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers.
- Hinrichs, D., Wetten, M. & Meuwissen, T. H. (2006). An algorithm to compute optimal genetic contributions in selection programs with large numbers of candidates. J Anim Sci 84, 3212–3218. [DOI] [PubMed] [Google Scholar]
- Hohle, T. H. & O'Brian, M. R. (2009). The mntH gene encodes the major Mn2+ transporter in Bradyrhizobium japonicum and is regulated by manganese via the Fur protein. Mol Microbiol 72, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung, J. M., Jones, H. A. & Perry, R. D. (1996). The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem – protein complexes as iron sources. Mol Microbiol 20, 725–739. [DOI] [PubMed] [Google Scholar]
- Humann, J. L., Schroeder, B. K., Mortimer, M. W., House, B. L., Yurgel, S. N., Maloney, S. C., Ward, K. L., Fallquist, H. M., Ziemkiewicz, H. T. & Kahn, M. L. (2008). Construction and expression of sugar kinase transcriptional gene fusions by using the Sinorhizobium meliloti ORFeome. Appl Environ Microbiol 74, 6756–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humann, J. L., Ziemkiewicz, H. T., Yurgel, S. N. & Kahn, M. L. (2009). Regulatory and DNA repair genes contribute to the desiccation resistance of Sinorhizobium meliloti Rm1021. Appl Environ Microbiol 75, 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R. A., Burgess, S. M. & Hirsh, D. (1986). Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci U S A 83, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch, J. K., Nedelmann, M., Kiel, K., Bartscht, K., Horstkotte, M. A., Dobinsky, S., Rohde, H. & Mack, D. (2003). Establishment of an arbitrary PCR for rapid identification of Tn917 insertion sites in Staphylococcus epidermidis: characterization of biofilm-negative and nonmucoid mutants. Appl Environ Microbiol 69, 5812–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, D., O'Brien, J., Welch, T., Clarke, P., Cuiv, P. O., Crosa, J. H. & O'Connell, M. (2001). Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J Bacteriol 183, 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade, H. M., Long, S. R., Ruvkun, G. B., Brown, S. E. & Ausubel, F. M. (1982). Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Ngok-Ngam, P., Ruangkiattikul, N., Mahavihakanont, A., Virgem, S. S., Sukchawalit, R. & Mongkolsuk, S. (2009). Roles of Agrobacterium tumefaciens RirA in iron regulation, oxidative stress response, and virulence. J Bacteriol 191, 2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noya, F., Arias, A. & Fabiano, E. (1997). Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J Bacteriol 179, 3076–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persmark, M., Pittman, P., Buyer, J. S., Schwyn, B., Gill, P. R., Jr & Neilands, J. B. (1993). Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J Am Chem Soc 115, 3950–3956. [Google Scholar]
- Platero, R., Peixoto, L., O'Brian, M. R. & Fabiano, E. (2004). Fur is involved in manganese-dependent regulation of mntA (sitA) expression in Sinorhizobium meliloti. Appl Environ Microbiol 70, 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, P. S., Blyth, A., Reid, C. J. & Walters, K. (1994). myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 140, 2787–2795. [Google Scholar]
- Rees, D. C. & Howard, J. B. (2000). Nitrogenase: standing at the crossroads. Curr Opin Chem Biol 4, 559–566. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sangwan, I. & O'Brian, M. R. (1992). Characterization of delta-aminolevulinic acid formation in soybean root nodules. Plant Physiol 98, 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. (2003). SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31, 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Priefer, U. & Pühler, A. (1983). Vector Plasmids for In-Vivo and In-Vitro Manipulations of Gram-Negative Bacteria. Edited by A. Pühler. Berlin: Springer.
- Stojiljkovic, I. & Hantke, K. (1994). Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol 13, 719–732. [DOI] [PubMed] [Google Scholar]
- Todd, J. D., Wexler, M., Sawers, G., Yeoman, K. H., Poole, P. S. & Johnston, A. W. (2002). RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology 148, 4059–4071. [DOI] [PubMed] [Google Scholar]
- Viguier, C., Cuív, P. Ó., Clarke, P. & O'Connell, M. (2005). RirA is the iron response regulator of the rhizobactin 1021 biosynthesis and transport genes in Sinorhizobium meliloti 2011. FEMS Microbiol Lett 246, 235–242. [DOI] [PubMed] [Google Scholar]
- Wexler, M., Yeoman, K. H., Stevens, J. B., de Luca, N. G., Sawers, G. & Johnston, A. W. (2001). The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol Microbiol 41, 801–816. [DOI] [PubMed] [Google Scholar]
- Wolk, C. P., Cai, Y. & Panoff, J. M. (1991). Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci U S A 88, 5355–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Sangwan, I., Lindemann, A., Hauser, F., Hennecke, H., Fischer, H. M. & O'Brian, M. R. (2006). Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol Microbiol 60, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurgel, S. N. & Kahn, M. L. (2005). Sinorhizobium meliloti dctA mutants with partial ability to transport dicarboxylic acids. J Bacteriol 187, 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]