Abstract
Optical maps were generated for 33 uropathogenic Escherichia coli (UPEC) isolates. For individual genomes, the NcoI restriction fragments aligned into a unique chromosome map for each individual isolate, which was then compared with the in silico restriction maps of all of the sequenced E. coli and Shigella strains. All of the UPEC isolates clustered separately from the Shigella strains as well as the laboratory and enterohaemorrhagic E. coli strains. Moreover, the individual strains appeared to cluster into distinct subgroups based on the dendrogram analyses. Phylogenetic grouping of these 33 strains showed that 32/33 were the B2 subgroup and 1/33 was subgroup A. To further characterize the similarities and differences among the 33 isolates, pathogenicity island (PAI), haemolysin and virulence gene comparisons were performed. A strong correlation was observed between individual subgroups and virulence factor genes as well as haemolysis activity. Furthermore, there was considerable conservation of sequenced-strain PAIs in the specific subgroups. Strains with different antibiotic-resistance patterns also appeared to sort into separate subgroups. Thus, the optical maps distinguished the UPEC strains from other E. coli strains and further subdivided the strains into distinct subgroups. This optical mapping procedure holds promise as an alternative way to subgroup all E. coli strains, including those involved in infections outside of the intestinal tract and epidemic strains with distinct patterns of antibiotic resistance.
INTRODUCTION
Uropathogenic Escherichia coli (UPEC) are the number one cause of urinary tract infections (UTIs), responsible for up to 90 % of the seven million cases that occur each year in the USA (Foxman, 2002; Hooton & Stamm, 1997). Approximately 50–60 % of all women will have a UTI during their lifetime (Foxman, 2002; Kunin, 1994). In the USA, a staggering $1.6 billion per year in healthcare costs is associated with UPEC infections (Foxman et al., 2000; Hooton & Stamm, 1997). Many people suffer from multiple UTIs that are typically caused by the strain originally isolated in the first UTI (Foxman et al., 1995). To initiate and sustain these UTIs, the UPEC use various virulence factors. Specific virulence factors and O : K : H serotypes are present more often in cystitis-causing isolates than in isolates found as normal flora in the intestinal tract of humans (Johnson, 2003). Multiple virulence factor genes are found on genetic elements termed pathogenicity islands (PAIs; Blum et al., 1994; Hacker et al., 1990), mobile genetic elements inserted adjacent to tRNA genes. In UPEC, five PAIs have been identified and their role in UPEC pathogenesis has been demonstrated (Hacker et al., 1997).
Besides comparisons of PAI prevalence, molecular epidemiological comparisons (Guttman & Dykhuizen, 1994; Kanamaru et al., 2006; Zhang et al., 2000) and genotypic-phenotypic statistical analysis of many E. coli isolates (Johnson, 2003) have been performed to achieve a grouping scheme for UPEC strains. Phylogenetic grouping of the E. coli strains has been another popular way to group E. coli strains. Four phylogenetic groups have been characterized: A, B1, B2 and D (Ochman & Selander, 1984). Extraintestinal E. coli responsible for UTIs belong chiefly to phylogenetic group B2 and less so to group D, whereas commensal E. coli (faecal) typically fall into groups A and B1 (Picard et al., 1999). Clermont et al. (2000) have devised a multiplex PCR for three genetic markers (chuA, yjaA and TSPE4.12) to separate E. coli strains into these four basic phylogenetic groups. The multiplex PCR analysis has provided a first step in separating E. coli strains, but most of the UPEC strains fall into only one subgroup using this technique. Both ribotyping and multilocus sequence typing (MLST) have been used to further separate the UPEC strains (Bingen-Bidois et al., 2002; Clermont et al. 2001; Tartof et al., 2005), but these techniques are complex methods of grouping UPEC strains.
In addition to phylogenetic grouping, pathotyping is another way to separate strains of a species, and is based on a particular set of virulence factors shared by some strains that assist in the pathogenesis process (Marrs et al., 2005). Although the E. coli that cause diarrhoea in humans have been separated into subgroups or pathotypes (Nataro & Kaper, 1998), the same has not been done for UPEC, even though this group of E. coli strains has been shown to be quite diverse (Zhang et al., 2000). Some attempts have been made to place the UPEC strains into pathotypes (Foxman et al., 1995; Marrs et al., 2005), but critical genetic differences that may define specific pathotypes remain to be elucidated. As a consequence of these genetic limitations, several E. coli genomes have been sequenced to uncover which set of genes may be contributing to UPEC pathogenesis. Because of the limitations outlined above, a clear UPEC pathotype subgrouping scheme has remained elusive.
In this study, we have used an optical mapping scheme to sort UPEC strains into distinct subgroups based on similarities in their restriction fragment patterns across the entire genome. This is believed to be the first time that this technology has been applied to group any bacteria. We have shown that UPEC strains are distinct from both enterohaemorrhagic and laboratory strains of E. coli. In addition, we demonstrate that there are two major UPEC subgroups. Furthermore, we show that strain separation by optical mapping correlates with haemolysis, the presence of PAIs, the distribution of several virulence factor genes, and antibiotic resistance.
METHODS
Bacterial strains.
All of the E. coli strains are listed in Table 1. E. coli strains NU149 and NU14 were obtained from women who had a UTI, and have been previously characterized (Hultgren et al., 1986; Schaeffer et al., 1987). Strain 536 (Hacker et al., 1983) was obtained from Jörg Hacker, Universität Würzburg. E. coli CFT073 (Mobley et al., 1990) was a kind gift from Rod Welch, University of Wisconsin-Madison. Strain UTI89 (Mulvey et al., 2001) was received from Scott Hultgren, Washington University. The W3110 (K-12) strain was taken from the University of Wisconsin-La Crosse culture collection. ATCC 25922 was obtained from the American Type Culture Collection. The 16 GLMC strains were collected in a consecutive manner from patients with UTIs at Gundersen Lutheran Medical Center, WI. They were all biochemically identified as E. coli. All 13 of the CV strains were obtained from Converge and were isolated from patients with UTIs in a consecutive manner around Boston, MA.
Table 1.
Bacterial strains used in this study
| Strain | Source or reference |
|---|---|
| E. coli strains | |
| GLMC 9 | Gundersen Lutheran Medical Center |
| GLMC 10 | Gundersen Lutheran Medical Center |
| GLMC 15 | Gundersen Lutheran Medical Center |
| GLMC 21 | Gundersen Lutheran Medical Center |
| GLMC 100 | Gundersen Lutheran Medical Center |
| GLMC 101 | Gundersen Lutheran Medical Center |
| GLMC 103 | Gundersen Lutheran Medical Center |
| GLMC 106 | Gundersen Lutheran Medical Center |
| GLMC 503 | Gundersen Lutheran Medical Center |
| GLMC 505 | Gundersen Lutheran Medical Center |
| GLMC 506 | Gundersen Lutheran Medical Center |
| GLMC 508 | Gundersen Lutheran Medical Center |
| GLMC 509 | Gundersen Lutheran Medical Center |
| GLMC 513 | Gundersen Lutheran Medical Center |
| GLMC 514 | Gundersen Lutheran Medical Center |
| GLMC 515 | Gundersen Lutheran Medical Center |
| CV1117292 | Converge |
| CV1117298 | Converge |
| CV1117326 | Converge |
| CV1117621 | Converge |
| CV1118322 | Converge |
| CV1118891 | Converge |
| CV1119429 | Converge |
| CV1119511 | Converge |
| CV1119857 | Converge |
| CV1121147 | Converge |
| CV1123039 | Converge |
| CV1125109 | Converge |
| CV1131494 | Converge |
| ATCC 25922 | ATCC |
| ATCC 8739 | ATCC |
| W3110 (K-12) | University of Wisconsin-La Crosse collection |
| DH10B (K-12) | Durfee et al. (2008) |
| UTI89 | Scott Hultgren (Chen et al., 2006b) |
| CFT073 | Rod Welch (Welch et al., 2002) |
| 536 | Jörg Hacker (Brzuszkiewicz et al., 2006) |
| NU14 | Hultgren et al. (1986) |
| NU149 | Schaeffer et al. (1987) |
| APEC 01 | Johnson et al. (2007) |
| Sakai | Hayashi et al. (2001) |
| EDL933 | Perna et al. (2001) |
| EC4115 | GenBank |
| E24377A | Rasko et al. (2008) |
| SMS-3-5 | Fricke et al. (2008) |
| HS | Rasko et al. (2008) |
| SE11 | Oshima et al. (2008) |
| S. boydii strains | |
| CDC 3083-94 | GenBank |
| Sb227 | Yang et al. (2005) |
| S. dysenteriae strain | |
| Sd197 | Yang et al. (2005) |
| S. flexneri strains | |
| 2457T | Wei et al. (2003) |
| 301 | Jin et al. (2002) |
| 8401 | Nie et al. (2006) |
| S. sonnei strain | |
| Ss046 | Yang et al. (2005) |
Haemolysis.
All of the E. coli strains were tested for their ability to lyse sheep red blood cells by plating on sheep blood agar plates (Becton Dickinson) and incubating for 24 h at 37 °C. All of the clinical isolates were tested for haemolysis using bacteria that were freshly isolated from infected urine. A zone of haemolysis around the bacterial colonies was considered a positive reaction for haemolysis.
Distribution of virulence factor genes.
The UPEC isolates were tested for nine virulence factor genes characteristic of UPEC: cnf, papGII, papGIII, hlyC, sfa, fimH, usp, aer and fyuA. Each PCR was carried out in a 50 μl volume using 50 pmol of each primer, 200 μM of each dNTP, 2.5 U Taq polymerase (Promega) and 1.5 μl bacterial lysate. Detection of the aer, cnf, sfa and hlyC genes used the primers designed by Yamamoto et al. (1995). Amplification of the papGII and papGIII genes used the method of Johnson & Brown (1996). PCR detection of fimH was done with primers used by Tseng et al. (2001). Primers for usp were as denoted by Takahashi et al. (2006). The primer set of Johnson & Stell (2000) were utilized for amplifying fyuA. All PCR runs were carried out in a Perkin Elmer 9700 thermocycler under the following conditions: initial denaturation at 94 °C for 5 min; 30 cycles of 94 °C for 10 s, 55 °C for 20 s and 72 °C for 30 s; and a final denaturation at 72 °C for 7 min. Each PCR run was repeated at least twice.
Optical map production and analysis.
Purified chromosomal DNA was deposited onto an optical mapping surface (derivatized glass coverslip), using a microfluidic device. The DNA was encased in a thin layer of acrylamide and incubated with the restriction enzyme NcoI in a humidified chamber at 37 °C for 60–120 min. The digested DNA was labelled with fluorescent JOJO-1 (Invitrogen) and the individual molecules were imaged by fluorescence microscopy. Digital images were collected by a fully automated image-acquisition system and image files were processed to create single-molecule optical maps using OpGen software programs (Zhou et al., 2004). Individual molecule restriction maps were overlapped by optical map assembler software. Briefly, this assembler works by comparing single-molecule restriction maps and estimating the probability that two molecules arose from overlapping genomic locations given a description of the likelihood of possible errors resulting from incomplete digestion of the molecule, spurious cuts of the molecule, and errors in the sizing estimates of the resulting fragments. Through repeated overlapping of molecules, the assembler reconstructs the ordered restriction map of the genome. This technique has been previously applied to map other bacterial genomes (Chen et al., 2006a; Kotewicz et al., 2007), as well as mapping enterohaemorrhagic E. coli (EHEC) strains (Kotewicz et al., 2007, 2008).
The Shigella sequences (Shigella boydii CDC 3083-94, GenBank accession no. CP001063; S. boydii Sb227, CP000036; Shigella flexneri 2a 2457T, AE014073; S. flexneri 2a 301, AE005674; S. flexneri 5 8401, CP00026; Shigella sonnei Ss046, CP000038; Shigella dysenteriae Sd197, CP000034) and the sequences for E. coli strains HS (GenBank accession no. CP000802), DH10B (K-12) (CP000948), W3110 (K-12) (AP009048), E24377A (CP000800), Sakai (BA000007), EDL933 (AE005174), SMS-3-5 (CP000970), UTI89 (CP000243), APEC 01 (CP000468), CFT073 (CP000247), ATCC 8739 (CP000946) and 536 (CP000247) came from the published NCBI microbial genomes. The maps were then constructed in silico by identifying the location of restriction sites in the sequences.
To construct the similarity cluster, each pair of maps was aligned using a dynamic programming algorithm based upon published methods (Myers & Huang, 1992; Pevzner et al., 2001; Waterman et al., 1984). This method finds the optimal alignment of two restriction maps according to a scoring model that incorporates fragment sizing errors, false and missing cuts, and missing small fragments. For a given alignment, the score is proportional to the log of the length of the alignment, penalized by the differences between the two maps, such that longer, better-matching alignments will have higher scores. This method has been used before to compare E. coli genomes (Chen et al., 2006a). From these alignments, adding up the lengths of the unmatched regions from both maps and dividing this by the sum of the lengths of both maps in the pair calculated a dissimilarity score for a pair of maps. A matrix of these pairwise scores was used as input to agnes, an agglomerative clustering method implemented in the R statistical package, which creates dendrograms using the unweighted pair group method with arithmetic mean (UPGMA). An analogous clustering method using sequence information has been shown to produce trees that match existing phylogeny data (Henz et al., 2005); however, we make no specific evolutionary claims based upon our trees.
Phylogenetic grouping.
We determined the phylogenetic groups of selected isolates in our study using the PCR-based method described in Clermont et al. (2000).
MIC determinations.
A VITEK machine was used to determine the antibiotic susceptibility of the 32 recent clinical isolates.
RESULTS
Genome-wide optical mapping comparisons of UPEC isolates
The NcoI restriction fragment optical maps of 33 UPEC isolates were arrayed and compared with each other as well as with in silico maps of sequenced strains of E. coli and Shigella deposited in GenBank (Fig. 1). Based on the map clustering, S. sonnei, S. flexneri and S. boydii were found in the E. coli cluster, whereas S. dysenteriae was the most dissimilar of any of the genomes that were mapped. The cluster is very good at highlighting the genomic diversity of these strains, but the exact cut-off for classifying these isolates into distinct groups is not clear. Using a 30 % dissimilarity cut-off, all of the E. coli isolates fell into five separate subgroups, yet the clinical UPEC isolates fell into two separate major subgroups, one more diverse than the other. The other three subgroups were EHEC, E. coli K-12 and other laboratory strains, and an environmental isolate from a heavy metal-contaminated site (Selander et al., 1987). Moreover, GLMC isolates from Wisconsin mapped side by side with the CV isolates from Massachusetts, demonstrating that geographical distribution contributes to only slight genomic variations between different strains. Although the isolates became segregated into subgroups, optical mapping was able to discriminate between each of the individual isolates because no two isolates were identical. Thus, optical mapping was effective at displaying the versatility and flexibility of the E. coli genome.
Fig. 1.
Genome similarity clustering using UPGMA for the 33 UPEC isolates and the in silico maps of sequenced E. coli and Shigella strains.
By using a 15 % dissimilarity cut-off, all of the E. coli isolates separated into 10 subgroups. Three of these subgroups aligned with the three sequenced UPEC strains (CFT073, UTI89 and 536). The GLMC 101 UPEC isolate aligned with no other isolates at this cut-off.
Detailed view of map and sequence similarity among several clinical isolates
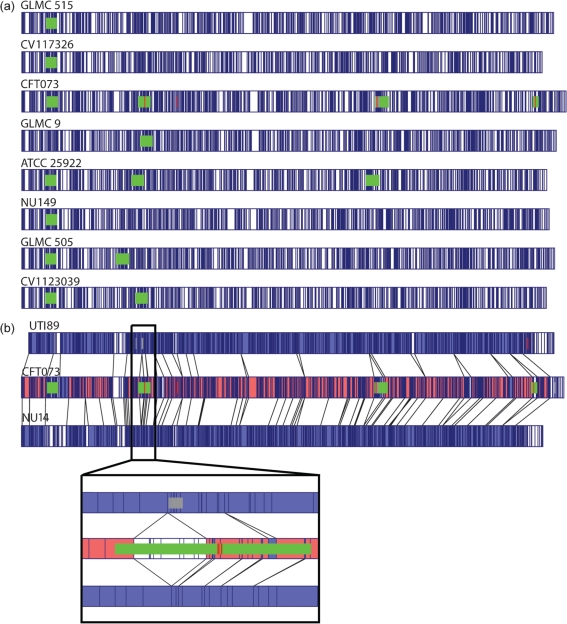
Optical mapping of the entire E. coli genome allowed segregation into several subgroups. A closer examination of the sequence alignments of part of one of the subgroups showed the similarities and differences between the isolates (Fig. 2a). The previously sequenced strain CFT073 had the largest genome (5.231 Mb) and the CV1117326 isolate had the smallest genome (5.000 Mb). A strain from the mid 1980s, NU149 (Schaeffer et al., 1987), and the ATCC 25922 strain were both contained in the CFT073 subgroup. Within the CFT073 subcategory of strains (CFT073, GLMC 9, ATCC 25922, NU149, GLMC 505, CV1123039), there appeared to be a conservation of the CFT073 PAIs. All of the isolates except GLMC 9 had PAI III (PAI-CFT073-aspV), and all of the isolates except NU149 had PAI-CFT073-serX, although the GLMC 505 isolate appeared to have the PAI-CFT073-serX inserted upstream from its position in the other genomes. An alignment of the subgroup containing strain 536 showed a similar pattern of conservation of PAIs among those isolates (data not shown).
Fig. 2.
(a) Overview of optical maps of E. coli strains from the CFT073 subgroup. Comparison of optical maps with the in silico map of CFT073. The alignment software highlights restriction fragment differences shown in white between isolates, whereas homologous regions are highlighted in blue. (b) A closer look at three strains (UTI89, CFT073, NU14), with sequence alignments overlaid on top of the maps. The vertical lines represent matching NcoI restriction sites. Green rectangles represent some of the PAIs. The insert shows the PAI-CFT073-serX in strain CFT073 that is absent in both the UTI89 and the NU14 strains.
A more detailed comparison was undertaken between two sequenced UPEC strains, UTI89 and CFT073, as well as strain NU14, a strain that is phylogenetically close to strain UTI89. Fig. 2(b) shows the alignment of the optical maps of NU14 and UTI89 to CFT073. Coloured regions indicate a match, and the white segments indicate regions where the optical maps could not be aligned. A great degree of similarity exists between the genome of a strain collected in the mid 1980s (NU14) and one collected in the late 1990s (UTI89). From the inset to Fig. 2(b), a close-up view of part of the genome for all three strains demonstrated the PAI III-CFT073-aspV present in CFT073, but absent in both UTI89 and NU14. Moreover, the NcoI restriction patterns between NU14 and UTI89 matched well, but did not match strain CFT073.
Genome similarity clustering versus phylogenetic grouping, haemolytic characterization, and distribution of several virulence factor genes
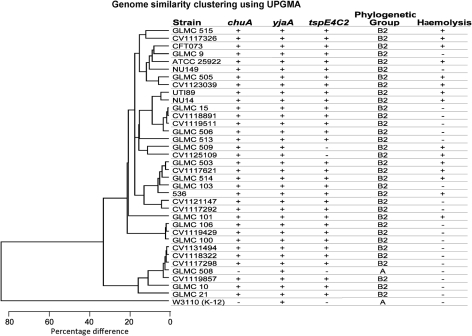
Optical mapping sorted the clinical isolates into subgroups. To add another layer to the characterization of the isolates, each isolate was phylogenetically typed using the method of Clermont et al. (2000). Genomic DNAs from the 30 recent clinical isolates, the two clinical isolates from the mid 1980s (NU14 and NU149), as well as the sequenced reference strains were PCR-amplified with primers specific for chuA, yjaA and tspE4C2. Of the 33 clinical isolates, 32/33 (97 %) typed as phylogenetic group B2. One isolate, GLMC 508, was phylogenetically typed as group A (Fig. 3), even though the phenotype and optical mapping data placed it more closely with the UPEC isolates in group B2. The K-12 laboratory strain of E. coli (W3110) displayed the A grouping, as described by Clermont et al. (2000). Thus, the phylogenetic grouping data did not add much additional information to further separate the strains.
Fig. 3.
Genome similarity clustering using UPGMA for E. coli isolates found in subgroups one and two, as well as phylogenetic grouping and haemolysis patterns.
In addition to the phylogenetic grouping of each isolate, haemolysis of sheep blood was also assessed. Twelve out of 33 isolates displayed haemolysis of sheep blood. Moreover, there was a haemolysis pattern for most of the isolates tested. Although strains NU149 and GLMC 9 did not lyse sheep blood, the remaining strains in the CFT073 subgroup demonstrated haemolysis. Both isolates at the top of the dendrogram in Fig. 3 also showed a haemolysis-positive phenotype, as did UTI89 and NU14. The closely related strains GLMC 503 and CV1117621 displayed haemolysis, and four out of five isolates found as part of the strain 536 subgroup were haemolytic. The stand-alone strain GLMC 101 also showed the ability to lyse sheep blood. The combination of phylogenetic grouping and the ability to lyse sheep blood added additional information to the optical maps of all of the isolates.
As an additional step to correlate the optical mapping with something tangible, each E. coli isolate was screened for the presence of several virulence factor genes that included aer, cnf, fimH, fyuA, papGII, papGIII, hlyC, sfa and usp. All of the strains in the one major UPEC subgroup (based on the 30 % dissimilarity cut-off) had identical virulence factor gene distributions, except for the usp gene missing in the GLMC 508 isolate and the aer and fyuA genes absent in strain GLMC 21 (Table 3). No cnf, papG, hlyC or sfa gene was detected in any of these strains. However, they all possessed the fimH gene, and all but one had the aer and fyuA genes. Although the distribution of the virulence factor genes in the other major UPEC subgroup (based on a 30 % dissimilarity cut-off) was more diverse, they aligned fairly closely with the subgroups from the 15 % dissimilarity cut-off; UPEC isolates within the CFT073 subgroup were quite similar, except for strain NU149, which lacked the papGII, hlyC and sfa genes, which is probably tied to the loss of the PAI in this strain. The UPEC strains found within the 536 subgroup were also typically conserved. Moreover, all of the strains that exhibited phenotypic haemolysis were positive for the presence of the hlyC gene. In addition, strain GLMC 103 also displayed amplification of the hlyC gene, which aligned this strain perfectly with the other strains in that subgroup.
Table 3.
Virulence factor gene distribution among uropathogenic E. coli isolates
+, Present; −, absent.
| E. coli isolate | Virulence factor gene | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| cnf | papGII | papGIII | hlyC | sfa | fimH | usp | aer | fyuA | |
| GLMC 515 | + | – | + | + | + | + | + | – | + |
| CV1117326 | + | – | + | + | + | + | + | – | + |
| CFT073 | – | + | – | + | + | + | + | + | + |
| GLMC 9 | – | + | – | – | + | + | + | + | + |
| ATCC 25922 | – | + | – | + | + | + | + | + | + |
| NU149 | – | – | – | – | – | + | + | + | + |
| GLMC 505 | + | – | + | + | + | + | + | – | + |
| CV1123039 | + | – | – | + | + | + | + | – | + |
| UTI89 | + | – | + | + | + | + | + | – | + |
| NU14 | + | – | – | + | + | + | + | – | + |
| GLMC 15 | – | + | – | – | – | + | + | – | + |
| CV1118891 | – | + | – | – | + | + | + | – | + |
| CV1119511 | – | + | – | – | – | + | + | – | + |
| GLMC 506 | – | + | – | – | – | + | + | – | + |
| GLMC 513 | – | – | – | – | + | + | + | – | + |
| GLMC 509 | + | + | – | + | + | + | + | – | + |
| CV1125109 | + | + | + | + | + | + | + | – | + |
| GLMC 503 | + | – | + | + | + | + | + | – | + |
| CV1117621 | + | – | + | + | + | + | + | – | + |
| GLMC 514 | + | – | + | + | + | + | + | – | + |
| GLMC 103 | + | – | + | + | – | + | + | + | + |
| 536 | – | – | + | + | + | + | + | – | + |
| CV1121147 | – | – | – | – | + | + | + | – | + |
| CV1117292 | – | – | – | – | – | + | + | – | + |
| GLMC 101 | – | + | – | + | – | + | + | + | + |
| GLMC 106 | – | – | – | – | – | + | + | + | + |
| CV1119429 | – | – | – | – | – | + | – | + | + |
| GLMC 100 | – | – | – | – | – | + | + | + | + |
| CV1131494 | – | – | – | – | – | + | + | + | + |
| CV1118322 | – | – | – | – | – | + | + | + | + |
| CV1117298 | – | – | – | – | – | + | + | + | + |
| GLMC 508 | – | – | – | – | – | + | – | + | + |
| CV1119857 | – | – | – | – | – | + | + | + | + |
| GLMC 10 | – | – | – | – | – | + | + | + | + |
| GLMC 21 | – | – | – | – | – | + | + | – | – |
| W3110 (K-12) | – | – | – | – | – | + | + | – | – |
Antibiotic-sensitivity patterns of the recent clinical isolates
Since UPEC strains continue to emerge that are resistant to front-line antibacterial drugs, the 30 recent clinical isolates from Wisconsin and Massachusetts were assayed for their antibiotic-sensitivity patterns against the front-line antibiotics used to treat E. coli urinary tract and systemic infections, then compared with the optical mapping patterns for these isolates. Eighteen of 30 isolates were resistant to ampicillin. The distribution for resistance to this antibiotic was well dispersed among the isolates; however, all of the isolates that grouped from CV1121147 to GLMC 21 found at the bottom of the dendrogram in Fig. 3 had the same ampicillin-resistance pattern. Four isolates had resistance to piperacillin, and eight demonstrated resistance to ampicillin/sulbactam. All of the strains tested showed susceptibility to piperacillin/tazobactam, cefazolin, cefotetan, ceftriaxone, cefepime, meropenem and nitrofurantoin (Table 2). Thus, no isolate displayed resistance to the cephalosporin class of drugs. Only seven isolates showed resistance to the sulfa drugs. For thesedrugs, the resistance patterns were spread out among different optical map subgroups. On the other hand, resistance to ciprofloxacin, levofloxacin and gentamicin was conserved among one subgroup from Fig. 3 that included strains CV1131494, CV1118322, CV1117298 and CV1119857. The grouping of fluoroquinolone-resistant isolates suggests that optical mapping could be useful in gauging outbreak strains of some antibiotic-resistant UPEC.
Table 2.
Antibacterial drug susceptibilities of 30 recent uropathogenic E. coli isolates and ATCC strain 25922
| E. coli isolate | Antibacterial drug* | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | SAM | PIP | TZP | CZ | CTT | CRO | FEP | MEM | CIP | LVX | SXT | F/M | NN | GM | |
| GLMC 515 | S† | S | S | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1117326 | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| GLMC 9 | R | R | R | S | S | S | S | S | S | S | – | S | – | – | S |
| ATCC 25922 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 505 | S | S | S | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1123039 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 15 | R | R | I | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1118891 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| CV1119511 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 506 | R | I | S | S | S | S | S | S | S | S | – | S | S | – | S |
| GLMC 513 | S | S | S | S | S | S | S | S | S | S | – | S | S | – | S |
| GLMC 509 | R | S | R | S | I | S | S | S | S | S | – | S | S | – | S |
| CV1125109 | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S |
| GLMC 503 | S | S | S | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1117621 | R | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 514 | S | S | S | S | S | S | S | S | S | S | – | S | S | – | S |
| GLMC 103 | S | S | S | S | S | S | S | S | S | S | – | R | S | – | S |
| CV1121147 | R | S | S | S | S | S | S | S | S | S | S | R | S | S | S |
| CV1117292 | R | I | I | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 101 | R | R | S | S | S | S | S | S | S | S | – | S | S | – | S |
| GLMC 106 | R | I | I | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1119429 | R | I | S | S | S | S | S | S | S | S | S | S | S | S | S |
| GLMC 100 | R | R | S | S | S | S | S | S | S | S | – | S | S | – | S |
| CV1131494 | R | I | S | S | S | S | S | S | S | R | R | S | S | I | R |
| CV1118322 | R | I | S | S | S | S | S | S | S | R | R | S | S | I | R |
| CV1117298 | R | I | R | S | S | S | S | S | S | R | R | R | S | S | S |
| GLMC 508 | R | R | I | S | R | S | S | S | S | R | – | R | S | – | S |
| CV1119857 | R | R | I | S | R | S | S | S | S | R | S | S | S | S | R |
| GLMC 10 | R | I | S | S | S | S | S | S | S | S | – | R | S | – | S |
| GLMC 21 | R | R | I | S | S | S | S | S | S | S | – | S | S | – | S |
*The following antibacterial drugs were used: AM, ampicillin; SAM, ampicillin/sulbactam; PIP, piperacillin; TZP, piperacillin/tazobactam; CZ, cefazolin; CTT, cefotetan; CRO, ceftriaxone; FEP, cefepime; MEM, meropenem; CIP, ciprofloxacin; LVX, levofloxacin; SXT, trimethoprim/sulfa; F/M, nitrofurantoin; NN, tobramycin; GM, gentamicin.
†S, Susceptible; I, intermediate; R, resistant; –, not determined.
DISCUSSION
Separating UPEC strains is contingent on the phylogenetic background, the acquisition of antibiotic resistance, and the presence or absence of virulence factor genes. In the last 4 years, the genomes of three UPEC strains have been sequenced (Brzuszkiewicz et al., 2006; Chen et al., 2006b; Welch et al., 2002). Each of these sequencing endeavours has collectively added a tremendous amount of information about the genetic makeup of UPEC strains, but has done little to sort E. coli into distinct subgroups because of the lack of comparative analysis. Genome sequencing of more E. coli strains would close this gap, but optical mapping is a more rapid and cost-effective technique that could provide the necessary information to assess the prevalence of virulence factor genes and changes that have led to antibiotic resistance, as well as help group the UPEC into distinct subgroups.
In this study, we have optically mapped 33 clinical isolates and compared their maps with in silico maps of sequenced strains of E. coli, representing what is believed to be the first study in which optical mapping has served as the basis for clustering bacterial species as shown in Fig. 1. From our analysis, the E. coli isolates divided into five subgroups using UPGMA alignments and a cut-off of 30 % dissimilarity. All of the sequenced UPEC strains (536, CFT073, UTI89) fell into the first subgroup, which also held most of the recent UPEC isolates and was the more diverse of the two UPEC subgroups. Our optical mapping scheme placed the APEC 01 strain in the same subgroup as the UTI89 strain. Both strains have similar MLST patterns (Johnson et al., 2007). It is possible that some minimal set of virulence factors is found in the genomes of these strains that allows them to initiate infections outside of the intestinal tract. Within the second subgroup, the seven isolates exhibited less diversity at the genetic level than the first subgroup. However, none of the isolates were identical. Past studies have not shown the separation of UPEC strains into major subgroups that we have demonstrated in this study. Subgroup three had the K-12 laboratory substrains of E. coli, W3110 and DH10B. The K-12 strain has lost many of the virulence factors associated with E. coli and has been altered to accept foreign DNA more readily. EHEC strains were found in the fourth subgroup. Two groups have successfully used optical mapping to distinguish between individual strains of ETEC H10407 and EHEC O157 : H7, but strain clustering was not performed (Chen et al., 2006a; Kotewicz et al., 2007, 2008). The fifth subgroup harboured an environmental strain of E. coli isolated from a heavy metal-contaminated site in South Carolina. Surprisingly, the UPEC isolates were more different from the other non-UPEC isolates as compared with the differences between Shigella isolates and non-UPEC strains.
The only significant difference between the tree based upon sequence (Henz et al., 2005) and the tree based upon optical maps was the relative position of the Shigella strains. A sequence-based tree will focus exclusively upon relative changes in conserved genes, whereas an optical map-based tree examines the overall structure of the genome. Since there are numerous genomic rearrangements between E. coli and Shigella (Yang et al., 2005), focusing only on the conserved genes will tend to mask many of the differences between these bacteria. On the other hand, optical mapping gives an unbiased look at the whole genome as compared with a sequence-based clustering and will pick up the structural changes in the genome more readily.
A high degree of similarity of UPEC isolates from disparate locations was found across the dataset, as isolates from Boston, Massachusetts, clustered closely with isolates collected in Wisconsin. From this optical mapping dataset, the geographical location did not correlate with the backbone structure of the E. coli isolates. Thus, optical mapping shows that the geographical structure appears to account for only a small fraction of the genomic variations of our isolates, which is consistent with an earlier study (Selander et al., 1987).
Because optical mapping shows a genome-wide snapshot of each isolate at a fraction of the cost and time needed to sequence a genome, this technology could potentially be used to sort UPEC isolates into pathotypes. Prior attempts at pathotyping have been hampered by the limited genetic information available and the laborious process of screening for specific virulence factor genes (Marrs et al., 2005; Foxman et al., 1995). A confounding factor for UPEC is their high mutation rate (Guttman & Dykhuizen, 1994; Denamur et al., 2002), so virulence factor genes may be missed because of the heterogeneity that results from mutations that prevent primer binding to a site. Because of these genetic limitations, several UPEC strains have been sequenced to study their pathogenesis: CFT073 (Mobley et al., 1990; Welch et al., 2002), UTI89 (Mulvey et al., 2001; Chen et al., 2006b) and 536 (Hacker et al., 1983). In this study, we have shown that E. coli isolates sort into distinct subgroups with similar haemolytic profiles. Our optical maps also demonstrate that PAIs can be identified among the isolates using optical mapping, and this includes differences in PAI positioning within the genome. Acquisition of virulence factors, including those found on PAIs, is independent of the main backbone structure of E. coli (Lloyd et al., 2007, 2009; Pupo et al., 1997, 2000), and the optical mapping illustrates that fact. The sorting of the E. coli isolates by optical mapping into a few distinct groups offers some hope that the technique will assist in the pathotyping of UPEC strains. In this regard, the screening for virulence factor genes among the 33 UPEC isolates also showed strong correlations with the subgrouping represented by the optical mapping analysis. An examination of virulence factor gene distribution has been applied before to UPEC strains (Bidet et al., 2005; Bingen-Bidois et al., 2002; Johnson & Stell, 2000; Sabate et al., 2006; Takahashi et al., 2006). These studies have shown a correlation between specific virulence factor genes and PAIs. We have also demonstrated this same correlation when comparing the data from Fig. 2 with the data provided in Table 3. Subgroups of UPEC sorted by the optical mapping technology also sorted along similar lines based on the presence or absence of nine virulence factor genes. Although the UPEC isolates were quite diverse, an even more striking observation was that the one major subgroup based on a 30 % dissimilarity cut-off was very uniform for the absence of most of these key virulence factor genes. Thus, optical mapping separated the UPEC isolates along similar lines to those that would be associated with their pathotype.
Besides an assessment of PAIs and other virulence factor-related genetic elements, this study also points to the utility of using optical mapping for tracking antibiotic-resistance patterns among UPEC strains. The emergence and spread of antibiotic-resistant E. coli strains is a serious health concern, particularly with respect to resistance to front-line drugs such as fluoroquinolones. Although plasmid-based resistance genes are one way to disseminate the resistance genes, more common are transposon or prophage insertion and the integration of the mobile genetic elements known as integrons, which have site-specific recombination systems built in (Rowe-Magnus et al., 2002). Several groups have tried to correlate the phylogenetic background with antibiotic resistance, with mixed results (Barl et al., 2008; Bruant et al., 2006; Graziani et al., 2009; Piatti et al., 2008; Rijavec et al., 2006; Yu et al., 2004). The most successful techniques used DNA microarrays (Barl et al., 2008; Bruant et al., 2006; Yu et al., 2004), but they are limited to the known genetic sequences available. In this study, all of the ciprofloxacin-resistant strains clustered together. A recent study has also shown that ciprofloxacin-resistant strains possess fewer virulence factor genes than ciprofloxacin-sensitive strains (Graziani et al., 2009). The potential utility of optical mapping has been demonstrated for tracking antibiotic-resistant outbreak strains.
Although optical mapping has shown a lot of commonality among the isolates, the technique was able to identify each individual isolate. The relative ease with which E. coli is able to gain and lose genetic elements, such as prophages and PAIs, means that each strain can be tagged as unique. Because optical mapping can track the large-scale changes mentioned above, it is capable of both clustering E. coli strains into subgroups based on greater commonality, as shown in this study, and distinguishing between different isolates, as described elsewhere (Kotewicz et al., 2007, 2008). Thus, we have shown for the first time that optical mapping is a viable way to subgroup bacteria and that it could be used as an alternative methodology to ribotyping or MLST analyses. In fact, Kotewicz et al. (2007) used optical mapping to track mobile genetic elements. Overall, we see optical mapping as a powerful tool able to analyse whole-genome structure for a fraction of the cost and time needed for whole-genome sequencing.
Acknowledgments
We wish to thank Jörg Hacker (Universität Würzburg), Scott Hultgren (Washington University) and Rod Welch (University of Wisconsin-Madison) for strains used in this study. The laboratory personnel at Gundersen Lutheran Medical Center are also thanked for identifying the GLMC strains. This work was supported by NIH grant AI065432-01A2 to W. R. S.
Abbreviations
EHEC, enterohaemorrhagic Escherichia coli
MLST, multilocus sequence typing
PAI, pathogenicity island
UPEC, uropathogenic Escherichia coli
UPGMA, unweighted pair group method with arithmetic mean
UTI, urinary tract infection
References
- Barl, T., Dobrindt, U., Yu, X., Katcoff, D. J., Sompolinsky, D., Bonacorsi, S., Hacker, J. & Bachmann, T. T. (2008). Genotyping DNA chip for the simultaneous assessment of antibiotic resistance and pathogenic potential of extraintestinal pathogenic Escherichia coli. Int J Antimicrob Agents 32, 272–277. [DOI] [PubMed] [Google Scholar]
- Bidet, P., Bonacorsi, S., Clermont, O., De Montille, C., Brahimi, N. & Bingen, E. (2005). Multiple insertional events, restricted by the genetic background, have led to acquisition of pathogenicity island IIJ96-like domains among Escherichia coli strains of different clinical origins. Infect Immun 73, 4081–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingen-Bidois, M., Clermont, O., Bonacorsi, S., Terki, M., Brahimi, N., Barraud, D. & Bingen, E. (2002). Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect Immun 70, 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, G., Ott, M., Lischewski, A., Ritter, A., Imrich, H., Tschape, H. & Hacker, J. (1994). Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun 62, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruant, G., Maynard, C., Bekal, S., Gaucher, I., Masson, L., Brousseau, R. & Harel, J. (2006). Development and validation of an oligonucleotide microarray for detection of multiple virulence and antimicrobial resistance genes in Escherichia coli. Appl Environ Microbiol 72, 3780–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzuszkiewicz, E., Bruggemann, H., Liesegang, H., Emmerth, M., Olschlager, T., Nagy, G., Albermann, K., Wagner, C., Buchreiser, C., Emody, L., Gottschalk, G., Hacker, J. & Dobrindt, U. (2006). How to become a uropathogen: a comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A 103, 12879–12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., Savarino, S. J. & Venkatesan, M. M. (2006a). Subtractive hybridization and optical mapping of the enterotoxigenic Escherichia coli H10407 chromosome: isolation of unique sequences and demonstration of significant similarity to the chromosome of E. coli K-12. Microbiology 152, 1041–1054. [DOI] [PubMed] [Google Scholar]
- Chen, S. L., Hung, C. S., Xu, J., Reigstad, C. S., Magrini, V., Sabo, A., Blasiar, D., Bieri, T., Meyer, R. R. & other authors (2006b). Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103, 5977–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont, O., Bonacorsi, S. & Bingen, E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66, 4555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont, O., Cordevant, C., Bonacorsi, S., Marecat, A., Lange, M. & Bingen, E. (2001). Automated ribotyping provides rapid phylogenetic subgroup affiliation of clinical extraintestinal pathogenic Escherichia coli strains. J Clin Microbiol 39, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur, E., Bonacorsi, S., Giraud, A., Duriez, P., Hilali, F., Amorin, C., Bingen, E., Andremont, A., Picard, B. & other authors (2002). High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J Bacteriol 184, 605–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Nelson, R., Baldwin, S., Plunkett, G., III, Burland, V., Mau, B., Petrosino, J. F., Qin, X., Muzny, D. M. & other authors (2008). The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190, 2597–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxman, B. (2002). Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 113 (Suppl. 1A), 5S–13S. [DOI] [PubMed] [Google Scholar]
- Foxman, B., Zhang, L., Tallman, P., Palin, K, Rode, C., Bloch, C., Gillespie, B. & Marrs, C. F. (1995). Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis 172, 1536–1541. [DOI] [PubMed] [Google Scholar]
- Foxman, B., Barlow, R., D'Arcy, H., Gillespie, B. & Sobel, J. D. (2000). Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol 10, 509–515. [DOI] [PubMed] [Google Scholar]
- Fricke, W. F., Wright, M. S., Lindell, A. H., Harkin, D. M., Baker-Austin, C., Ravel, J. & Stepanauskas, R. (2008). Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J Bacteriol 190, 6779–6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani, C., Luzzi, I., Corro, M., Tomei, F., Parisi, G., Giufre, M., Morabito, S., Caprioli, A. & Cerquetti, M. (2009). Phylogenetic background and virulence genotype of ciprofloxacin-susceptible and ciprofloxacin-resistant Escherichia coli strains of human and avian origin. J Infect Dis 199, 1209–1217. [DOI] [PubMed] [Google Scholar]
- Guttman, D. S. & Dykhuizen, D. (1994). Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266, 1380–1383. [DOI] [PubMed] [Google Scholar]
- Hacker, J., Knapp, S. & Goebel, W. (1983). Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J Bacteriol 154, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker, J., Bender, L., Ott, M., Wingender, J., Lund, B., Marre, R. & Goebel, W. (1990). Deletions of chromosomal regions coding for fimbriae and hemolysins occur in vitro and in vivo in various extraintestinal Escherichia coli isolates. Microb Pathog 8, 213–225. [DOI] [PubMed] [Google Scholar]
- Hacker, J., Blum-Oehler, G., Muhldorfer, I. & Tschape, T. (1997). Pathogenicity islands of virulent bacteria: structure, function, and impact on microbial evolution. Mol Microbiol 23, 1089–1097. [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Hayashi, T., Makino, K., Ohnishi, M., Kurokawa, K., Ishii, K., Yokoyama, K., Han, C. G., Ohtsubo, E. & other authors (2001). Complete genome sequence of enterohemorrhagic Escherichia coli O157 : H7 and genomic comparison with a laboratory strain K-12. DNA Res 8, 11–22. [DOI] [PubMed] [Google Scholar]
- Henz, S. R., Huson, D. H., Auch, A. F., Nieselt-Struwe, K. & Schuster, S. C. (2005). Whole-genome prokaryotic phylogeny. Bioinformatics 21, 2329–2335. [DOI] [PubMed] [Google Scholar]
- Hooton, T. M. & Stamm, W. E. (1997). Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin North Am 11, 551–581. [DOI] [PubMed] [Google Scholar]
- Hultgren, S. J., Schwan, W. R., Schaeffer, A. J. & Duncan, J. L. (1986). Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun 54, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q., Yuan, Z., Xu, J., Wang, Y., Shen, Y., Lu, W., Wang, J., Liu, H., Yang, J. & other authors (2002). Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res 30, 4432–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. R. (2003). Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect Dis Clin North Am 17, 261–278. [DOI] [PubMed] [Google Scholar]
- Johnson, J. R. & Brown, J. J. (1996). A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1–4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis 173, 920–926. [DOI] [PubMed] [Google Scholar]
- Johnson, J. R. & Stell, A. L. (2000). Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181, 261–272. [DOI] [PubMed] [Google Scholar]
- Johnson, T. J., Kariyawasam, S., Wannemuehler, Y., Mangiamele, P., Johnson, S. J., Doetkott, C., Skyberg, J. A., Lynne, A. M., Johnson, J. R. & Nolan, L. K. (2007). The genome sequence of avian pathogenic Escherichia coli strain O1 : K1 : H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 189, 3228–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamaru, S., Kurazano, H., Nakano, M., Terai, A., Ogawa, O. & Yamamoto, S. (2006). Subtyping of uropathogenic Escherichia coli according to the pathogenicity island encoding uropathogenic-specific protein: comparison with phylogenetic groups. Int J Urol 13, 754–760. [DOI] [PubMed] [Google Scholar]
- Kotewicz, M. L., Jackson, S. A., LeClerc, J. E. & Cebula, T. A. (2007). Optical maps distinguish individual strains of Escherichia coli O157 : H7. Microbiology 153, 1720–1723. [DOI] [PubMed] [Google Scholar]
- Kotewicz, M. L., Mammel, M. K., LeClerc, J. E. & Cebula, T. A. (2008). Opical mapping and 454 sequencing of Escherichia coli O157 : H7 isolates linked to the US 2006 spinach-associated outbreak. Microbiology 154, 3518–3528. [DOI] [PubMed] [Google Scholar]
- Kunin, C. M. (1994). Urinary tract infections in females. Clin Infect Dis 18, 1–10. [DOI] [PubMed] [Google Scholar]
- Lloyd, A. L., Rasko, D. A. & Mobley, H. L. T. (2007). Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189, 3532–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A. L., Henderson, T. A., Vigil, P. D. & Mobley, H. L. T. (2009). Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol 191, 3469–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs, C. F., Zhang, L. & Foxman, B. (2005). Escherichia coli mediated urinary tract infections: are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol Lett 252, 183–190. [DOI] [PubMed] [Google Scholar]
- Mobley, H. L., Green, D. M., Trifillis, A. L., Johnson, D. E., Chippendale, G. R., Lockatell, C. V., Jones, B. D. & Warren, J. W. (1990). Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58, 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey, M. A., Schilling, J. D. & Hultgren, S. J. (2001). Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69, 4572–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, E. W. & Huang, X. (1992). An O (N2 log N) restriction map comparison and search algorithm. Bull Math Biol 54, 599–618. [DOI] [PubMed] [Google Scholar]
- Nataro, J. P. & Kaper, J. B. (1998). Diarrheagenic Escherichia coli. Clin Microbiol Rev 11, 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie, H., Yang, F., Zhang, X., Yang, J., Chen, L., Wang, J., Xiong, Z., Peng, J., Sun, L. & other authors (2006). Complete genome sequence of Shigella flexneri 5b and comparison with Shigella flexneri 2a. BMC Genomics 7, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H. & Selander, R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157, 690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima, K., Toh, H., Ogura, Y., Sasamoto, H., Morita, H., Park, S. H., Ooka, T., Iyoda, S., Taylor, T. D. & other authors (2008). Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res 15, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna, N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J. & other authors (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157 : H7. Nature 409, 529–533. [DOI] [PubMed] [Google Scholar]
- Pevzner, P. A., Tang, H. & Waterman, M. S. (2001). An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A 98, 9748–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti, G., Mannini, A., Balistreri, M. & Schito, A. M. (2008). Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. J Clin Microbiol 46, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, B., Garcia, J. S., Gouriou, S., Duriez, P., Brahimi, N., Bingen, E., Elion, J. & Denamur, E. (1999). The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67, 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo, G. M., Karaolis, D. K., Lan, R. & Reeves, P. R. (1997). Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun 65, 2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo, G. M., Lan, R. & Reeves, P. R. (2000). Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A 97, 10567–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko, D. A., Rosovitz, M. J., Myers, G. S., Mongodin, E. F., Fricke, W. F., Gajer, P., Crabtree, J., Sebaihia, M., Thomson, N. R. & other authors (2008). The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190, 6881–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijavec, M., Erjavec, M. S., Avgustino, J. A., Reissbrodt, R., Pruth, A., Krizan-Hergouth, V. & Zgur-Bertok, D. (2006). High prevalence of multidrug resistance and random distribution of mobile genetic elements among uropathogenic Escherichia coli (UPEC) of the four major phylogenetic groups. Curr Microbiol 53, 158–162. [DOI] [PubMed] [Google Scholar]
- Rowe-Magnus, A. D., Davies, J. & Mazel, D. (2002). Impact of integrons and transposons on the evolution of resistance and virulence. Curr Top Microbiol Immunol 264, 167–188. [PubMed] [Google Scholar]
- Sabate, M., Moreno, E., Perez, T., Andreu, A. & Prats, G. (2006). Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect 12, 880–886. [DOI] [PubMed] [Google Scholar]
- Schaeffer, A. J., Schwan, W. R., Hultgren, S. J. & Duncan, J. L. (1987). Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect Immun 55, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander, R. K., Caugant, D. A. & Whittam, T. S. (1987). Genetic structure and variation in natural populations of Escherichia coli. In Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology, pp. 1625–1648. Edited by J. L. Ingraham & F. C. Neidhardt. Washington, DC: American Society for Microbiology.
- Takahashi, A., Kanamaru, S., Kurazono, H., Kunishima, Y., Tsukamoto, T., Ogawa, O. & Yamamoto, S. (2006). Escherichia coli isolates associated with uncomplicated and complicated cystitis and asymptomatic bacteruria possess similar phylogenies, virulence genes, and O-serogroup profiles. J Clin Microbiol 44, 4589–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof, S. Y., Solberg, O. D., Manges, A. R. & Riley, L. W. (2005). Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol 43, 5860–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, C. C., Huang, J. J., Ko, W. C., Yan, J. J. & Wu, J. J. (2001). Decreased predominance of papG class II allele in Escherichia coli strains isolated from adults with acute pyelonephritis and urinary tract abnormalities. J Urol 166, 1643–1646. [PubMed] [Google Scholar]
- Waterman, M. S., Smith, T. F. & Katcher, H. L. (1984). Algorithms for restriction map comparisons. Nucleic Acids Res 12, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., Goldberg, M. B., Burland, V., Venkatesan, M. M., Deng, W., Fournier, G., Mayhew, G. F., Plunkett, G., III, Rose, D. J. & other authors (2003). Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 71, 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, R. A., Burland, V., Plunkett, G., III, Redford, P., Roesch, P., Rasko, D., Buckles, E. L., Liou, S.-R., Boutin, A. & other authors (2002). Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99, 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., Terai, A., Yuri, K., Kurazono, H., Takeda, Y. & Yoshida, O. (1995). Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12, 85–90. [DOI] [PubMed] [Google Scholar]
- Yang, F., Yang, J., Zhang, X., Chen, L., Jiang, Y., Yan, Y., Tang, X., Wang, J., Xiong, Z. & other authors (2005). Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res 33, 6445–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Susa, M., Knabbe, C., Schmid, R. D. & Bachmann, T. T. (2004). Development and validation of a diagnostic DNA microarray to detect quinolone-resistant Escherichia coli among clinical isolates. J Clin Microbiol 42, 4083–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Foxman, B., Manning, S. M., Tallman, P. & Marrs, C. F. (2000). Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect Immun 68, 2009–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S., Kile, A., Bechner, M., Place, M., Kvikstad, E., Deng, W., Wei, J., Severin, J., Runnheim, R. & other authors (2004). Single-molecule approach to bacterial genomic comparisons via optical mapping. J Bacteriol 186, 7773–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]