Abstract
The Campylobacter jejuni flagellin protein is O-glycosylated with structural analogues of the nine-carbon sugar pseudaminic acid. The most common modifications in the C. jejuni 81-176 strain are the 5,7-di-N-acetylated derivative (Pse5Ac7Ac) and an acetamidino-substituted version (Pse5Am7Ac). Other structures detected include O-acetylated and N-acetylglutamine-substituted derivatives (Pse5Am7Ac8OAc and Pse5Am7Ac8GlnNAc, respectively). Recently, a derivative of pseudaminic acid modified with a di-O-methylglyceroyl group was detected in C. jejuni NCTC 11168 strain. The gene products required for Pse5Ac7Ac biosynthesis have been characterized, but those genes involved in generating other structures have not. We have demonstrated that the mobility of the NCTC 11168 flagellin protein in SDS-PAGE gels can vary spontaneously and we investigated the role of single nucleotide repeats or homopolymeric-tract-containing genes from the flagellin glycosylation locus in this process. One such gene, Cj1295, was shown to be responsible for structural changes in the flagellin glycoprotein. Mass spectrometry demonstrated that the Cj1295 gene is required for glycosylation with the di-O-methylglyceroyl-modified version of pseudaminic acid.
INTRODUCTION
The Gram-negative, microaerophilic bacterium Campylobacter jejuni is one of the most common causes of gastroenteritis in humans. Cells of C. jejuni are motile due to the action of single bipolar flagella, and functional flagella are required for both intestinal colonization of animal models (Aguero-Rosenfeld et al., 1990; Nachamkin et al., 1993; Pavlovskis et al., 1991; Wassenaar et al., 1993) and adherence to and invasion of cultured eukaryotic cells (Grant et al., 1993; Wassenaar et al., 1991; Yao et al., 1994). The flagellum filament is formed from polymeric flagellin proteins FlaA and FlaB. These proteins display antigenic variation (Alm et al., 1992; Harris et al., 1987; Logan et al., 2002) and are post-translationally modified by glycosylation (Doig et al., 1996). The glycan component of the flagellin glycoprotein is O-linked to multiple (up to 19 in C. jejuni strain 81-176) serine/threonine residues in the central domain of the flagellin protein (Thibault et al., 2001). The glycan was initially thought to be N-acetyl neuraminic acid (Doig et al., 1996; Guerry et al., 1996) but is now known to be composed of the related sugar 5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-l-mannononulosonic acid or pseudaminic acid (Pse5Ac7Ac) (Thibault et al., 2001). As well as Pse5Ac7Ac, the flagellin from C. jejuni 81-176 can also be modified with structural analogues of this basal sugar such as an acetamidino variant (Pse5Am7Ac), as well as variants substituted with acetyl (Pse5Ac7Ac8OAc), propionyl (Pse5Pr7Pr) or N-acetyl glutamine structures (Pse5Am7Ac8GlnNAc) (Schirm et al., 2005; Thibault et al., 2001). In C. jejuni NCTC 11168, recent structural analyses have identified further novel flagellin modifications consisting of the related sugar legionaminic acid and derivatives, along with di-O-methyl glyceric acid derivatives of Pse5Ac7Ac and Pse5Ac7Am (Logan et al., 2009).
The genes involved in Campylobacter flagellin glycosylation cluster in a single locus that also contains the tandemly arranged flaA and flaB genes (Szymanski et al., 2003b; Szymanski & Wren, 2005). The genes encoding the enzymes required for the biosynthesis of cytidine monophosphate-activated pseudaminic acid (CMP-Pse5Ac7Ac) from N-acetylglucosamine (GlcNAc) have been identified in both Campylobacter and Helicobacter pylori (Chou et al., 2005; Creuzenet, 2004; Goon et al., 2003; Ishiyama et al., 2006; Liu & Tanner 2006; Obhi & Creuzenet, 2005; Schirm et al., 2003; Schoenhofen et al., 2006a, b). However, these genes constitute only a subset of those forming the Campylobacter flagellin glycosylation locus (Guerry et al., 2006; McNally et al., 2006) and the genes involved in generating the array of structural analogues of pseudaminic acid that are detected on the Campylobacter flagellin remain unidentified.
One mechanism for generating structural variation at the bacterial cell surface is phase variation, defined as the random and reversible switching on/off of gene expression. Phase variation in C. jejuni and a number of bacterial pathogens such as Neisseria meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae and H. pylori, is due to the presence of short, often intragenic, nucleotide repeats (Moxon et al., 2006). Such repeats are relatively unstable due to their inherent susceptibility to a process termed slipped strand mispairing that occurs during DNA replication. The resultant insertion/deletion events produce frame-shifting and populations of cells that differ in their ability to produce full-length proteins. In Campylobacter coli, this process mediates a motile to non-motile phenotypic switch due to phase variation of a single nucleotide repeat or homopolymeric tract within the flhA gene (Park et al., 2000). The genome sequence of C. jejuni NCTC 11168 revealed an abundance of hypervariable homopolymeric tracts of G/C residues (Parkhill et al., 2000). Genes containing such homopolymeric tracts are mostly clustered in three loci involved in biosynthesis of the surface-located structures capsular polysaccharide and lipo-oligosaccharide (LOS), as well as the flagellin glycosylation locus (Parkhill et al., 2000). These homopolymeric tracts have been shown to have a high frequency of insertion/deletion mutations resulting in polymorphic populations of C. jejuni cells (Parkhill et al., 2000; Wassenaar et al., 2002). In glycosyltransferase-encoding genes from the LOS biosynthetic locus, phase variation associated with homopolymeric G/C tracts generates LOS structural variation (Gilbert et al., 2002; Linton et al., 2000). Structural analysis of C. jejuni capsular polysaccharide identified variation in the glycan component of the glycolipid (Szymanski et al., 2003a), but the role of homopolymeric-tract-containing genes from the capsular biosynthetic locus has not been identified. Translational frameshifting at G/C homopolymeric tracts within maf genes from the C. jejuni NCTC 11168 flagellin glycosylation locus was shown to partially restore motility in a non-motile maf gene mutant (Karlyshev et al., 2002), and more recently, mutation of a homopolymeric-tract-containing maf gene resulted in altered flagellin glycosylation (van Alphen et al., 2008). It has also been reported that phase variation of flagellar biosynthesis can be mediated by mutation at single nucleotide repeats within the genes encoding the FlgSR two-component system (Hendrixson, 2006, 2008).
It has been known for some time that Campylobacter flagellin proteins can undergo spontaneous antigenic variation (Harris et al., 1987). More recently, considerable structural variation of the flagellin glycan has been demonstrated; however, the genetic basis of this structural variation has not been determined. In this report, we demonstrate that the homopolymeric-tract-containing Cj1295 gene, encoding a protein of unknown function, is responsible for generating a defined structural variant of the glycan component of the C. jejuni flagellin glycoprotein.
METHODS
Bacterial strains and growth conditions.
All C. jejuni strains were grown at 37 °C on 7 % horse blood (TCS Biosciences) agar supplemented, as required, with kanamycin (50 μg ml−1) and chloramphenicol (20 μg ml−1) in a microaerobic chamber (Don Whitley) with an atmosphere of 10 % carbon dioxide, 5 % oxygen and 85 % nitrogen. Motility of C. jejuni strains was analysed by inoculating 0.4 % agar Mueller–Hinton plates. Escherichia coli strains were grown in Luria–Bertani (LB) broth or on LB agar at 37 °C supplemented, as required, with ampicillin (100 μg ml−1), kanamycin (50 μg ml−1) and chloramphenicol (20 μg ml−1).
Insertional mutagenesis of the C. jejuni NCTC 11168 Cj1295 gene.
A 1308 bp PCR product was amplified from C. jejuni NCTC 11168 genomic DNA with primers 1295mutf (5′-GTG GAT AAG CTA GAC TTT AGA-3′) and 1295mutr (5′-TCA TTT TAC CAG TCC ATA AAA-3′) and cloned into plasmid pGEM-T Easy (Invitrogen). The resultant plasmid (p1295) was cut at the single BclI site located at nt 789–794 inclusive of the 1308 bp insert generated by PCR; a kanamycin cassette (Trieu-Cuot et al., 1985) with BamHI termini was ligated into this site to generate plasmid p1295 : : aphA. Following transformation into E. coli TOP 10 cells (Invitrogen), transformants were selected on kanamycin-containing media and restriction digestion was used to identify plasmids with the kanamycin cassette in the same orientation as the Cj1295 gene. This plasmid was electroporated into C. jejuni NCTC 11168 using standard methods (van Vliet et al., 1998) and transformants were verified by PCR.
Complementation of the Cj1295 gene knockout mutant.
In order to complement the Cj1295 knockout mutant, we integrated a functional copy of the Cj1295 gene onto the chromosome. To minimize disruption of other genes we targeted a region annotated as a pseudogene (Cj0223) in the C. jejuni NCTC 11168 genome sequence and a vector designed to integrate functional genes onto the chromosome was designed. This was based on a 2179 bp fragment corresponding to nt 205297–207475 inclusive of the C. jejuni NCTC 11168 genome sequence, cloned into pUC18. In order to facilitate insertion of genes into the centre of this region, a SpeI restriction site was created using site-directed mutagenesis. The primer 5′-GGT GTA GTA AGT ACT AGT AAT TGT AAT GTC C-3′ and its exact complement were employed to generate the SpeI site (underlined).
In parallel, a 1411 bp PCR product was amplified from C. jejuni NCTC 11168 genomic DNA using primers 1295F2 (5′-GCT GAA CTT AGT ATA CCT TGT-3′) and 1295mutr (see above) with Pfu proof-reading polymerase (Stratagene). This amplified DNA from 103 bp upstream of the putative start codon of the Cj1295 gene through to the putative stop codon. This PCR product was A-tailed with Taq polymerase (Bioline) and cloned into pGEM-T Easy (Invitrogen). The single nucleotide repeat of G residues was then altered using site-directed mutagenesis employing a pair of complementary primers: 1295mutf (5′-TAA ATA AAA CTC TGG GAG GGG GTA TAC TC-3′) and the exact complement 1295mutr.
A chloramphenicol resistance cassette was amplified from plasmid pAV35 (van Vliet et al., 1998) using primers catFAatIISpeI (5′-TGA CTG GAC GTC ACT AGT TGC TCG GCG GTG TTC CTT TCA AG-3′) and catRSacII (5′-TCA CTG CCG CGG TTA TTT ATT CAG CAA GTC TTG TAA-3′), digested with AatII and SacII and cloned into AatII/SacII-digested plasmid p1295QC. The chloramphenicol resistance cassette and full-length Cj1295 gene with an interrupted G/C tract were cut out from this plasmid with SpeI and ligated into the SpeI site of the pseudogene-containing pUC18 based plasmid (see above) to create plasmid pCj1295comp.
Following sequence verification of the Cj1295 gene, this construct was electroporated into C. jejuni NCTC 11168 and chloramphenicol-resistant colonies were isolated. Genomic DNA was extracted from a chloramphenicol-resistant strain and checked for insertion of the full-length Cj1295 gene with interrupted G/C tract along with the chloramphenicol resistance cassette into the pseudogene Cj0223 by PCR. Genomic DNA from this strain was used to naturally transform strain C. jejuni NCTC 11168 Cj1295 : : aphA to chloramphenicol resistance using established methods (van Vliet et al., 1998). Kanamycin- and chloramphenicol-resistant colonies were isolated and the site of insertion of the functional Cj1295 gene with interrupted G/C tract was verified by PCR and sequencing.
Analysis of flagellin mobility by protein electrophoresis and Western blotting.
Whole-cell protein extracts were electrophoresed through 12 % Bis/Tris polyacrylamide gels (Invitrogen), according to the manufacturer's instructions. Gels were either stained with colloidal Coomassie stain (Pierce) or electroblotted to PVDF membranes. Flagellin proteins were visualized using a polyclonal anti-flagellin antibody (kindly supplied by Dr Shaun Cawthraw and Professor Diane Newell) and anti-rabbit IgG peroxidase-coupled secondary antibody (Sigma) with DAB staining (Vector).
Partial purification of flagellin protein and analysis by mass spectrometry.
Flagellin protein was purified using the method described by Power et al. (1994). Purified flagellin was separated by SDS-PAGE and stained using Novex colloidal blue reagent (Invitrogen) and the desired protein was excised, lyophilized and digested with trypsin (E.C.3.4.21.4, Promega) overnight. Peptides were then extracted from gel pieces and analysed by nano-liquid chromatography/electrospray ionization (LC-ES)–MS/MS using a reverse-phase nano-HPLC system (Dionex) connected to a quadrupole time-of-flight mass spectrometer (Q-STAR Pulsar I, MDS Sciex). The digests were separated by a binary nano-HPLC gradient generated by an Ultimate pump fitted with a Famos autosampler and a Switchos microcolumn switching module (LC Packings). An analytical C18 nanocapillary (75 μm inside diameter×15 cm; PepMap) and a micro precolumn C18 cartridge were employed for online peptide separation. The digest was first loaded onto the precolumn and eluted with 0.1 % formic acid (Sigma) in water (HPLC grade; Purite) for 4 min. The eluant was then transferred onto an analytical C18 nanocapillary HPLC column and eluted at a flow rate of 150 nl min−1. The following gradient was employed: 99 % solvent A from 0 to 5 min, 99–90 % A from 5 to 10 min, 90–60 % A from 10 to 70 min, 60–50 % A from 70 to 71 min, 50–5 % A from 71 to 75 min, 5 % A from 75 to 85 min, 5–95 % A from 85 to 86 min, and 95 % A from 86 to 90 min. Solvent A consisted of 0.05 % (v/v) formic acid in a 95 : 5 (v/v) water/acetonitrile mixture, whilst solvent B consisted of 0.04 % formic acid in a 95 : 5 (v/v) acetonitrile/water mixture. Data acquisition was performed using Analyst QS software with an automatic information-dependent-acquisition function.
RESULTS
Instability of homopolymeric-tract-containing genes is associated with altered flagellin mobility
Whilst investigating flagella-mediated motility in C. jejuni NCTC 11168, two single colony-derived motile variants with a minor difference in whole-cell protein profile between the 50 kDa and 75 kDa markers were obtained by chance (boxed in Fig. 1a). In order to determine whether this was due to heterogeneity of the approximately 60 kDa flagellin protein, Western blotting was carried out using a flagellin-specific antiserum (Fig. 1b). This clearly demonstrated that the flagellin proteins from the two variants, termed v1 and v2, had different electrophoretic mobilities.
Fig. 1.
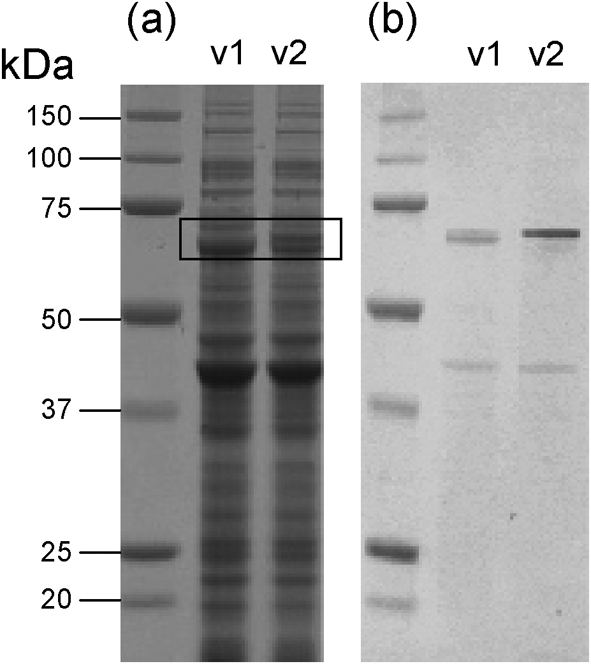
Altered mobility of flagellin protein from motile variants of C. jejuni NCTC 11168. (a) Whole-cell protein profiles of variants v1 and v2 of C. jejuni NCTC 11168. The boxed region outlines a region of the gel where there is apparent variation. (b) Western blot (a) probed with anti-flagellin antiserum. A clear flagellin protein mobility shift is evident when comparing v1 and v2. The faint band observed between 37 and 50 kDa is due to non-specific binding of antibody to the major outer-membrane protein.
We hypothesized that these flagellin mobility variants arose due to changes in hypervariable homopolymeric tracts, more specifically 12 homopolymeric G/C tracts from those identified by Parkhill et al. (2000) that are potentially involved in flagellin modification (Table 1). Within the C. jejuni NCTC 11168 flagellin glycosylation locus, there are ten homopolymeric tracts of G/C residues, nine of which are intragenically located, indicating potentially phase variable genes, along with a single intergenic homopolymeric tract of G residues located just upstream of the Cj1321 gene. Two further homopolymeric G/C tracts located outside of the flagellin glycosylation locus were also included as they were located within genes (Cj0617 and Cj0170/1) that had significant levels of sequence similarity to homopolymeric tract-containing genes from the flagellin glycosylation locus (Table 1). The repeat containing regions from each of the 12 homopolymeric tract-containing genes associated with flagellin glycosylation were amplified by PCR and the tract was sequenced. Comparison of the on/off status of the corresponding genes identified three flagellin glycosylation-associated genes that differed in their translational on/off status between the flagellin mobility variants NCTC 11168 v1 and v2. Two of these, the adjacent Cj1305 and Cj1306 genes, belong to the Cj0617 paralogous gene family encoding putative proteins of unknown function. In the v1 variant, the Cj1305 gene is in-frame and Cj1306 is out-of-frame, whilst in the v2 variant, the Cj1305 gene is out-of-frame and Cj1306 is in-frame. The third gene that differed in translational on/off status between flagellin mobility variants NCTC 11168 v1 and v2 is coding sequence Cj1295. In the v1 variant, Cj1295 is out-of-frame with eight G residues, whilst in the v2 variant, the Cj1295 gene is in-frame with nine G residues (Table 1). Given the complexity of the Cj0617 paralogous gene family and that Cj1305 and Cj1306 both differed in translational status between v1 and v2, we decided to investigate the role of the homopolymeric-tract-containing Cj1295 gene in flagellin modification.
Table 1.
Sequence of homopolymeric tracts associated with the C. jejuni NCTC 11168 flagellin glycosylation locus in flagellin mobility variants
| CDS | Putative function | Length of single nucleotide repeat (bp) and consequent translational status* | |
|---|---|---|---|
| Variant 1 | Variant 2 | ||
| Cj1295 | Aminopeptidase | 8 (−) | 9 (+) |
| Cj1296/7 | N-Acetyltransferase | 8 (−) | 8 (−) |
| Cj1305 | Unknown (Cj0617 family) | 9 (+) | 10 (−) |
| Cj1306 | Unknown (Cj0617 family) | 8 (−) | 9 (+) |
| Cj1310 | Unknown (Cj0617 family) | 9 (+) | 9 (+) |
| Cj1342 | Unknown (Cj0617 family) | 9 (+) | 9 (+) |
| Cj1325/6 | Unknown | 10 (−) | 10 (−) |
| Cj0617 | Unknown (Cj0617 family) | 10 (+) | 10 (+) |
| Upstream of Cj1321 | Transferase | 10 | 10 |
| Cj1317/8 | Unknown (Cj1318 family) | 9 (−) | 9 (−) |
| Cj1334 | Unknown (Cj1318 family) | 9 (−) | 9 (−) |
| Cj0170/1 | Unknown | 10 (−) | 9 (−) |
*+, In-frame; −, out-of-frame.
Analysis of the C. jejuni Cj1295 gene
The C. jejuni NCTC 11168 Cj1295 predicted coding sequence encodes a putative protein of 435 aa (49.9 kDa) that lacks both a predicted signal peptide sequence and transmembrane regions. This putative protein belongs to the Protein Information Resource PIRSF015244 family with the tentative function of ‘polysaccharide biosynthesis protein with an aminopeptidase-like domain’. Members of this protein family are found in phylogenetically diverse bacterial species, including Frankia and Streptomyces, from the phylum Actinobacteria; Bradyrhizobium and Brucella, from the Alphaproteobacteria; Hydrogenivirga from the Aquificae; C. jejuni, C. coli, Campylobacter lari, Campylobacter fetus and Campylobacter upsaliensis from the Delta-/Epsilonproteobacteria; Clostridium acetobutylicum from the phylum Firmicutes; and Aeromonas from the Gammaproteobacteria. In the majority of species, the coding sequences encoding Cj1295-like proteins are located within clusters of putative genes involved in sugar biosynthesis/modification or sugar transferases. A number of these clusters also contain genes annotated as sialic acid synthases. For Campylobacter spp. and C. acetobutylicum, Cj1295 homologue-containing gene clusters encode proteins involved in sugar biosynthesis/modification and flagellum biosynthesis.
In C. jejuni, the Cj1295 coding sequence is part of a large cluster of genes, a number of which encode enzymes known to be involved in glycosylation of the flagellin protein. This locus varies in gene content among C. jejuni strains; however, Cj1295-like coding sequences are present in all ten C. jejuni strains for which sequence data are available, along with C. coli, C. lari, C. fetus and C. upsaliensis. Comparison of C. jejuni Cj1295-like coding sequences from different strains reveals high levels of nucleotide sequence identity apart from at the homopolymeric tract of G/C residues located in the 5′ region of the gene.
Construction of ‘knock-out’ and ‘knock-in’ Cj1295 genes
In order to investigate Cj1295 gene product function, an insertional knockout mutant was created in C. jejuni NCTC 11168 to create C. jejuni NCTC 11168 Cj1295 : : aphA. For comparison, a Cj1295 gene was constructed with an interrupted homopolymeric tract. Full-length Cj1295 was cloned and site-directed mutagenesis was employed to alter the homopolymeric tract of G residues so that (i) the gene was in-frame, (ii) the run of G residues was interrupted and (iii) there was no change to the amino acid sequence. This was achieved by inserting an adenosine (A) residue to replace a G residue so that the resulting gene contained a region consisting of three G residues followed by a single A residue followed by five G residues rather than a continuous run of nine G residues.
In order to construct a C. jejuni strain expressing a Cj1295 gene with an interrupted homopolymeric tract, the engineered variant was recombined onto the chromosome at a site distant from the flagellin glycosylation locus. To minimize possible secondary effects of gene insertion, the site chosen was within a pseudogene (annotated as Cj0223). Expression of the Cj1295 gene with an interrupted homopolymeric tract was under the control of the promoter associated with a chloramphenicol resistance gene placed immediately upstream and in the same transcriptional orientation. Insertion of the adjacent chloramphenicol resistance and Cj1295 genes within the Cj0223 pseudogene on the C. jejuni chromosome was verified by PCR (data not shown).
Cj1295 gene function is associated with reduced electrophoretic mobility of the flagellin glycoprotein
The possibility that the instability of other homopolymeric-tract-containing genes might contribute to any phenotypic changes observed when studying Cj1295 gene function necessitated the following approach. Genomic DNA from NCTC 11168 with a Cj1295 gene with an interrupted homopolymeric tract inserted into the pseudogene Cj0223 (see above), was used to naturally transform C. jejuni NCTC 11168 Cj1295 : : aphA. From the transformation mixture, both singly (kanamycin) and doubly (kanamycin and chloramphenicol) resistant colonies were isolated. The kanamycin-resistant colonies lack a functional Cj1295 gene whilst the doubly kanamycin- and chloramphenicol-resistant colonies contain a functional Cj1295 gene. The mobility of the flagellin protein was determined directly from three kanamycin-resistant and three kanamycin/chloramphenicol-resistant single colonies. In agreement with data obtained from variants v1 and v2 (see above), the flagellin glycoprotein from the kanamycin-resistant colonies lacking a functional Cj1295 gene had decreased mobility in SDS-PAGE gels compared with flagellin from the kanamycin/chloramphenicol-resistant colonies containing a functional Cj1295 gene (Fig. 2). This experiment was repeated three times and in each case, a similar shift in flagellin mobility was observed. These data indicate that Cj1295 function is involved in modification of the flagellin protein and that inherent instability of the homopolymeric tract would result in distinct populations of C. jejuni NCTC 11168 with structurally variant flagellin glycoprotein. Despite these variant flagellins, both strains were fully motile (as assessed by point-inoculating reduced agar plates), both were flagellated (as assessed by electron microscopy) and there was no difference in their capacity to autoagglutinate (data not shown).
Fig. 2.
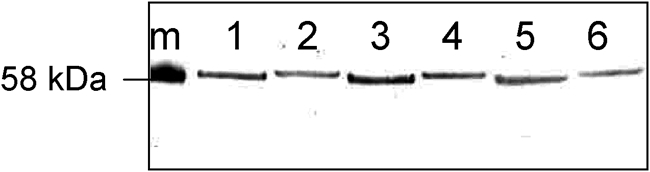
Flagellin mobility shift due to Cj1295 gene function. Three kanamycin-resistant colonies with non-functional Cj1295 genes (lanes 1, 3 and 5) and three kanamycin- and chloramphenicol-resistant colonies with functional Cj1295 genes (lanes 2, 4 and 6) were analysed by Western blotting with an anti-flagellin antibody. Lane m, molecular mass marker.
Cj1295 gene function is associated with the di-O-methyl-glyceric acid modification of pseudaminic acid
Flagellin proteins from C. jejuni NCTC 11168 1295 : : aphA and the complemented strain, were partially purified by SDS-PAGE and analysed by mass spectrometry. Gel pieces corresponding to flagellin were subjected to in-gel trypsin digestion and the resulting peptides were eluted and analysed by LC-ES-MS/MS. The data were analysed for the presence of oxonium ions at m/z 317 and 316, which correspond to the flagellin glycan Pse5Ac7Ac and its acetamidino derivative Pse5Am7Ac, respectively, previously demonstrated to modify the flagellin of C. jejuni 81-176 (Thibault et al., 2001). Additionally, it has recently been reported that the flagellin of C. jejuni NCTC 11168 is modified with a di-O-methyl-glyceric acid derivative of pseudaminic acid (Logan et al., 2009). Accordingly, the data were also analysed for the presence of oxonium ions at m/z 391 and 390, which are indicative of the presence of this pseudaminic acid derivative on flagellin glycopeptides.
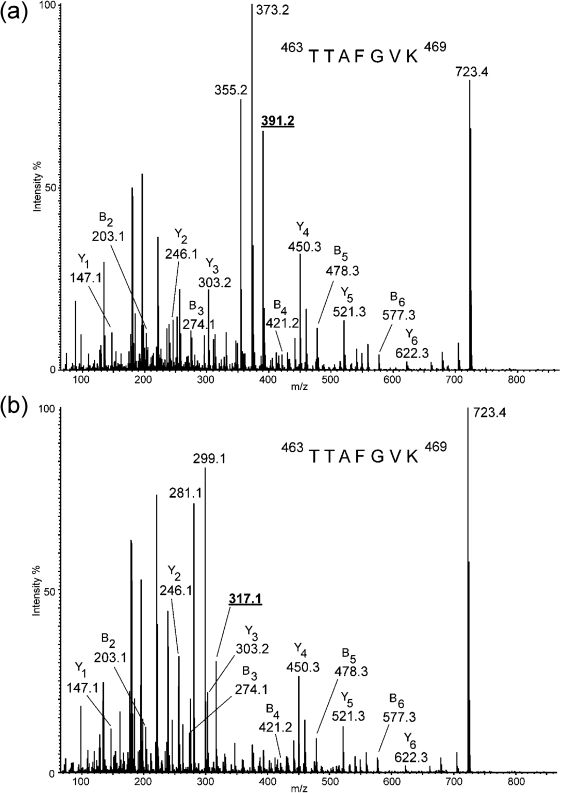
The data from the LC-ES-MS/MS analysis of the flagellin from C. jejuni NCTC 11168 Cj1295 : : aphA cmCj1295 strain indicated that the most abundant modification found on the flagellin glycopeptides was the dimethylglyceric acid derivative of Pse5Ac7Ac (oxonium ion at m/z 391; Fig. 3a and Table 2). Glycopeptides were also detected that were modified with its related acetamidino derivative (oxonium ion at m/z 390) and with Pse5Ac7Ac, respectively (Table 2).
Fig. 3.
NanoLC-ES-MS/MS analysis of the tryptic digest of C. jejuni 11168 flagellin. The spectra show the glycopeptide 463TTAFGVK469 modified with a single glycan. (a) The doubly charged ion at m/z 557.3 derived from the NCTC 11168 Cj1295 : : aphA cmCj1295 strain expressing full-length Cj1295 with an interrupted homopolymeric tract. The characteristic oxonium ion at m/z 391 for the dimethylglyceric acid analogue of pseudaminic acid is underlined. (b) The doubly charged ion at m/z 520.3 derived from the NCTC 11168 Cj1295 : : aphA strain that does not express the Cj1295 gene product. The characteristic oxonium ion at m/z 317 for pseudaminic acid is underlined.
Table 2.
LC-ES-MS/MS analysis of flagellin-derived tryptic glycopeptides from the NCTC 11168 Cj1295 : : aphA cmCj1295 strain expressing a full-length Cj1295 gene with an interrupted homopolymeric tract (Cj1295+) and the NCTC 11168 Cj1295 : : aphA strain (Cj1295−)
| Peptide | Observed glycopeptide signal (m/z) | Glycan oxonium ion | Deduced [M+H]+ | Peptide sequence | Modification | Flagellin |
|---|---|---|---|---|---|---|
| Cj1295+ | ||||||
| T463-469 | 557.3 [M+2H]2+ | 391 | 1113.5 | TTAFGVK | Pse5dimethylglycerate7Ac | FlaA, FlaB |
| T179-190 | 843.4 [M+2H]2+ | 391 | 1685.8 | ISSSGEVQFTLK | Pse5dimethylglycerate7Ac | FlaB |
| T179-190 | 850.4 [M+2H]2+ | 391 | 1699.8 | ISTSGEVQFTLK | Pse5dimethylglycerate7Ac | FlaA |
| T203-222 | 1167.6 [M+2H]2+ | 391 | 2334.2 | VVISTSVGTGLGALADEINK | Pse5dimethylglycerate7Ac | FlaA |
| 1130.6 [M+2H]2+ | 317 | 2260.2 | Pse5Ac7Ac | |||
| T203-222 | 783.4 [M+3H]3+; 1174.3 [M+2H]2+ | 391 | 2348.2 | VVISTSVGTGLGALAEEINK | Pse5dimethylglycerate7Ac | FlaB |
| T338-365 | 1202.2 [M+3H]3+ | 390 | 3604.7 | DILISGSNLSSAGFGATQFISQASVSLR | Pse5dimethylglycerate7Am ×2 | FlaA, FlaB |
| Cj1295− | ||||||
| T463-469 | 520.3 [M+2H]2+ | 317 | 1039.4 | TTAFGVK | Pse5Ac7Ac | FlaA, FlaB |
| T179-190 | 806.4 [M+2H]2+ | 317 | 1611.7 | ISSSGEVQFTLK | Pse5Ac7Ac | FlaB |
| T179-190 | 813.4 [M+2H]2+ | 317 | 1625.7 | ISTSGEVQFTLK | Pse5Ac7Ac | FlaA |
| T203-222 | 1130.6 [M+2H]2+ | 317 | 2260.1 | VVISTSVGTGLGALADEINK | Pse5Ac7Ac | FlaA |
| T203-222 | 1137.6 [M+2H]2+ | 317 | 2274.1 | VVISTSVGTGLGALAEEINK | Pse5Ac7Ac | FlaB |
| T338-365 | 1153.6 [M+3H]3+ | 316 | 3456.7 | DILISGSNLSSAGFGATQFISQASVSLR | Pse5Ac7Am x 2 | FlaA, FlaB |
In contrast, the LC-ES-MS/MS analysis following trypsin digestion of the flagellin from NCTC 11168 Cj1295 : : aphA strain indicated the distinct absence of glycopeptides modified with dimethylglyceric acid pseudaminic acid derivative. The glycopeptides observed were modified with Pse5Ac7Ac or its acetamidino derivative (oxonium ions at m/z 317 or 316, respectively; Fig. 3b and Table 2). The increased mass of the dimethylglyceric acid derivatives of pseudaminic acid relative to Pse5Ac7Ac/Pse5Ac7Am correlates with the decreased mobility in SDS-PAGE gels of the flagellin protein modified with this sugar given that there are multiple sites of glycosylation on the flagellin protein.
DISCUSSION
The identification of a large number of homopolymeric tracts of G/C residues, many of which were shown to be unstable, within genes from chromosomal loci involved in biosynthesis of capsular polysaccharide, LOS and the flagellin glycoprotein was the most significant finding of the C. jejuni NCTC 11168 genome sequencing project (Parkhill et al., 2000). The resultant on/off switching in the translational status of homopolymeric-tract-containing genes is thought to mediate generation of structural diversity of the bacterial cell surface. Indeed, the presence of many homopolymeric-tract-containing genes in the flagellin glycosylation locus is congruent with the observed structural heterogeneity of the glycan component of the flagellin glycoprotein (Logan et al., 2009; Schirm et al., 2005; Thibault et al., 2001). However, despite considerable progress in defining the genes from this locus that encode the enzymes required for biosynthesis of Pse5Ac7Ac, the most commonly detected flagellin modification (Chou et al., 2005; Creuzenet, 2004; Goon et al., 2003; Ishiyama et al., 2006; Liu & Tanner 2006; Obhi & Creuzenet 2005; Schirm et al., 2003; Schoenhofen et al., 2006b), little progress has been made in determining how the observed structural variants of Pse5Ac7Ac are generated and what role homopolymeric tract-containing genes may have. We have tested the hypothesis that homopolymeric-tract-containing genes from the flagellin glycosylation locus are involved in generating flagellin glycan diversity by identifying spontaneously generated intra-strain variation in mobility of the C. jejuni NCTC 11168 flagellin glycoprotein and associating this with differences in the translational status of homopolymeric-tract-containing genes Cj1305, Cj1306 and Cj1295 from the flagellin glycosylation locus. We have gone on to define more precisely the role of Cj1295 in generating variant flagellins of differing mobility and demonstrated that this is due to altered flagellin glycan structure. This study is therefore an important first step towards defining the components of the flagellin glycosylation machinery that are involved in generating structural diversity in the Campylobacter flagellin glycan.
The approach employed to investigate Cj1295 gene function involved construction of a C. jejuni NCTC 11168 Cj1295 ‘knock-out’ strain by standard insertional knockout mutagenesis along with a so-called ‘knock-in’ strain. The latter was generated by interrupting the homopolymeric tract of G residues and then introducing this gene into the genome of the ‘knock-out’ strain within a pseudogene (see Methods). This strategy ensured direct comparison of isogenic strains differing only in their capacity to produce a functional Cj1295 gene product. When flagellin proteins from these strains were compared, there was a clear difference in their mobility in SDS-PAGE gels so that a functional Cj1295 gene was associated with a flagellin glycoprotein of reduced mobility. When conducting such experiments, it is important that the contribution of other unstable homopolymeric-tract-containing genes from the flagellin glycosylation locus is considered, as their spontaneous frame-shifting may also lead to modification of the flagellin glycan with consequent mobility shifting. Therefore, we repeated the transformation experiments three times and analysed the flagellin mobility of multiple single colonies directly from the primary transformation plates, i.e. without subsequent subculture. In all cases, a consistent flagellin mobility shift was observed. We further investigated the cause of this mobility shift by mass spectrometry, showing that the Cj1295 gene function was associated with the presence of a di-O-methyl glyceric acid-derivative of pseudaminic acid (Logan et al., 2009). The full complement of NCTC 11168 flagellin glycosylation sites has not yet been identified, but our data (Table 2) suggest that the majority are modified with the di-O-methyl glyceric acid derivative of pseudaminic acid when the Cj1295 gene is functional.
The Cj1295 gene is located at one end of the large cluster of genes known as the flagellin glycosylation locus. A number of genes from this locus (including Cj1293 and Cj1294 located immediately upstream of Cj1295) are involved in the biosynthesis of pseudaminic acid (Goon et al., 2003; Obhi & Creuzenet, 2005), the principal glycan component of the flagellin glycoprotein. Indeed, all genes necessary for the biosynthesis of cytidine monophosphate-activated pseudaminic acid (CMP-Pse5Ac7Ac) from GlcNAc have been identified (Chou et al., 2005; Creuzenet, 2004; Goon et al., 2003; Ishiyama et al., 2006; Liu & Tanner 2006; Obhi & Creuzenet, 2005; Schirm et al., 2003; Schoenhofen et al., 2006a, b), and the corresponding gene products are capable of synthesizing CMP-Pse5Ac7Ac from GlcNAc in vitro (Schoenhofen et al., 2006a). However, analysis of the Campylobacter flagellin glycan from strains 81-176 and NCTC 11168 has clearly shown that, as well as Pse5Ac7Ac, a number of structurally related analogues are present (Logan et al., 2009; Schirm et al., 2005; Thibault et al., 2001). The predicted amino acid sequence of the Cj1295 gene product contains an aminopeptidase-like domain, yet the Cj1295 gene product activity is associated with changes not to the flagellin polypeptide but to the flagellin glycan, more specifically, presence of a pseudaminic acid-like sugar of 389–390 Da. In a very recent report, this modification in NCTC 11168 flagellin was identified by mass spectrometry, NMR and metabolomics as CMP-7-acetamido-5-(2,3-di-O-methylglyceroyl)amino-3,5,7,9-tetradeoxy-l-glycero-α-l-manno-nonulosonic acid (Logan et al., 2009). We hypothesize that the Cj1295 gene product is directly involved in generating this modification. Our current working hypothesis is, that in cells where the Cj1295 gene product is produced, the protein acts to cleave the acetamido group on carbon 5 of the basal Pse5Ac7Ac/Pse5Ac7Am sugar. The presence of an aminopeptidase-like domain in Cj1295 suggests that the covalent bond linking the amide group, which resembles a peptide bond, may be the cleavage site enabling substitution of the acetamido group with a methylglyceroyl group. Knockout mutation of genes involved in flagellin glycosylation that result in altered flagellin charge train patterns have also been shown to alter autoagglutination properties of the corresponding C. jejuni cells (Guerry et al., 2006; van Alphen et al., 2008). We observed no change in autoagglutination associated with the presence or absence of a functional Cj1295 gene, presumably because the associated glycan variation did not result in altered charge.
The Cj1295 gene is one of ten homopolymeric-tract-containing genes found within the flagellin glycosylation locus of strain NCTC 11168. Of particular interest are the Cj0617 gene family in which all five members (four located in the flagellin glycosylation locus) contain homopolymeric tracts. Also of interest are a second paralogous gene family known as the motility accessory factor or maf genes (Karlyshev et al., 2002). Of the seven maf genes present in the C. jejuni NCTC 11168 flagellin glycosylation locus, two contain homopolymeric tracts. Both the Cj0617 and maf gene family, along with other homopolymeric-tract-containing genes of the flagellin glycosylation locus are likely to contribute to generating diversity in the flagellin glycan, although their specific contributions will require further detailed study. Comparison of flagellin glycosylation loci from different strains of C. jejuni reveals considerable diversity in gene content, but the Cj1295 gene is present in all strains for which genome sequence data are available and in each case contains a homopolymeric tract.
Phase variable expression of short sequence repeat-containing genes encoding glycosyltransferases leads to variant glycans of meningococcal and gonococcal pilin glycoproteins (Aas et al., 2007; Banerjee et al., 2002; Power et al., 2003). In a similar manner, the Cj1295 homopolymeric-tract-containing gene generates variant flagellin glycosylation and it is interesting to speculate on such diversity in C. jejuni. One possible explanation is that during colonization, the random generation of structural diversity in the flagellin glycan is important in evading host immune responses. The flagellin protein is an immunodominant antigen in C. jejuni-colonized chickens (Cawthraw et al., 1994) and in chickens immunized with Campylobacter antigens (Widders et al., 1998). Flagellin is also recognized by antibodies from the sera of humans infected with C. jejuni (Cawthraw et al., 2002; Nachamkin & Hart, 1985; Wenman et al., 1985). Alternatively, the generation of such diversity may be involved in preventing the binding of flagellatropic bacteriophages (Coward et al., 2006).
Acknowledgments
This work was funded by the award of a Wellcome Trust Career Development Fellowship (D. L.), UK Biotechnology and Biological Science Research Council Core Support Grant B19088 (A. D. and H. R. M.), BBSRC Integrative Systems Biology Grant BBC5196701 (P. H. and A. D.) and BBSRC DTA studentship (J. A. B.). We thank Professors Ian S. Roberts (University of Manchester) and Brendan W. Wren (London School of Hygiene and Tropical Medicine) for their advice and support.
Abbreviations
GlcNAc, N-acetylglucosamine
LB, Luria–Bertani
LC-ES, liquid chromatography/electrospray ionization
LOS, lipo-oligosaccharide
References
- Aas, F. E., Vik, A., Vedde, J., Koomey, M. & Egge-Jacobsen, W. (2007). Neisseria gonorrhoeae O-linked pilin glycosylation: functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol 65, 607–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguero-Rosenfeld, M. E., Yang, X. H. & Nachamkin, I. (1990). Infection of adult Syrian hamsters with flagellar variants of Campylobacter jejuni. Infect Immun 58, 2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm, R. A., Guerry, P., Power, M. E. & Trust, T. J. (1992). Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J Bacteriol 174, 4230–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A., Wang, R., Supernavage, S. L., Ghosh, S. K., Parker, J., Ganesh, N. F., Wang, P. G., Gulati, S. & Rice, P. A. (2002). Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J Exp Med 196, 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthraw, S., Ayling, R., Nuijten, P., Wassenaar, T. & Newell, D. G. (1994). Isotype, specificity, and kinetics of systemic and mucosal antibodies to Campylobacter jejuni antigens, including flagellin, during experimental oral infections of chickens. Avian Dis 38, 341–349. [PubMed] [Google Scholar]
- Cawthraw, S. A., Feldman, R. A., Sayers, A. R. & Newell, D. G. (2002). Long-term antibody responses following human infection with Campylobacter jejuni. Clin Exp Immunol 130, 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, W. K., Dick, S., Wakarchuk, W. W. & Tanner, M. E. (2005). Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J Biol Chem 280, 35922–35928. [DOI] [PubMed] [Google Scholar]
- Coward, C., Grant, A. J., Swift, C., Philp, J., Towler, R., Heydarian, M., Frost, J. A. & Maskell, D. A. (2006). Phase-variable surface structures are required for infection of Campylobacter jejuni by bacteriophages. Appl Environ Microbiol 72, 4638–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creuzenet, C. (2004). Characterization of Cj1293, a new UDP-GlcNAc C6 dehydratase from Campylobacter jejuni. FEBS Lett 559, 136–140. [DOI] [PubMed] [Google Scholar]
- Doig, P., Kinsella, N., Guerrry, P. & Trust, T. J. (1996). Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol 19, 379–387. [DOI] [PubMed] [Google Scholar]
- Gilbert, M., Karwaski, M.-F., Bernatchez, S., Young, N. M., Taboada, E., Michniewicz, J., Cunningham, A. M. & Wakarchuk, W. W. (2002). The genetic basis for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. J Biol Chem 277, 327–337. [DOI] [PubMed] [Google Scholar]
- Goon, S., Kelly, J. F., Logan, S. M., Ewing, C. P. & Guerry, P. (2003). Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol 50, 659–671. [DOI] [PubMed] [Google Scholar]
- Grant, C. C., Konkel, M. E., Cieplak, W. & Tompkins, L. S. (1993). Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun 61, 1764–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry, P., Doig, P., Alm, R. A., Burr, D. H., Kinsella, N. & Trust, T. J. (1996). Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol Microbiol 19, 369–378. [DOI] [PubMed] [Google Scholar]
- Guerry, P., Ewing, C. P., Schirm, M., Lorenzo, M., Kelly, J., Pattarini, D., Majam, G., Thibault, P. & Logan, S. (2006). Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, L. A., Logan, S. M., Guerry, P. & Trust, T. J. (1987). Antigenic variation in Campylobacter flagella. J Bacteriol 169, 5066–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson, D. R. (2006). A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol Microbiol 61, 1646–1659. [DOI] [PubMed] [Google Scholar]
- Hendrixson, D. R. (2008). Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol Microbiol 70, 519–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama, N., Creuzenet, C., Miller, W. L., Demendi, M., Anderson, E. M., Harauz, G., Lam, J. S. & Berghuis, A. M. (2006). Structural studies of FlaA1 from Helicobacter pylori reveal the mechanism for inverting 4,6-dehydratase activity. J Biol Chem 281, 24489–24495. [DOI] [PubMed] [Google Scholar]
- Karlyshev, A. V., Linton, D., Gregson, N. A. & Wren, B. W. (2002). A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148, 473–480. [DOI] [PubMed] [Google Scholar]
- Linton, D., Gilbert, M., Hitchen, P. G., Dell, A., Morris, H. R., Wakarchuk, W. W., Gregson, N. A. & Wren, B. W. (2000). Phase variation of a β-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol Microbiol 37, 501–514. [DOI] [PubMed] [Google Scholar]
- Liu, F. & Tanner, M. E. (2006). PseG of pseudaminic acid biosynthesis, a UDP-sugar hydrolase as a masked glycosyltransferase. J Biol Chem 281, 20902–20909. [DOI] [PubMed] [Google Scholar]
- Logan, S. M., Kelly, J. F., Thibault, P., Ewing, C. P. & Guerry, P. (2002). Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol Microbiol 46, 587–597. [DOI] [PubMed] [Google Scholar]
- Logan, S. M., Hui, J. P. M., Vinogradov, E., Aubry, A. J., Melanson, J. E., Kelly, J. F., Nothaft, H. & Soo, E. C. (2009). Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J 276, 1014–1023. [DOI] [PubMed] [Google Scholar]
- McNally, D. J., Hui, J. P. M., Aubry, A. J., Mui, K. K. K., Guerry, P., Brisson, J.-R., Logan, S. M. & Soo, E. C. (2006). Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81–176 using a focused metabolomics approach. J Biol Chem 281, 18489–18498. [DOI] [PubMed] [Google Scholar]
- Moxon, R., Baylis, C. & Hood, D. (2006). Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet 40, 307–333. [DOI] [PubMed] [Google Scholar]
- Nachamkin, I. & Hart, A. M. (1985). Western blot analysis of the human antibody response to Campylobacter jejuni cellular antigens during gastrointestinal infection. J Clin Microbiol 21, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachamkin, I., Yang, X. H. & Stern, N. J. (1993). Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol 59, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obhi, R. K. & Creuzenet, C. (2005). Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aninotransferase that generates UDP-4-amino-4,6-dideoxy-GalNAc. J Biol Chem 280, 20902–20908. [DOI] [PubMed] [Google Scholar]
- Park, S. F., Purdy, D. & Leach, S. (2000). Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J Bacteriol 182, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., Chillingworth, T., Davies, R. M., Feltwell, T. & other authors (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. [DOI] [PubMed] [Google Scholar]
- Pavlovskis, O. R., Rollins, D. M., Haberberger, R. L., Jr., Green, A. E., Habash, L., Strocko, S. & Walker, R. I. (1991). Significance of flagella in colonization resistance of rabbits immunized with Campylobacter spp. Infect Immun 59, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, M. E., Guerry, P., McCubbin, W. D., Kay, C. M. & Trust, T. J. (1994). Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J Bacteriol 176, 3303–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, P. M., Roddam, L. F., Rutter, K., Fitzpatrick, S. Z., Srikhanta, Y. N. & Jennings, M. P. (2003). Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol 49, 833–847. [DOI] [PubMed] [Google Scholar]
- Schirm, M., Soo, E. C., Aubry, A. J., Austin, J., Thibault, P. & Logan, S. M. (2003). Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol 48, 1579–1592. [DOI] [PubMed] [Google Scholar]
- Schirm, M., Schoenhofen, I. C., Logan, S. M., Waldron, K. C. & Thibault, P. (2005). Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal Chem 77, 7774–7782. [DOI] [PubMed] [Google Scholar]
- Schoenhofen, I. C., McNally, D. J., Brisson, J.-R. & Logan, S. M. (2006a). Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16, 8C–14C. [DOI] [PubMed] [Google Scholar]
- Schoenhofen, I. C., Lunin, V. L., Julien, J.-P., Li, Y., Ajamian, E., Matte, A., Cygler, M., Brisson, J. R., Aubry, A. & other authors (2006b). Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem 281, 8907–8916. [DOI] [PubMed] [Google Scholar]
- Szymanski, C. M. & Wren, B. W. (2005). Protein glycosylation on bacterial mucosal pathogens. Nat Rev Microbiol 3, 225–237. [DOI] [PubMed] [Google Scholar]
- Szymanski, C. M., St. Michael, F., Jarrell, H. C., Li, J., Gilbert, M., Larocque, S., Vinogradov, E. & Brisson, J. R. (2003a). Detection of N-linked glycans and phase variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J Biol Chem 278, 24509–24520. [DOI] [PubMed] [Google Scholar]
- Szymanski, C. M., Logan, S. M., Linton, D. & Wren, B. W. (2003b). Campylobacter – a tale of two protein glycosylation systems. Trends Microbiol 11, 233–238. [DOI] [PubMed] [Google Scholar]
- Thibault, P., Logan, S. M., Kelly, J. F., Brisson, J.-R., Ewing, C. P., Trust, T. J. & Guerry, P. (2001). Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem 276, 34862–34870. [DOI] [PubMed] [Google Scholar]
- Trieu-Cuot, P., Gerbaud, G., Lambet, T. & Courvalin, P. (1985). In vivo transfer of genetic information between Gram-positive and Gram-negative bacteria. EMBO J 4, 3583–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen, L. B., Wuhrer, M., Bleumink-Pluym, N. M. C., Hensbergen, P. J., Deeelder, A. M. & van Putten, J. P. M. (2008). A functional Campylobacter jejuni maf4 gene results in novel glycoforms on flagellin and altered autoagglutination behaviour. Microbiology 154, 3385–3397. [DOI] [PubMed] [Google Scholar]
- van Vliet, A. H. M., Wood, A. C., Henderson, J., Wooldridge, K. & Ketley, J. M. (1998). Genetic manipulation of enteric Campylobacter species. In Methods in Microbiology, vol. 271, Bacterial Pathogenesis, pp. 407–419. Edited by P. Williams, J. Ketley & G. Salmond. San Diego: Academic Press.
- Wassenaar, T. M., Bleumink-Pluym, N. M. & van der Zeijst, B. A. (1991). Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J 10, 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar, T. M., van der Zeijst, B. A., Ayling, R. & Newell, D. G. (1993). Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J Gen Microbiol 139, 1171–1175. [DOI] [PubMed] [Google Scholar]
- Wassenaar, T. M., Wagenaar, J. A., Rigter, A., Fearnley, C., Newell, D. G. & Duim, B. (2002). Homonucleotide stretches in chromosomal DNA of Campylobacter jejuni display high frequency polymorphism as detected by direct PCR analysis. FEMS Microbiol Lett 212, 77–85. [DOI] [PubMed] [Google Scholar]
- Wenman, W. M., Chai, J., Louie, T. J., Goudreau, C., Lior, H., Newell, D. G., Pearson, A. D. & Taylor, D. E. (1985). Antigenic analysis of Campylobacter flagellar protein and other proteins. J Clin Microbiol 21, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widders, P. R., Thomas, L. M., Long, K. A., Tokhi, M. A., Panaccio, M. & Apos, E. (1998). The specificity of antibody in chickens immunised to reduce intestinal colonisation with Campylobacter jejuni. Vet Microbiol 64, 39–50. [DOI] [PubMed] [Google Scholar]
- Yao, R., Burr, D. H., Doig, P., Trust, T. J., Niu, H. & Guerry, P. (1994). Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol 14, 883–893. [DOI] [PubMed] [Google Scholar]