Abstract
Pseudomonas aeruginosa (Pa) and Burkholderia cepacia complex (Bcc) lung infections are responsible for much of the mortality in cystic fibrosis (CF). However, little is known about the ecological interactions between these two, often co-infecting, species. This study provides what is believed to be the first report of the intra- and interspecies bacteriocin-like inhibition potential of Pa and Bcc strains recovered from CF patients. A total of 66 strains were screened, and shown to possess bacteriocin-like inhibitory activity (97 % of Pa strains and 68 % of Bcc strains showed inhibitory activity), much of which acted across species boundaries. Further phenotypic and molecular-based assays revealed that the source of this inhibition differs for the two species. In Pa, much of the inhibitory activity is due to the well-known S and RF pyocins. In contrast, Bcc inhibition is due to unknown mechanisms, although RF-like toxins were implicated in some strains. These data suggest that bacteriocin-based inhibition may play a role in governing Pa and Bcc interactions in the CF lung and may, therefore, offer a novel approach to mediating these often fatal infections.
INTRODUCTION
Individuals with cystic fibrosis (CF) face a lifelong battle with chronic bacterial lung infections. Pseudomonas aeruginosa (Pa) is the predominant infectious agent in the lungs of adult CF patients and most individuals are infected shortly after birth (Cystic Fibrosis Foundation, 2007). Alginate-producing mucoid variants emerge over the course of several years, and form dense bacterial biofilms in the CF lung. Once established, eradication of these infections is generally not possible (Govan & Deretic, 1996; Hentzer et al., 2001).
In the 1970s, members of the Burkholderia cepacia complex (Bcc) of species were first identified in the airways of CF patients (Govan & Deretic, 1996). Although few in number (∼3 %), the clinical manifestations of these infections can be severe (Cystic Fibrosis Foundation, 2007; Kalish et al., 2006). Colonization may be asymptomatic, or result in progressive decline in lung function. A smaller number of infections result in ‘cepacia syndrome’, which is a fatal pneumonia that results in death (Jones et al., 2004; Kalish et al., 2006; Tablan et al., 1985). Given that CF patients are initially infected with Pa, Bcc strains must either compete with or act in synergy with established Pa biofilms (Al-Bakri et al., 2004). How such interactions occur is unknown. In fact, little is known about the process of Bcc invasion and its outcomes. Do the strains from the two species co-exist peacefully? Do Bcc strains out-compete and displace pre-existing Pa strains? Few studies have sought to address how these two bacterial species interact in the CF lung (Al-Bakri et al., 2004; McKenney et al., 1995; Weaver & Kolter, 2004).
One factor known to mediate bacterial interactions is the production of potent toxins known as bacteriocins (Riley & Gordon, 1999). Unlike traditional antibiotics, many bacteriocins have a relatively narrow killing range. They have been implicated in intraspecific competition brought on by limited nutrients (Riley & Wertz, 2002b). In some cases, however, bacteriocins are also able to kill more broadly, and they have been implicated as a primary mechanism for mediating microbial diversity (Kerr et al., 2002).
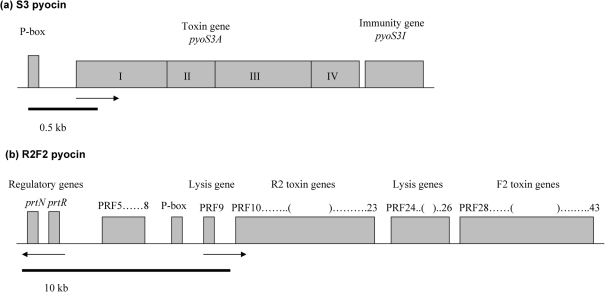
Much is already known about pyocins, the bacteriocins produced by Pseudomonas spp. In fact, more than 90 % of all Pa strains examined to date produce one or more of three pyocin types: S, R and F (Fyfe et al., 1984). S pyocins are high-molecular-mass proteins that resemble the well-known colicins produced by Escherichia coli. They are protease sensitive, and most kill by DNA degradation (Michel-Briand & Baysse, 2002). Four S pyocins (S1, S2, S3 and AP41) have been studied extensively (Duport et al., 1995; Sano & Kageyama, 1981; Sano et al., 1990, 1993a, b) (Fig. 1a). All S-pyocin operons share the presence of two genes: the larger gene (toxin gene) provides the killing activity, while the smaller (immunity gene) provides immunity against killing by that pyocin. The toxin gene comprises four domains: I produces receptor-binding activity, which enables a pyocin to recognize a specific target; II has an unknown function; III enables translocation of the toxin through the plasma membrane; and IV produces the killing (DNase) activity (Michel-Briand & Baysse, 2002).
Fig. 1.
Genetic organization of exemplar pyocin genes (adapted from Michel-Briand & Baysse, 2002). (a) S3 pyocin: P-box refers to the binding site for PrtN (not shown); the toxin gene is divided into four domains: I–IV. (b) R2F2 pyocin. Arrows indicate the direction of transcription.
The R and F pyocins, which resemble bacteriophage tails, are resistant to nuclease and protease digestion, and they kill by depolarizing the cell membrane. The genes encoding these pyocins are found in a cluster (which is why they are often referred to as RF pyocins), and they are located between trpE (anthranilate synthase component I) and trpG (anthranilate synthase component II) (Nakayama et al., 2000; Shinomiya et al., 1983). Fig. 1(b) shows the genetic organization of pyocin R2F2 genes. The open reading frames (PRF3–PRF43) include regulatory, lysis, and R and F pyocin genes. The lysis genes (PRF9, PRF24, PRF25 and PRF26) are similar in sequence to the lysis cassettes of P2 and lambda phages (Nakayama et al., 2000). There are 16 R and F pyocin genes (PRF10–PRF23 and PRF28–PRF43) (Nakayama et al., 2000). The encoded proteins show high levels of sequence similarity to tail genes of P2 and lambda phage (Nakayama et al., 2000). The function of PRF5–PRF8 is unknown.
The S, R and F pyocins are controlled by the same ptrN and ptrR regulatory genes (Fig. 1b) (Michel-Briand & Baysse, 2002; Nakayama et al., 2000). PtrN is a transcriptional activator that is repressed by PtrR (Matsui et al., 1993). Under DNA damage, RecA protein cleaves the PtrR repressor protein, which leads to expression of ptrN. PtrN binds to the regulatory sequence (P-box), thus inducing transcription of pyocin genes (Matsui et al., 1993).
Stress conditions, such as might be encountered in a CF lung, may induce pyocin production. Microarray analysis of Pa genomes has revealed that pyocin transcription is upregulated by hydrogen peroxide and ciprofloxacin (Brazas & Hancock, 2005; Chang et al., 2005). Waite & Curtis (2009) investigated the effect of pyocins in mixed-culture biofilms under aerobic and anaerobic conditions, and revealed that sensitivity to pyocin increased under anaerobic conditions. Further, Heo et al. (2007) revealed that R pyocins provide a competitive advantage during growth in planktonic conditions.
Far less is known about cepaciacins, which are the bacteriocins produced by Burkholderia. Approximately 30 % of Burkholderia cepacia (formerly Pseudomonas cepacia) strains produce cepaciacins (Govan & Harris, 1985). The few characterized cepaciacins resemble R-type pyocins, as they are resistant to trypsin digestion, and appear phage-tail-like when viewed under the electron microscope (Govan & Harris, 1985). Nothing further is known about the abundance, diversity or molecular biology of these toxins.
The goal of the present study was to determine whether clinical strains of Pa and Bcc isolated from the CF lung produce bacteriocins, and, if so, whether these play a role in mediating intra- and interspecific bacterial interactions in the CF lung. To this end, a collection of clinical Pa and Bcc strains from CF patients was screened for bacteriocin production and sensitivity. This collection consisted of strains isolated as paired (one strain of Pa and one strain of Bcc isolated from the same lung) or unpaired strains. Phenotypic screens for bacteriocin production, coupled with typing methods to identify individual bacteriocins, revealed a diversity of inhibitory mechanisms. Further, the patterns of within- and between-species inhibition suggested a role for these toxins in mediating bacterial interactions in the CF lung.
METHODS
Bacterial strains.
The bacterial strains used in this study included a total of 66 clinical strains of Pa (38 strains) and Bcc (28 strains) obtained from the Children's Hospital, Boston, MA, USA. The study protocol was approved by the Committee on Human Research at the Children's Hospital, and written informed consent was obtained. The clinical collection can be divided into two groups. The first was composed of 14 Pa and seven Bcc paired isolates procured from seven patients with CF. Each pair consisted of one Pa strain and one Bcc strain isolated from the same CF patient. The second group was composed of 24 Pa and 21 Bcc unpaired isolates, which were isolated from 31 patients. The Bcc strains included four genomovars (1 Burkholderia vietnamiensis, 12 Burkholderia multivorans, 11 Burkholderia dolosa and 4 Burkholderia cenocepacia). The reference strain collection included bacteriocin-producer strains, which are Pa strains known to produce the following pyocins: S1 and R4 (PML28), AP41 and F3 (PAF41), S2, R and F (NIH18), and S2, R2 and F2 (PAO1) (Ito et al., 1970; Kuroda & Kageyama, 1981; Nakayama et al., 2000; Sano et al., 1990; Seo & Galloway, 1990). The reference collection also included indicator strains sensitive to various combinations of pyocins: PML1516d (S1SS2SAP41S), NIH3 (S1SS2SAP41S), NIH3S1R (S1RS2SAP41S), NIH3S2R (S1SS2RAP41S), NIH3AP41R (S1SS2SAP41R), 3295 (AP41S), 3012 (AP41SF3S), 7NSK2 (S1SS3S), 7NSK2-fpvA (S1SS3R), PML14 (S1SR1SR2SR3SR4SR5S), 13s (S1SR1SR2SR3SR4SR5S) and NIH5 (ATCC 25317) (F1SF2SF3S) (de Chial et al., 2003; Kageyama et al., 1979; Kuroda & Kageyama, 1981; Sano & Kageyama, 1981; Sano et al., 1993a; Williams et al., 2008). Four cloned S pyocins (S1, S2, S3 and AP41) were employed in this bacteriocin identification scheme (Duport et al., 1995; Sano & Kageyama, 1993; Sano et al., 1993b). There is no corresponding set of Bcc reference strains.
Bacteriocin production and sensitivity screen.
The patch assay was used to identify bacteriocin-like inhibition, which involves overnight growth in 10 ml Luria–Bertani (LB) broth at 37 °C, with shaking at 250 r.p.m. A 6 ml volume of LB top agar (0.6 %, w/v), mixed with 3 μl mitomycin C (0.5 μg ml−1; Sigma) and 100 μl indicator cells (108 cells), was plated as a lawn on an LB agar plate (Pugsley & Oudega, 1987). The producer strain (∼106 cells) was spotted on the indicator overlay with a toothpick. Two spots per strain were applied. After overnight incubation at 37 °C, the plates were scored. If a strain produced an inhibition factor, such as a bacteriocin, active against the lawn, a zone of inhibition appeared. The clinical collection was screened in an all-by-all assay, i.e. each strain was used as a potential inhibitor producer and indicator. The all-by-all patch assay was done in duplicate. All positive results were tested a third time.
A one-tailed Mann–Whitney U test was used for non-parametric comparison of inhibitory activities in order to identify significant differences in inhibition frequencies (Zar, 1999).
Phenotypic bacteriocin identification.
The clinical collection was screened against the indicator strains using the patch assay. In addition, a cell-free extract of each putative bacteriocin-producing strain was subjected to trypsin digestion, filtration and freezing to distinguish between protease-sensitive and phage-like bacteriocins (Pugsley & Oudega, 1987; Riley et al., 2003). Cell-free extracts of putative producer strains were digested with trypsin (5 mg ml−1) for 30 min at 25 °C (to inactivate protease-sensitive bacteriocins), frozen at −70 °C (which tends to fracture phage-tail-type bacteriocins), and filtered in a 100 kDa Microcon YM-100 centrifugal filter device (Millipore) using centrifugation at 14 000 r.p.m. for 12 min (which tends to retain phage-tail-like bacteriocins). The treated cell-free extracts were tested on the lawns of pyocin-sensitive reference and clinical strains.
Molecular screening.
Genomic DNA was isolated using a DNeasy Tissue kit (Qiagen), and the Gram-negative protocol was applied to 2×109 cells from an overnight culture. A standard PCR protocol was used to screen genomes for the presence of all previously characterized S-type pyocin genes (encoding S1, S2, S3 and AP41) and their corresponding immunity genes (Duport et al., 1995; Sano & Kageyama, 1993; Sano et al., 1993b). Multiple primer sets were designed, based on existing pyocin sequences, to amplify different regions of each S pyocin gene and the corresponding immunity genes (see Supplementary Table S1, available with the online version of this paper). Further, primer sets specific to the regulatory region (P-box) of pyocins S1, S2 and AP41 were employed. Primers designed to amplify the RF-type pyocins, targeting PRF10, PRF31 and PRF38 genes, were taken from the literature (Nakayama et al., 2000). NCBI accession numbers of the sequenced pyocins, primer positions, primer sequences, amplicon sizes and identities are given in Table S1.
RESULTS
Inhibitory activity in Pa and Bcc strains
A phenotypic screen for inhibitory activity and sensitivity was conducted on a collection of 66 clinical strains of Pa (38 strains) and Bcc (28 strains) isolated from CF patients. The collection consisted of strains isolated as pairs, i.e. one Pa strain and one Bcc strain isolated from the same patient (taken from 7 patients, and resulting in 14 pairs; 14 Pa strains paired with 7 Bcc strains), and unpaired strains (24 Pa and 21 Bcc strains). Bacteriocin production and sensitivity was assayed in an all-by-all comparison, i.e. each strain was used as a putative producer and sensitive strain.
In this screen, Pa and Bcc strains showed different levels of inhibitory activity. The majority of the Pa strains (97 %) were inhibitory (Table 1). Most (76 %) inhibited both species, resulting in similar levels of intra- versus interspecific inhibition (92 % versus 81 %). Only 3 % of Pa strains showed no inhibitory activity.
Table 1.
Inhibitory activity of Pa and Bcc strains from the CF lung
PI, paired isolates; UPI, unpaired isolates; na, not applicable.
| Inhibitory activity | No. of Pa isolates (%) | No. of Bcc isolates (%) | ||||
|---|---|---|---|---|---|---|
| PI, n=14 | UPI, n=24 | Total, N=38 | PI, n=7 | UPI, n=21 | Total, N=28 | |
| No inhibition | 0 (0) | 1 (4) | 1 (3) | 4 (57) | 5 (24) | 9 (32) |
| Inhibits only Pa | 1 (7) | 5 (21) | 6 (16) | 0 (0) | 4 (19) | 4 (14) |
| Inhibits only Bcc | 1 (7) | 1 (4) | 2 (5) | 0 (0) | 3 (14) | 3 (11) |
| Inhibits both Pa and Bcc | 12 (86) | 17 (71) | 29 (76) | 3 (43) | 9 (43) | 12 (43) |
| Total inhibition | 14 (100) | 23 (96) | 37 (97) | 3 (43) | 16 (76) | 19 (68) |
| Paired inhibition | 2 (14) | na | 2 (5) | 0 (0) | na | 0 (0) |
The Bcc strains showed lower levels of overall inhibitory activity (68 %) (Table 1). The one-tailed Mann–Whitney U test confirmed significantly higher levels of total inhibition by Pa versus Bcc strains (P<0.0005). Similar to Pa, most Bcc strains (43 %) inhibited both species, again resulting in similar levels of intra- versus interspecific inhibition (54 % versus 57 %). A much larger fraction of Bcc strains (32 %) displayed no inhibitory activity.
The CF strain collection could be further divided into strains isolated as paired strains or unpaired strains. The paired and unpaired Pa strains showed similar levels of total inhibitory activity (Table 1). All paired and most unpaired (96 %) isolates possessed inhibitory activity. Most of the paired strains (86 %) inhibited both species, while the remainder inhibited either Bcc (7 %) or Pa (7 %). Two of the paired Pa strains inhibited their Bcc pair mates. The inhibitory unpaired Pa strains showed similar patterns of inhibitory activity: 71 % inhibited both species, and the remainder inhibited either Bcc (4 %) or Pa (21 %).
The paired Bcc strains showed inhibitory activity patterns that were significantly different from their Pa pair mates (Table 1). Only 43 % of paired Bcc isolates possessed inhibitory activity, at significantly lower levels than for paired Pa isolates (0.025<P<0.05), and all of these strains inhibited both species, but not their pair mates. The unpaired Bcc isolates showed quite similar inhibitory activity patterns compared with those of unpaired Pa strains. Most of the unpaired Bcc strains were inhibitory (76 %), and most of these (43 %) inhibited both species. The remainder inhibited only Bcc (14 %) or only Pa strains (19 %).
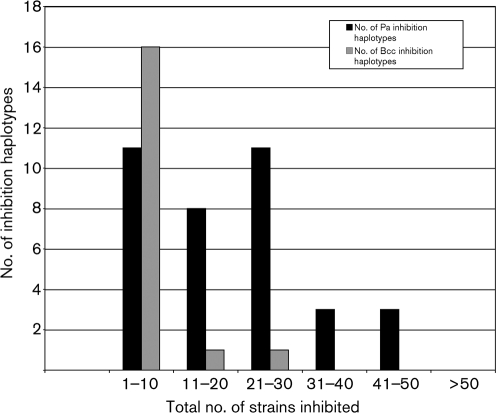
Inhibition haplotypes were determined for each producer strain. The 37 Pa and 19 Bcc producers displayed 36 and 18 haplotypes, respectively (Fig. 2). Further, all Pa isolates inhibited multiple Pa and Bcc strains; 12 Pa inhibited 1–10 strains (11 inhibition haplotypes), 8 inhibited 11–20 strains (8 inhibition haplotypes), 11 inhibited 21–30 strains (11 inhibition haplotypes), and 3 inhibited 31–40 strains (3 inhibition haplotypes). Three of the paired Pa isolates were able to inhibit most of the strains in the collection (41–50 strains). The Bcc isolates had a more limited inhibitory activity range. The majority (17 strains) inhibited 1–10 strains (16 inhibition haplotypes). Two Bcc strains were more broadly inhibitory, acting against 13 or 24 strains.
Fig. 2.
Inhibition haplotypes of clinical Pa and Bcc strains.
Bacteriocins of clinical Pa and Bcc strains are a source of inhibitory activity
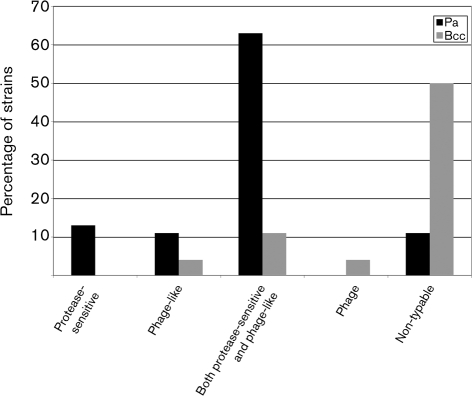
A series of phenotypic assays were employed to assess whether the observed inhibitory activities were due to bacteriocin or bacteriophage production. Cell-free extracts of each producer strain were subjected to filtration (to retain phage-like bacteriocins), protease digestion (to digest protein moieties) and freezing (to fracture phage-like bacteriocins). These assays revealed that 13 % of the Pa strains produced substances that appeared to be protease-sensitive bacteriocins, 11 % produced phage-like bacteriocins, and 63 % produced both (Fig. 3). Finally, 11 % of the Pa strains could not be characterized because of loss of inhibitory activity in the cell-free extract.
Fig. 3.
Inhibition types produced by clinical Pa and Bcc strains.
Surprisingly, all of the Bcc producer strains lost inhibitory activity in their cell-free extracts. This result was further explored by concentrating the extracts 20-fold. One strain (4 %) showed phage-like bacteriocin activity (Fig. 3), none showed only protease-sensitive bacteriocin activity, and 11 % showed both types of activity. One Bcc strain (4 %) produced phage plaques, indicating that the inhibitory activity was due to lytic activity of a bacteriophage. Finally, 50 % could not be characterized because of complete loss of inhibitory activity, even after 20-fold concentration of the extract (Fig. 3).
Clinical strains have the potential to produce multiple bacteriocins
A set of Pa reference strains was employed to identify the source of some or all of the inhibition identified above. These strains are either producers of, or are sensitive to, specific S, R and F pyocins (de Chial et al., 2003; Ito et al., 1970; Kageyama et al., 1979; Kuroda & Kageyama, 1981; Nakayama et al., 2000; Sano et al., 1990; Seo & Galloway, 1990; Williams et al., 2008). To permit an even finer degree of bacteriocin identification, cloned pyocins (S1, S2, S3 and AP41) (Duport et al., 1995; Sano & Kageyama, 1981; Sano et al., 1993b) were employed as references. There are no corresponding reference strains for Bcc bacteriocins.
All strains were assayed for bacteriocin-like inhibitory activity against the Pa reference collection. The bacteriocin phenotypes predicted from these inhibition haplotypes are given in Table 2. Among the 37 Pa producer strains, 76 % inhibited one or more of the reference strains. Strains predicted to possess the most bacteriocins (Pa I: S1, S2, S3, AP41, R2, R4, F2, F3; and Pa II: S1, S2, AP41, R2, R4, F2, F3) were the most common phenotypes encountered in the clinical Pa strains. These two phenotypes inhibited an average of 31 and 23 strains, respectively. Pa II strains, possessing all but the S3 phenotype (PaIIS3−), showed a 42 % decrease in Bcc inhibition in comparison with Pa I. The Pa III (S1, R2, R4, F2) phenotype was found in only one strain; however, it inhibited the greatest number of clinical strains, an average of 44 strains. Pa strains with the Pa IV (S1, R2, R4, F2, F3) and Pa V (S1, S3, AP41, R2, R4, F2, F3) phenotypes inhibited the least number of clinical strains, an average of 6.5 and 9, respectively. Strains with the Pa VI (S1, S2, AP41) phenotype inhibited a mean of 17 clinical strains. Nine Pa producer strains (Pa VII) did not inhibit any of the reference strains, and had the lowest level of inhibition, with an average of 2.4 clinical strains inhibited. Among the 19 Bcc producer strains, only three (16 %) inhibited any of the Pa reference strains. These three strains possessed the Bcc I (S1, S2, AP41, F3), Bcc II (F3) and Bcc III (R2, F2) phenotypes (Table 2). Sixteen Bcc producer strains (Bcc IV) did not inhibit any of the reference strains, and they inhibited an average of 5.8 clinical strains.
Table 2.
Bacteriocin phenotypes of clinical Pa and Bcc strains
| Producer species | Phenotype designation | Bacteriocin phenotype* | No. of strains | Mean no. of Pa strains inhibited | Mean no. of Bcc strains inhibited | Total no. of strains inhibited |
|---|---|---|---|---|---|---|
| Pa | Pa I | S1, S2, S3, AP41, R2, R4, F2, F3 | 10 | 19 | 12 | 31 |
| Pa II | S1, S2, AP41, R2, R4, F2, F3 | 12 | 16 | 7 | 23 | |
| Pa III | S1, R2, R4, F2 | 1 | 18 | 26 | 44 | |
| Pa IV | S1, R2, R4, F2, F3 | 2 | 5.5 | 1 | 6.5 | |
| Pa V | S1, S3, AP41, R2, R4, F2, F3 | 1 | 6 | 3 | 9 | |
| Pa VI | S1, S2, AP41 | 2 | 8.5 | 8.5 | 17 | |
| Pa VII | – | 9 | 1.8 | 0.6 | 2.4 | |
| Bcc | Bcc I | S1, S2, AP41, F3 | 1 | 6 | 4 | 10 |
| Bcc II | F3 | 1 | 4 | 2 | 6 | |
| Bcc III | R2, F2 | 1 | 1 | 2 | 3 | |
| Bcc IV | – | 16 | 2.8 | 3 | 5.8 |
*S-type pyocins: S1, S2, S3 and AP41; RF-type pyocins: R2, R4, F2 and F3.
Clinical strains show sensitivity to multiple bacteriocins
The clinical strains were exposed to cloned S pyocins (Duport et al., 1995; Sano & Kageyama, 1981; Sano et al., 1993b), and 89 % were sensitive (Table 3). Sixty-two per cent of these strains were sensitive to a single pyocin, and 38 % were sensitive to multiple pyocins. Finally, none of the Bcc strains were sensitive to the cloned S pyocins (Table 3).
Table 3.
S-pyocin sensitivity of clinical Pa and Bcc strains
Cloned S pyocins were used for these assays. None of the 28 Bcc strains were sensitive to the cloned S pyocins.
| Pyocin-sensitivity phenotype* | No. of sensitive Pa strains (n=38) |
|---|---|
| S1 | 3 |
| S2 | 0 |
| S3 | 13 |
| AP41 | 5 |
| S1, S2 | 6 |
| S1, S3 | 0 |
| S1, AP41 | 1 |
| S2, S3 | 0 |
| S2, AP41 | 0 |
| S3, AP41 | 3 |
| S1, S2, AP41 | 1 |
| S1, S3, AP41 | 2 |
| S2, S3, AP41 | 0 |
| S1, S2, S3, AP41 | 0 |
| Total sensitivity | 34 (89 %) |
*Phenotypes are according to sensitivity to S-pyocin producing clones; e.g. S1: only S1 sensitivity; S1, S2: S1 and S2 sensitivity.
Source of the intra- and interspecific inhibitory activity of clinical strains
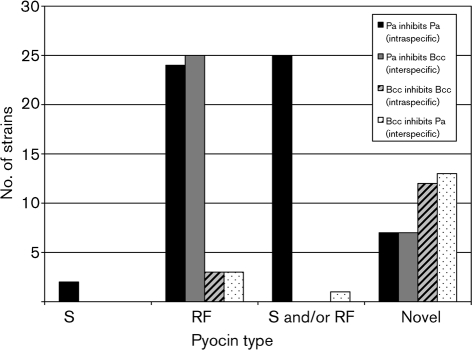
Based on inhibition haplotypes, 28 Pa strains were predicted to produce combinations of S- and RF-type pyocins, which kill within and between species (Fig. 4). Two strains inhibited intraspecifically with S pyocins, 24 with RF-type pyocins, and 25 with S and/or RF pyocins. Twenty-five strains acted interspecifically, all with RF-type pyocins. Finally, 14 produced novel bacteriocins that acted intraspecifically (50 %) or interspecifically (50 %).
Fig. 4.
Source of the intra- and interspecific inhibitory activity of clinical Pa and Bcc strains.
In contrast, the majority of Bcc strains produced novel bacteriocin-like activity (Fig. 4). Three strains produced RF-like factors, and 12 produced novel bacteriocins, all of which acted intraspecifically. Further, three produced RF-like pyocins, one produced S- and/or RF-like pyocins, and the remainder (13) produced novel inhibitors that inhibited interspecifically (Fig. 4).
Molecular screening: clinical Pa strains possess multiple pyocin genes
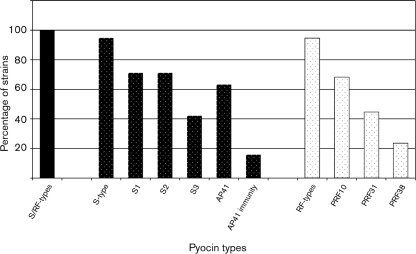
PCR primers specific to S-type (S1, S2, S3 and AP41) (Duport et al., 1995; Sano & Kageyama, 1981; Sano et al., 1993b) and RF-type (PRF10, PRF31 and PRF38) pyocin genes were employed to screen for the presence of known pyocin-encoding genes (Supplementary Table S1). All Pa strains were amplification positive for one or more of the pyocin genes (Fig. 5). Ninety-five per cent of the strains possessed one or more S-pyocin genes: 71 % amplified from the S1 gene, 71 % from the S2 gene, 42 % from the S3 gene, and 63 % from the AP41 gene. A further 16 % amplified from the pyocin AP41 immunity gene, but not the corresponding toxin gene. Ninety-five per cent of the strains were amplification positive for one or more RF-pyocin genes; 68 % for PRF10, 45 % for PRF31, and 24 % for PRF38 (Fig. 5). Finally, none of the Bcc strains were amplification positive for any of the pyocin genes (data not shown).
Fig. 5.
Pyocin gene distribution in the genomes of clinical Pa strains.
DISCUSSION
Individuals with CF face a lifelong battle with chronic bacterial lung infections (Govan & Deretic, 1996). Pa is the most prevalent species in the CF lung, and is associated with high mortality (Cystic Fibrosis Foundation, 2007; Emerson et al., 2002). CF patients are first colonized with non-mucoid Pa in childhood, which is replaced by mucoid variants in adulthood (Gibson et al., 2003). Mucoid Pa form biofilms that are virtually impossible to eradicate, even with aggressive antibiotic therapy (Hentzer et al., 2001). Secondary bacterial infections, such as with B. cepacia, although less common, are of particular concern because they are often associated with faster decline in lung function, and may result in cepacia syndrome, which is a rapidly progressing and fatal pneumonia (Huang et al., 2001; Lambiase et al., 2006). What is remarkable about these secondary infections is that they require a strain that is able to compete with, and perhaps even displace, the established Pseudomonas strain (Lambiase et al., 2006; Ledson et al., 1998, 2002; McManus et al., 2004). Further, the time interval between exposure and invasion may be as short as days to weeks (Ledson et al., 1998; Tablan et al., 1985; Whiteford et al., 1995). This ability of Bcc strains to compete with, or even displace, the resident Pa strains stands in sharp contrast with our seeming inability to significantly impact long-standing Pseudomonas lung infections with even the most aggressive use of broad-spectrum antibiotics.
Bacteriocins, narrow-spectrum antimicrobials, are recognized as one of the most common mechanisms by which bacteria mediate population- and community-level interactions (Kerr et al., 2002; Riley & Gordon, 1999; Riley & Wertz, 2002b). It is already well known that Pa is one of the most prolific bacteriocin producers. Indeed, studies indicate that nearly all clinical, and only slightly fewer environmental (∼70 %) Pa strains produce typable pyocins (Bouhaddioui et al., 2002; Farmer & Herman, 1969; Jones et al., 1974; Zabransky & Day, 1969). Typing of a much smaller number of B. cepacia (previously known as P. cepacia) strains from clinical sources has revealed far fewer producers (30 %) of putative bacteriocin-like toxins (labelled cepaciacins) (Govan & Harris, 1985). What little we know about cepaciacins suggests that some may be similar to the phage-tail-like R pyocins (Govan & Harris, 1985).
In an attempt to understand how Pa and Bcc strains interact in the CF lung, isolates of both species were surveyed for the production of bacteriocins. Not surprisingly, both species were found to be prolific producers of bacteriocin-like toxins, with 97 % of Pa strains and 68 % of Bcc strains capable of inhibition (Table 1). For Pa, most of the inhibitory activity detected (76 %) was due to S and RF pyocins (Table 2). In contrast, for Bcc, most of the inhibition could not be characterized (Table 2), and only a small fraction (16 %) was attributable to bacteriocin-like proteins (Table 2).
More surprisingly, the levels of between-species inhibition were high: 81 % of the Pa producer strains and 57 % of the Bcc producers inhibited the other species (Table 1). Previous studies of Gram-negative bacteriocins have described bacteriocins as narrow-spectrum toxins, referring to the observation that they are active against members of the same species, and generally display restricted levels of inhibition outside of the producing species (Riley & Wertz, 2002a, b). For example, a sample of 122 closely related Klebsiella species possess only 20 % interspecific inhibitory activity (Riley et al., 2003).
The high levels of interspecific inhibition detected for Pa and Bcc may reflect the competitive interactions that occur between these species in the CF lung. Prior studies have suggested that these two species actively inhibit each other's growth. McKenney et al. (1995) revealed that the addition of cell-free Pa exo-products to the growth medium used to cultivate Bcc enhances the production of siderophores, lipases and proteases. Further, Weaver & Kolter (2004) examined the effect of cell-free extracts of Bcc on Pa gene expression, and revealed that most of the upregulated genes of Pa are normally induced under iron-limited conditions. The authors concluded that iron-limited conditions might be created in these pairwise growth conditions due to the iron chelator ornibactin produced by Bcc. Indeed, iron siderophores and S pyocins (S2, S3) share the same receptors (type I and II ferripyoverdines, respectively) (Denayer et al., 2007). Thus, S pyocins are better absorbed by sensitive strains under iron-limited conditions (Ohkawa et al., 1980). Given these facts, Bcc colonization in the CF lung may induce intraspecific Pa inhibitory activity. Clearly, the effect of iron on the interspecific inhibitory activity of pyocins and cepaciacins warrants further investigation. For these purposes, identification of cepaciacins and the corresponding cell surface receptors is required.
The majority of Pa interspecific inhibitory activity was due to RF-type pyocins (Fig. 4). In fact, R pyocins inhibit a variety of Gram-negative bacteria (Blackwell et al., 1979; Filiatrault et al., 2001). This study provides what is thought to be the first substantial report of the potential role of RF pyocins in inhibition of clinical Bcc strains. The remainder of the Pa interspecific inhibitory activity was due to novel virulence factors. The majority of Bcc interspecific inhibitory activity was due to novel factors (Fig. 4), which requires further investigation.
Another intriguing observation reported here is that most isolates of Pa from the CF lung produced multiple bacteriocins (Table 2, Fig. 5). In fact, the most frequently encountered bacteriocin types (Pa I–II) included essentially all characterized S and RF pyocins. The observation of such high levels of Pa bacteriocin diversity is good news for those interested in the potential use of bacteriocins as narrow-spectrum antimicrobials. Although the bacteriocin-like production patterns were complex, they offered a wide range of inhibition specificities.
Given the lack of effective antibiotic therapies for adult patients with CF, it is intriguing that the strains tested here were inhibited by a diversity of Pa and Bcc bacteriocins (real and putative). Bacteriocins may be an effective alternative for mediating some of these long-standing infections. One requirement would be the production of bacteriocin-sensitivity profiles to permit identification of the appropriate therapeutic bacteriocins. Of course, such treatment will select for bacteriocin resistance (e.g. protease sensitivity, LPS receptor modification) (Loutet et al., 2006). However, given the plethora of potential bacteriocins, cocktails of toxins could be created that would result in far lower rates of resistance evolution (M. A. Riley, unpublished observations). Bacteriocins are now frequently being considered for such therapeutic use (Gillor et al., 2005). For example, they are employed as antibiotics for treatment of mastitis in dairy cows (Diez-Gonzalez, 2007), and to inhibit growth of pathogenic E. coli in newborn piglets and foals (Gillor et al., 2005).
The most striking difference between the inhibitory patterns of the Pa and Bcc strains involved their breadth of activity. Almost all Pa isolates were able to inhibit a wide range of Pa and Bcc strains (Fig. 2). In contrast, Bcc strains, on average, inhibited a much more limited number of strains. Further, the Bcc inhibitory substances appeared to be phenotypically unique. For example, standard bacteriocin assays are highly successful when applied to most species of Gram-negative bacteria (Pugsley & Oudega, 1987; Riley et al., 2003), yet these methods were far less effective when applied to Bcc strains. Most intriguing was the complete loss of all inhibitory activity in Bcc cell-free extracts. Even with 20-fold concentrations, it was not possible to recover inhibitory activity, which had been clearly and repeatedly observed on plate-based screens. In fact, the most successful protocols for obtaining Bcc inhibition involved concomitant growth of both the producer and sensitive strains on a solid surface. Perhaps inhibition production requires severely restricted resources and/or high levels of competition. An alternative explanation could be the presence of unstable phage particles in lysates from producer strains (Summer et al., 2004). Although we were unable to propagate bacteriophage activity, one Bcc strain was clearly shown to inhibit via phage (Fig. 3). Recent studies on the bacteriophages of Bcc have revealed that bacteriophages are able to inhibit different Bcc genomovars. However, these phages have a relatively limited ability to inhibit Pa strains (Langley et al., 2003; Seed & Dennis, 2005). Clearly, this intriguing loss of inhibitory activity for the majority of Bcc producers requires further investigation.
Pa strains are usually the persistent resident bacterial species in the adult CF lung. A recent study revealed a clear example of how R pyocins may play a role in mediating intraspecific competition and succession of Pa strains (Heo et al., 2007). Further, pyocin production has been shown to impact intraspecific interactions in mixed biofilms (Waite & Curtis, 2009). It is rare for a Bcc strain to be able to invade these established populations. In the relatively infrequent cases of co-existence (i.e. the paired strains in this study), the Pa and Bcc strains are usually unable to inhibit each other. Only two Pa strains (via RF pyocins) and none of the Bcc strains were able to inhibit their pair mates (Table 1, Fig. 4). This observation may simply reflect the outcome of prior competition: if one or the other was able to inhibit the invader or resident strain, they would not be found to coexist. Clearly most unpaired Pa and Bcc strains were able to inhibit each other (75 % versus 62 %, respectively), even while the pair mates could not (Table 1).
Future efforts will focus on two significant features of bacteriocin biology revealed by this study. First, it is surprising that there have been no molecular investigations of cepaciacins. Given the importance of Bcc in human disease, the presence of these potent toxins should have generated prior interest. In contrast, numerous studies have focused on the characterization of pyocins. Preliminary studies designed to provide a molecular characterization of cepaciacins identified here caution us that this may not be a simple exercise. To date, our efforts have not resulted in a single cloned cepaciacin, or even a bacteriocin-like inhibitory function, from these Bcc strains. Second, the suggestion that bacteriocin-like inhibition may play a role in mediating strain dynamics in the CF lung serves as inspiration for future studies designed to characterize the potential of these narrow-spectrum antimicrobials to serve as future CF therapeutics. Clearly, such toxins are able to mediate interspecific interactions of Pa and Bcc. Whether we can employ these toxins for our own purposes in a clinical setting remains to be seen.
Acknowledgments
This work was supported by NIH grants RO1GM068657-01 and RO1 AI064588-01A2. We would like to thank Dr Yumiko Sano for kindly providing Pa indicator strains (NIH3, PML1516d, NIH3S1R, NIH3S2R, NIH3AP41R, 3012 and 3295) and cloned S pyocins (S1, S2 and AP41). We would also like to thank Dr Pierre Cornelis for providing pyocin S3 clone, Pa 7NSK2 and Pa 7NSK2-fpvA, Dr Dean School for Pa PML14 and Pa 13s strains, and Dr Fred Ausubel for providing Pa PAO1. We thank Meredith Little and Amanda Gellett for collection of clinical strains, and the patients for participating in this study. We also thank Shanika Collins for helping with the PCR study, Emma White for helping with the pilot bacteriocin screening study, and Dr Chris Vriezen, Dr Michelle Lizotte-Waniewski and Mike Valliere for intellectual support and critical review of the manuscript.
Abbreviations
Bcc, Burkholderia cepacia complex
CF, cystic fibrosis
Pa, Pseudomonas aeruginosa
Footnotes
A supplementary table of primers is available with the online version of this paper.
References
- Al-Bakri, A. G., Gilbert, P. & Allison, D. G. (2004). Immigration and emigration of Burkholderia cepacia and Pseudomonas aeruginosa between and within mixed biofilm communities. J Appl Microbiol 96, 455–463. [DOI] [PubMed] [Google Scholar]
- Blackwell, C. C., Young, H. & Anderson, I. (1979). Sensitivity of Neisseria gonorrhoeae to partially purified R-type pyocines and a possible approach to epidemiological typing. J Med Microbiol 12, 321–335. [DOI] [PubMed] [Google Scholar]
- Bouhaddioui, B., Ben Slama, K., Gharbi, S. & Boudabous, A. (2002). Epidemiology of clinical and environmental Pseudomonas aeruginosa strains. Ann Microbiol 52, 223–235. [Google Scholar]
- Brazas, M. D. & Hancock, R. E. (2005). Ciprofloxacin induction of a susceptibility determinant in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49, 3222–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W., Small, D. A., Toghrol, F. & Bentley, W. E. (2005). Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation (2007). Patient Registry 2007 Annual Data Report. Bethesda, MD: Cystic Fibrosis Foundation.
- de Chial, M., Ghysels, B., Beatson, S. A., Geoffroy, V., Meyer, J. M., Pattery, T., Baysse, C., Chablain, P., Parsons, Y. N. & other authors (2003). Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149, 821–831. [DOI] [PubMed] [Google Scholar]
- Denayer, S., Matthijs, S. & Cornelis, P. (2007). Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189, 7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Gonzalez, F. (2007). Use of bacteriocins in livestock. In Research and Applications in Bacteriocins, pp. 117–129. Edited by M. A. Riley & O. Gillor. Norwich, UK: Horizon Bioscience.
- Duport, C., Baysse, C. & Michel-Briand, Y. (1995). Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J Biol Chem 270, 8920–8927. [DOI] [PubMed] [Google Scholar]
- Emerson, J., Rosenfeld, M., McNamara, S., Ramsey, B. & Gibson, R. L. (2002). Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol 34, 91–100. [DOI] [PubMed] [Google Scholar]
- Farmer, J. J., III & Herman, L. G. (1969). Epidemiological fingerprinting of Pseudomonas aeruginosa by the production of and sensitivity of pyocin and bacteriophage. Appl Microbiol 18, 760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault, M. J., Munson, R. S., Jr & Campagnari, A. A. (2001). Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J Bacteriol 183, 5756–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe, J. A., Harris, G. & Govan, J. R. (1984). Revised pyocin typing method for Pseudomonas aeruginosa. J Clin Microbiol 20, 47–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, R. L., Burns, J. L. & Ramsey, B. W. (2003). Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168, 918–951. [DOI] [PubMed] [Google Scholar]
- Gillor, O., Nigro, L. M. & Riley, M. A. (2005). Genetically engineered bacteriocins and their potential as the next generation of antimicrobials. Curr Pharm Des 11, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Govan, J. R. & Deretic, V. (1996). Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60, 539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan, J. R. & Harris, G. (1985). Typing of Pseudomonas cepacia by bacteriocin susceptibility and production. J Clin Microbiol 22, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentzer, M., Teitzel, G. M., Balzer, G. J., Heydorn, A., Molin, S., Givskov, M. & Parsek, M. R. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183, 5395–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, Y. J., Chung, I. Y., Choi, K. B. & Cho, Y. H. (2007). R-type pyocin is required for competitive growth advantage between Pseudomonas aeruginosa strains. J Microbiol Biotechnol 17, 180–185. [PubMed] [Google Scholar]
- Huang, C. H., Jang, T. N., Liu, C. Y., Fung, C. P., Yu, K. W. & Wong, W. W. (2001). Characteristics of patients with Burkholderia cepacia bacteremia. J Microbiol Immunol Infect 34, 215–219. [PubMed] [Google Scholar]
- Ito, S., Kageyama, M. & Egami, F. (1970). Isolation and characterization of pyocins from several strains of Pseudomonas aeruginosa. J Gen Appl Microbiol 16, 205–214. [Google Scholar]
- Jones, L. F., Zakanycz, J. P., Thomas, E. T. & Farmer, J. J., III (1974). Pyocin typing of Pseudomonas aeruginosa: a simplified method. Appl Microbiol 27, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. M., Dodd, M. E., Govan, J. R., Barcus, V., Doherty, C. J., Morris, J. & Webb, A. K. (2004). Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax 59, 948–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, M., Shinomiya, T., Aihara, Y. & Kobayashi, M. (1979). Characterization of a bacteriophage related to R-type pyocins. J Virol 32, 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish, L. A., Waltz, D. A., Dovey, M., Potter-Bynoe, G., McAdam, A. J., Lipuma, J. J., Gerard, C. & Goldmann, D. (2006). Impact of Burkholderia dolosa on lung function and survival in cystic fibrosis. Am J Respir Crit Care Med 173, 421–425. [DOI] [PubMed] [Google Scholar]
- Kerr, B., Riley, M. A., Feldman, M. W. & Bohannan, B. J. (2002). Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174. [DOI] [PubMed] [Google Scholar]
- Kuroda, K. & Kageyama, M. (1981). Comparative study of F-type pyocins of Pseudomonas aeruginosa. J Biochem 89, 1721–1736. [DOI] [PubMed] [Google Scholar]
- Lambiase, A., Raia, V., Del Pezzo, M., Sepe, A., Carnovale, V. & Rossano, F. (2006). Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, R., Kenna, D. T., Vandamme, P., Ure, R. & Govan, J. R. (2003). Lysogeny and bacteriophage host range within the Burkholderia cepacia complex. J Med Microbiol 52, 483–490. [DOI] [PubMed] [Google Scholar]
- Ledson, M. J., Gallagher, M. J., Corkill, J. E., Hart, C. A. & Walshaw, M. J. (1998). Cross infection between cystic fibrosis patients colonised with Burkholderia cepacia. Thorax 53, 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledson, M. J., Gallagher, M. J., Jackson, M., Hart, C. A. & Walshaw, M. J. (2002). Outcome of Burkholderia cepacia colonisation in an adult cystic fibrosis centre. Thorax 57, 142–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet, S. A., Flannagan, R. S., Kooi, C., Sokol, P. A. & Valvano, M. A. (2006). A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol 188, 2073–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, H., Sano, Y., Ishihara, H. & Shinomiya, T. (1993). Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175, 1257–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney, D., Brown, K. E. & Allison, D. G. (1995). Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: evidence of interspecies communication. J Bacteriol 177, 6989–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus, T. E., McDowell, A., Moore, J. E. & Elborn, S. J. (2004). Organisms isolated from adults with cystic fibrosis. Ann Clin Microbiol Antimicrob 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand, Y. & Baysse, C. (2002). The pyocins of Pseudomonas aeruginosa. Biochimie 84, 499–510. [DOI] [PubMed] [Google Scholar]
- Nakayama, K., Takashima, K., Ishihara, H., Shinomiya, T., Kageyama, M., Kanaya, S., Ohnishi, M., Murata, T., Mori, H. & Hayashi, T. (2000). The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38, 213–231. [DOI] [PubMed] [Google Scholar]
- Ohkawa, I., Shiga, S. & Kageyama, M. (1980). Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J Biochem 87, 323–331. [DOI] [PubMed] [Google Scholar]
- Pugsley, A. P. & Oudega, B. (1987). Methods for studying colicins and their plasmids. In Plasmids: a Practical Approach, pp. 105–161. Edited by K. G. Hardy. Oxford: IRL.
- Riley, M. A. & Gordon, D. M. (1999). The ecological role of bacteriocins in bacterial competition. Trends Microbiol 7, 129–133. [DOI] [PubMed] [Google Scholar]
- Riley, M. A. & Wertz, J. E. (2002a). Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84, 357–364. [DOI] [PubMed] [Google Scholar]
- Riley, M. A. & Wertz, J. E. (2002b). Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56, 117–137. [DOI] [PubMed] [Google Scholar]
- Riley, M. A., Goldstone, C. M., Wertz, J. E. & Gordon, D. (2003). A phylogenetic approach to assessing the targets of microbial warfare. J Evol Biol 16, 690–697. [DOI] [PubMed] [Google Scholar]
- Sano, Y. & Kageyama, M. (1981). Purification and properties of an S-type pyocin, pyocin AP41. J Bacteriol 146, 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y. & Kageyama, M. (1993). A novel transposon-like structure carries the genes for pyocin AP41, a Pseudomonas aeruginosa bacteriocin with a DNase domain homology to E2 group colicins. Mol Gen Genet 237, 161–170. [DOI] [PubMed] [Google Scholar]
- Sano, Y., Matsui, H., Kobayashi, M. & Kageyama, M. (1990). Pyocins S1 and S2, bacteriocins of Pseudomonas aeruginosa. In Pseudomonas: Biotransformations, Pathogenesis, and Evolving Biotechnology, pp. 352–358. Edited by S. Silver, A. M. Chakrabarty, B. Iglewski & S. Kaplan. Washington, DC: American Society for Microbiology.
- Sano, Y., Kobayashi, M. & Kageyama, M. (1993a). Functional domains of S-type pyocins deduced from chimeric molecules. J Bacteriol 175, 6179–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y., Matsui, H., Kobayashi, M. & Kageyama, M. (1993b). Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J Bacteriol 175, 2907–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed, K. D. & Dennis, J. J. (2005). Isolation and characterization of bacteriophages of the Burkholderia cepacia complex. FEMS Microbiol Lett 251, 273–280. [DOI] [PubMed] [Google Scholar]
- Seo, Y. & Galloway, D. R. (1990). Purification of the pyocin S2 complex from Pseudomonas aeruginosa PAO1: analysis of DNase activity. Biochem Biophys Res Commun 172, 455–461. [DOI] [PubMed] [Google Scholar]
- Shinomiya, T., Shiga, S. & Kageyama, M. (1983). Genetic determinant of pyocin R2 in Pseudomonas aeruginosa PAO. I. Localization of the pyocin R2 gene cluster between the trpCD and trpE genes. Mol Gen Genet 189, 375–381. [DOI] [PubMed] [Google Scholar]
- Summer, E. J., Gonzalez, C. F., Carlisle, T., Mebane, L. M., Cass, A. M., Savva, C. G., LiPuma, J. & Young, R. (2004). Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J Mol Biol 340, 49–65. [DOI] [PubMed] [Google Scholar]
- Tablan, O. C., Chorba, T. L., Schidlow, D. V., White, J. W., Hardy, K. A., Gilligan, P. H., Morgan, W. M., Carson, L. A., Martone, W. J. & other authors (1985). Pseudomonas cepacia colonization in patients with cystic fibrosis: risk factors and clinical outcome. J Pediatr 107, 382–387. [DOI] [PubMed] [Google Scholar]
- Waite, R. D. & Curtis, M. A. (2009). Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J Bacteriol 191, 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, V. B. & Kolter, R. (2004). Burkholderia spp. alter Pseudomonas aeruginosa physiology through iron sequestration. J Bacteriol 186, 2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford, M. L., Wilkinson, J. D., McColl, J. H., Conlon, F. M., Michie, J. R., Evans, T. J. & Paton, J. Y. (1995). Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 50, 1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S. R., Gebhart, D., Martin, D. W. & Scholl, D. (2008). Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74, 3868–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabransky, R. J. & Day, F. E. (1969). Pyocine typing of clinical strains of Pseudomonas aeruginosa. Appl Microbiol 17, 293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar, J. H. (1999). Biostatistical Analysis, 4th edn. Upper Saddle River, NJ: Prentice Hall.