Abstract
As a part of our initiative aimed at a large-scale comparative analysis of fungal mitochondrial genomes, we determined the complete DNA sequence of the mitochondrial genome of the yeast Candida subhashii and found that it exhibits a number of peculiar features. First, the mitochondrial genome is represented by linear dsDNA molecules of uniform length (29 795 bp), with an unusually high content of guanine and cytosine residues (52.7 %). Second, the coding sequences lack introns; thus, the genome has a relatively compact organization. Third, the termini of the linear molecules consist of long inverted repeats and seem to contain a protein covalently bound to terminal nucleotides at the 5′ ends. This architecture resembles the telomeres in a number of linear viral and plasmid DNA genomes classified as invertrons, in which the terminal proteins serve as specific primers for the initiation of DNA synthesis. Finally, although the mitochondrial genome of C. subhashii contains essentially the same set of genes as other closely related pathogenic Candida species, we identified additional ORFs encoding two homologues of the family B protein-priming DNA polymerases and an unknown protein. The terminal structures and the genes for DNA polymerases are reminiscent of linear mitochondrial plasmids, indicating that this genome architecture might have emerged from fortuitous recombination between an ancestral, presumably circular, mitochondrial genome and an invertron-like element.
INTRODUCTION
A linear form of the mitochondrial genome capped with specific terminal structures dubbed mitochondrial telomeres occurs frequently in eukaryotic species from diverse phylogenetic lineages (Nosek & Tomaska, 2002; Nosek et al., 1998). Structurally different types of mitochondrial telomeres can be considered as independent solutions of the ‘end-replication problem’ (Olovnikov, 1973; Watson, 1972), indicating that linear genomes in mitochondria emerged repeatedly in evolution through different molecular mechanisms of telomere maintenance (Nosek & Tomaska, 2002, 2008). Several lines of evidence indicate that telomeric structures can be transferred between different genophores as structural and functional modules. Hence, a selfish genetic element, such as a linear plasmid, may recombine with a circular genome, resulting in the formation of linearized DNA molecules with plasmid-derived telomeres (Nosek & Tomaska, 2003).
Integration of linear DNA plasmids into mitochondrial genomes appears to be a relatively common event, as many organisms contain sequences of plasmid origin in their mitochondrial DNA (mtDNA). These events generate genomic rearrangements, eventually leading to genome linearization, and may be associated with a particular phenotypic trait (Handa, 2008; Klassen & Meinhardt, 2007). For example: (i) the integration of the linear plasmids S1 and S2 into the maize mitochondrial genome results in genome fragmentation, the formation of linearized mtDNA molecules with the plasmid copies at their termini, and the ‘cytoplasmic male sterility’ phenotype (Schardl et al., 1984); (ii) in the slime mould Physarum polycephalum, the linear plasmid mF leads to site-specific linearization of the genome and promotes mitochondrial fusion (Takano et al., 1994); and (iii) in the euascomycete Podospora anserina, the linear plasmid pAL2-1 is implicated in life-span control (Hermanns & Osiewacz, 1992). These linear mitochondrial plasmids are classified in the invertron family, which includes adenoviral and bacteriophage genomes (e.g. phi29, PRD1), linear DNA plasmids in Streptomyces, and cytoplasmic killer plasmids in yeasts. Their typical features are terminal inverted repeats (TIRs) and a terminal protein (TP, t-protein) covalently bound to the 5′ ends of linear DNA molecules and implicated in a unique strategy for solving the end-replication problem (Meinhardt & Klassen, 2007; Sakaguchi, 1990). Moreover, these genomes encode a family B DNA polymerase, which employs a TP with an attached deoxyribonucleoside monophosphate for protein-priming of DNA synthesis by a strand displacement mechanism (Salas, 1991).
Although plasmid sequences integrated in mtDNA occur frequently among plant and fungal species, only a few examples of stable linear mitochondrial genomes possessing plasmid-related telomeres have been reported. These include the chrysophyte alga Ochromonas danica (Burger et al., 2000; Coleman et al., 1991), the chytridiomycete Hyaloraphidium curvatum (Forget et al., 2002), the moon jelly Aurelia aurita (Shao et al., 2006), and the two hydra species Hydra oligactis (Kayal & Lavrov, 2008) and Hydra magnipapillata (Voigt et al., 2008). Of them, only A. aurita and O. danica mtDNAs contain ORFs encoding homologues of the family B DNA polymerases (Burger et al., 2000; Shao et al., 2006). However, these homologues appear to have truncated N-terminal domains; hence, they may encode inactive forms of the enzyme.
In this study, we determined and analysed the complete mtDNA sequence of the yeast Candida subhashii. This yeast was recently isolated from the peritoneal dialysis fluid of a patient with end-stage renal failure and was taxonomically classified as a novel Candida species closely related to the pathogenic yeasts from the ‘CTG clade’ of hemiascomycetes (Adam et al., 2009). The clade is monophyletic and has two major subgroups: the first is represented by species such as Debaryomyces hansenii, Clavispora lusitaniae and Pichia guilliermondii, and the second by Candida albicans, Candida dubliniensis, Candida parapsilosis, Candida tropicalis and Lodderomyces elongisporus (Fitzpatrick et al., 2006). Phylogenetic analysis based on D1/D2 and internal transcribed spacer (ITS) domains of the rDNA cluster indicates that the C. subhashii lineage branches at the base of the C. albicans–C. parapsilosis subgroup (Adam et al., 2009).
Earlier studies have revealed that the mitochondrial genome architecture varies in species belonging to the CTG clade. While C. albicans possesses a circular-mapping mtDNA (Anderson et al., 2001), C. parapsilosis mitochondria contain a linear DNA genome that terminates with arrays of tandem repeats (Nosek et al., 2004). Here we report another case of a linear mitochondrial genome in this clade. The linear mtDNA molecules found in C. subhashii terminate with TIRs and seem to possess a protein covalently bound to their 5′ termini. This is the first reported example of such a mitochondrial genome architecture in hemiascomycetes. In addition, the genome exhibits several additional interesting features, such as an unexpectedly high content of guanine and cytosine (G+C) residues, the absence of introns, and the presence of two ORFs encoding homologues of family B DNA polymerases. These features suggest that the evolutionary emergence of this genome might have resulted from a recombination event(s) of an ancestral circular-mapping mtDNA with a linear DNA plasmid.
METHODS
Yeast strains.
The type strain of the yeast C. subhashii CBS10753 (FR-392-06; Adam et al., 2009) was used in this work. Yeast cultures were grown in liquid YPDG medium (1 % yeast extract, 1 % peptone, 0.5 % glucose, 3 %, v/v, glycerol) with constant shaking at 28 °C until late exponential phase.
DNA sequence assembly, annotation and analysis.
Yeast mitochondria were isolated from 2 l of an overnight cell culture grown in YPDG medium, and the mtDNA was prepared from the crude mitochondrial fraction using anion-exchange chromatography, as previously described (Valach et al., 2008). The complete mtDNA sequence was determined by the dideoxy chain-termination method and the sequencing reactions were carried out at Macrogen (http://www.macrogen.com/). First, we cloned selected BamHI and HindIII mtDNA fragments into the pTZ18R vector and sequenced the termini of plasmid inserts. Then, we designed oligonucleotide primers from these sequences and used the primer-walking strategy for determination of the complete DNA sequence directly on the purified mtDNA template. The telomeric sequences were verified using sequence analysis of cloned terminal XbaI fragments (see below). The sequence reads were assembled and annotated using the Vector NTI Advance v. 10.1.1 (Invitrogen) and Geneious v. 4.8.5 (Biomatters) software packages. Coding sequences were identified through blastx and blastp searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and sequence alignments of deduced protein products with their homologues from closely related Candida species. The coding potential of unknown ORFs was predicted by Fickett's algorithm (Fickett, 1982) using the TestCode program (http://www.bioinformatics.org/sms/testcode.html). The sequences for tRNAs were detected using the tRNAscan-SE Search Server (http://lowelab.ucsc.edu/tRNAscan-SE/) using default search mode and the Mito/Chloroplast source (Lowe & Eddy, 1997). The rrnL and rrnS genes for rRNAs were annotated as follows. The blastn search localized the rrnS gene between the trnM and trnI sequences. Subsequently, this region was aligned with the sequence of Saccharomyces cerevisiae rrnS, which indicated the position of C. subhashii rrnS to be in the region 25 903–27 394. A similar approach was used for the identification of the C. subhashii rrnL gene in the region 18 583–21 414. In this case, the ends were defined by the trnA gene at the 3′ end and by comparison with the 5′ end of S. cerevisiae rrnL.
Base composition and cumulative GC skew were calculated using the DNA base composition analysis tool (http://molbiol-tools.ca/Jie_Zheng/dna.html). The phylogenetic analysis was performed using sequences of conserved mitochondrial proteins. The alignments of sequence homologues were constructed using muscle (Edgar, 2004), manually edited to remove gaps and concatenated (Atp6-Atp8-Atp9-Cob-Cox1-Cox2-Cox3). The dataset containing 1717 amino acid positions was analysed by a maximum-likelihood (ML) algorithm using the PhyML program (Guindon & Gascuel, 2003) with the JTT+Γ model and four discrete categories, and tested by performing 500 bootstrap replicates.
Cloning and analysis of the terminal mtDNA fragments.
The terminal restriction enzyme fragments generated by XbaI endonuclease (1.5 and 2.8 kb) were cloned essentially as described for termini of linear mitochondrial plasmids of P. anserina (Hermanns & Osiewacz, 1992) and Pichia kluyveri (Blaisonneau et al., 1999). Briefly, isolated mtDNA was treated with calf intestinal phosphatase (CIP) for 60 min at 37 °C. The enzyme was inhibited by incubation in the presence of 5 mM EDTA (pH 7.5) at 70 °C for 10 min, and mtDNA was then purified using a NucleoSpin Extract II column (Macherey-Nagel). Next, XbaI-generated mtDNA fragments were ligated into the pUC19 vector digested with the endonucleases SmaI and XbaI. Sequences of five plasmid clones containing the terminal XbaI fragments were determined.
The mtDNA termini were analysed for their sensitivity to exonucleases as follows. Approximately 1 μg mtDNA was treated with either exonuclease III (New England Biolabs) or BAL-31 nuclease (New England Biolabs) according to the manufacturer's instructions. After heat inactivation of the enzyme, the mtDNA was extracted, digested with the restriction endonuclease XbaI (Takara) and electrophoretically separated on a 0.9 % agarose gel. A linear 1040 bp long dsDNA fragment generated by digestion with PvuII endonuclease was added to the C. subhashii mtDNA sample as an internal control of BAL-31 activity.
PFGE.
Analysis of mtDNA by PFGE was performed essentially as described by Fukuhara et al. (1993). Briefly, whole-cell DNA samples were prepared in agarose blocks containing Zymolyase 20T (ICN, 0.25 mg ml−1) and incubated overnight in 10 mM Tris/HCl, 0.45 M EDTA (pH 8.0) and 7.5 % 2-mercaptoethanol at 37 °C. The agarose blocks were transferred into 0.45 M EDTA (pH 8.0) and 1 % N-lauroylsarcosine and incubated with or without proteinase K (1 mg ml−1) overnight at 50 °C. Separations were performed on a 1.5 % (w/v) agarose gel using the CHEF Mapper XA Chiller System (Bio-Rad) with pulse switching at 5 to 20 s (linear ramping and 12 ° angle between the electric fields) for 42 h at 5 V cm−1 in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) at 10 °C throughout.
Analysis of mtDNA–protein complexes.
The crude mitochondrial fraction prepared above was resuspended in 1 ml 50 mM Tris/HCl, 20 mM EDTA (pH 7.5), 1 % SDS and 0.2 mM PMSF and incubated for 30 min at 65 °C. Subsequently, 0.2 ml 5 M potassium acetate was added and the precipitate was centrifuged for 5 min at 10 000 g at 4 °C. Nucleic acids in the supernatant were precipitated with an equal volume of 2-propanol, centrifuged for 15 min at 10 000 g at 4 °C, washed with 70 % ethanol and dried under a vacuum. The precipitate containing mtDNA–TP complexes was resuspended in 150 μl 10 mM Tris/HCl and 1 mM EDTA (pH 7.5). Isolated mtDNA (2 μl) was digested in a final volume of 20 μl with the restriction enzymes ClaI or PvuI for 60 min, followed by treatment with proteinase K (100 μg ml−1) at 50 °C for 60 min. Restriction enzyme fragments were electrophoretically separated on a 1.2 % agarose gel in 0.5× TBE buffer. Samples not treated with proteinase K were used as controls.
Miscellaneous.
Enzymatic manipulations with DNA, cloning procedures, DNA labelling and Southern blot analyses were performed essentially as described by Sambrook & Russell (2001).
RESULTS
Organization of the C. subhashii mitochondrial genome
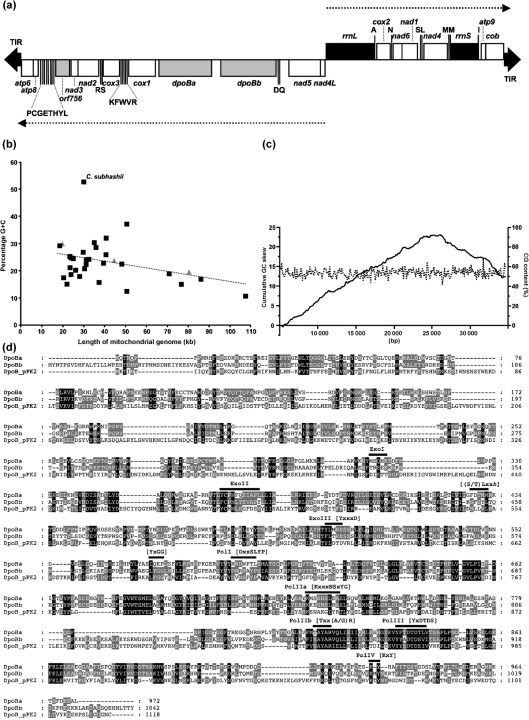
The complete DNA sequence of the C. subhashii mitochondrial genome was determined by the primer-walking strategy directly on a purified mtDNA template (Fig. 1a). The assembled contig was 29 795 bp long and its analysis indicated that the genome was represented by linear dsDNA molecules with 729 bp long TIRs. Base composition analysis revealed that the genome contained 52.7 % G+C residues, which is substantially different from all known yeast mtDNAs (Fig. 1b, Supplementary Table S1). The G+C residues were almost evenly distributed throughout the sequence (Fig. 1c). Non-coding intergenic regions contained a slightly higher G+C content (54.1 %) than annotated genes (52.4 %). Although guanine and cytosine residues were present at similar frequencies (27.4 % G vs 25.2 % C) in the complete mtDNA sequence, the presumed transcribed strand of the two transcription units (i.e. nad4L-to-left telomere and rrnL-to-right telomere; see below) was slightly biased toward cytosine (29.1 %) and adenine (26.5 %) residues. Codon usage analysis revealed that G+C-rich codons predominated in coding sequences (Supplementary Table S2), and we observed that cytosines and adenines were highly enriched specifically at the third codon position of protein-coding sequences (45.8 % C vs 23.4 % G and 23.4 % A vs 7.4 % T) (Supplementary Table S3).
Fig. 1.
Linear mitochondrial genome from the yeast C. subhashii. (a) Genetic map of the 29 795 bp linear mtDNA. ORFs encoding proteins (open rectangles), rRNAs (black rectangles), tRNAs (labelled by the single-letter codes for their cognate amino acids) and long TIRs (black triangles at the termini) are indicated. Dotted lines with arrowheads indicate the direction of gene transcription. The genes presumably derived from a plasmid are shown as grey rectangles. Dubious ORFs mentioned in the text were omitted. (b) The percentage of G+C residues was plotted against the length of the mitochondrial genome. Available complete DNA sequences of yeast mitochondrial genomes were downloaded from public databases (Supplementary Table S1). Archiascomycete and hemiascomycete species are shown as grey triangles and black squares, respectively. (c) G+C content and cumulative GC skew analyses of the C. subhashii mtDNA were performed with window/step settings of 100/100. (d) Amino acid sequence alignment of putative DNA polymerases encoded by the C. subhashii mtDNA (DpoBa and DpoBb) and the linear plasmid pPK2 from mitochondria of Pichia kluyveri (DpoB_pPK2) was performed using the muscle utility (Edgar, 2004) of the Geneious Pro 4.8.5 package (Drummond et al., 2009) and manually adjusted. The shading was performed using the GeneDoc program (Nicholas et al., 1997). The motifs conserved in the family B DNA polymerases are shown above the sequences.
Computer analysis of the C. subhashii mtDNA sequence revealed 14 ORFs encoding homologues of typical mitochondrial proteins, such as the subunits of the respiratory chain (nad1–nad6, nad4L, cox1–3 and cob) and ATP synthase (atp6, atp8 and atp9), as well as the genes for 24 tRNAs (trn) and two RNAs of the small (rrnS) and large (rrnL) subunits of the mitochondrial ribosome. We did not identify any introns in the C. subhashii mtDNA.
Next, we analysed the intergenic regions and identified six ORFs longer than 100 codons. The reading frames orf2916 and orf3126 encoded similar proteins. In both cases, blastp searches identified the family B DNA polymerase encoded by the Pichia kluyveri mitochondrial plasmid pPK2 as the best hit in the NCBI nr (non-redundant) database. A more detailed analysis of their sequences uncovered motifs similar to exonuclease and polymerase domains from bacteriophage DNA polymerases (Longas et al., 2006; Rodriguez et al., 2004). Therefore, we assigned these two ORFs as dpoBa and dpoBb, respectively (Fig. 1d). Four additional ORFs (i.e. orf327, orf354, orf522 and orf756) did not exhibit any significant similarity. We used the TestCode utility to predict whether these sequences encoded proteins (Fickett, 1982). The calculated TestCode value for orf756 was 1.222, which indicates that it might encode a protein product. Codon frequencies in orf756 exhibited a pattern similar to that found in the genes for conserved proteins, thus supporting this prediction. However, in the case of orf327, the TestCode analysis was not conclusive (the value was 0.828), and orf354 and orf522 were clearly classified as non-coding (the values were 0.609 and 0.576, respectively). Therefore, we considered the three ORFs dubious.
In closely related species such as C. parapsilosis, reading frames of the gene pairs nad2-nad3, nad4L-nad5 and nad6-nad1 partially overlap (Nosek & Fukuhara, 1994). These gene pairs were also linked in the C. subhashii mtDNA. We observed that the nad4L and nad5 ORFs were separated, but the nad6 and nad2 ORFs extended into the nad1 and nad3 reading frames by 1 and 37 nucleotides, respectively. However, the Nad3p sequence comparison revealed that its counterparts from related species of the CTG clade are shorter than the protein deduced from the full-length ORF of C. subhashii mtDNA by 12 amino acids. This indicates the intriguing possibility that translation of C. subhashii nad3 may start at the GUG codon that occurs a single base downstream of the nad2 termination codon. In addition, we found that the sequence for tRNAArg overlapped with the 5′ end of the predicted nad2 ORF by 27 nucleotides, indicating that the nad2-coding sequence may be shorter than the predicted ORF and that its translation may start at the second AUG codon. This possibility is supported by comparison of the amino acid sequence with its homologues. A similar analysis of apocytochrome b and cytochrome c oxidase subunit 1 suggested that their coding sequences were shorter by 78 and 45 nucleotides, respectively, than the full-length ORFs.
The trn genes were present either individually or in small clusters containing two to eight trn sequences. The mean G+C content in trn sequences was 47.2 %, which is higher than that observed in other yeast species (e.g. 36.6 % in C. parapsilosis). Nevertheless, the structures of all predicted tRNAs were essentially conserved with their counterparts from the closely related species C. albicans and C. parapsilosis. However, we detected two notable differences. First, the highly conserved motif GUUC in the pseudouridine (TψC) loop of tRNAAsp was replaced by GGUC. Second, the tRNAArg had UCG instead of ACG as the anticodon triplet, which may represent an adaptation associated with the difference in frequencies of corresponding codons for arginine. Namely, the CGA codon is present 41 times in predicted protein-coding sequences (21 times in a standard set) in C. subhashii but only once in C. parapsilosis (Supplementary Table S2). Our analysis of the sequences encoding conserved proteins showed that AUA, CUN and UGA are interpreted as isoleucine, leucine and tryptophan, respectively. Decoding of UGA codons is supported by the presence of tRNATrp with the UCA anticodon triplet.
The gene order indicated that the genome was organized into two transcription units (nad4L-atp6 and rrnL-cob). We assumed that the transcription of C. subhashii mtDNA could start from the region between nad4L and rrnL sequences and terminate at or near the telomeres, leading to two polycistronic primary transcripts processed into mature mRNAs presumably by excision of the tRNA and rRNA sequences. Non-coding sequences (including TIRs and dubious ORFs) represented 17.2 % of the genome, underlining its compact organization.
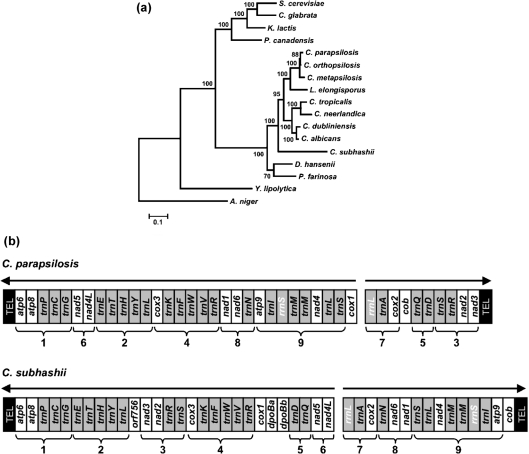
Next, we used a set of conserved proteins encoded by mtDNAs to analyse the phylogenetic relationship between C. subhashii and other species from the CTG clade. The phylogenetic tree topology, with statistically significant support, indicated that the C. subhashii lineage was basal to the C. albicans–C. parapsilosis subgroup of the clade (Fig. 2a). This confirms the results calculated from the sequences of the nuclear rRNA gene cluster (Adam et al., 2009). Indeed, the mitochondrial genomes in C. subhashii, C. parapsilosis and C. albicans exhibited a number of common features. They contained the same set of highly conserved genes (except for the unknown ORFs and dpoB genes present in C. subhashii), which were arranged in several conserved clusters, each containing two to eight genes. Two polycistronic transcription units were also observed in C. parapsilosis mtDNA (Nosek et al., 2004), and similar to C. subhashii, one transcription unit started with the rrnL-trnA-cox2 cluster and the second terminated with the trnG-trnC-trnP-atp8-atp6 cluster (Fig. 2b). On the other hand, the two genomes differ substantially in their G+C content, the presence of additional genes, and the structure of mitochondrial telomeres.
Fig. 2.
Phylogenetic and comparative analyses of the C. subhashii mtDNA. (a) Phylogenetic tree of hemiascomycetes calculated from the concatenated alignments of protein sequences encoded by the mtDNAs (i.e. Atp6-Atp8-Atp9-Cob-Cox1-Cox2-Cox3) using the ML method. The values of the bootstrap test (percentage from 500 replicates) are shown at the nodes, and the scale bar shows the evolutionary distance unit (calculated as the number of amino acid substitutions per site). (b) Comparison of the linear mitochondrial genomes from the yeasts C. subhashii and C. parapsilosis revealed nine clusters (numbered 1–9) with conserved gene order. Protein-coding genes (white rectangles), the genes for tRNAs and rRNAs (grey rectangles) and telomeres (black rectangles) are shown. Arrows indicate the direction of presumed polycistronic transcription units. Note that the telomeric structures of these mitochondrial genomes are different.
Mapping the termini of the linear mitochondrial genome
We confirmed the linearity of the C. subhashii mitochondrial genome using several approaches. First, we performed a sequencing reaction on the mtDNA template using the primer 5′-AGGAGACAGCAGTGGAGAA-3′, which binds to mtDNA in the region 254–272 bp from both ends of the genome contig. The determined sequence terminated with four cytosines and a single adenine residue. However, the terminal adenine residues were determined to be an artefact of direct sequencing on a linear mtDNA template. A similar artefact has been detected previously during the analysis of the linear mitochondrial plasmid pPK2 (Blaisonneau et al., 1999). To verify the terminal sequences of the linear mtDNA, we cloned both terminal XbaI fragments of mtDNA treated with alkaline phosphatase in plasmid pUC19 and determined the insert sequences in four clones with a 1.5 kb and one clone with a 2.8 kb XbaI fragment. In all cases, the sequence terminated at the 3′ ends exactly containing four cytosine residues without any additional nucleotides. As the T4 DNA ligase is known to catalyse the formation of a phosphodiester bond between juxtaposed 5′ phosphate and 3′ hydroxyl termini in duplex DNA, successful cloning of the terminal XbaI fragments into the vector digested with XbaI and SmaI endonucleases indicated that the linear mtDNA molecules were blunt-ended, and their DNA sequence analysis confirmed that both mtDNA ends had identical terminal nucleotides.
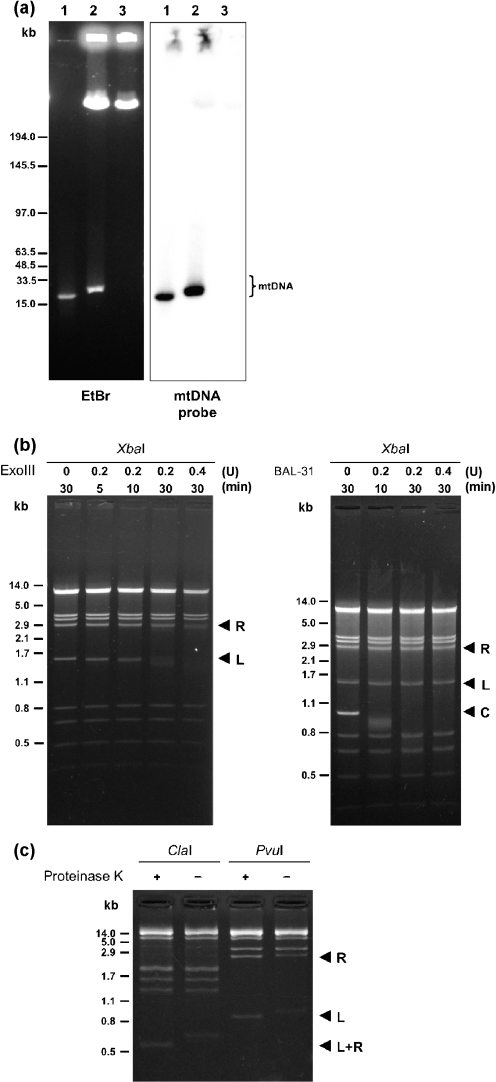
Next, we demonstrated the linearity of the genome using the PFGE approach. It has been previously shown that a linear mitochondrial genome migrates in PFGE as a distinct band (corresponding approximately to the genome size) ahead of nuclear chromosomal DNA, while mtDNA molecules from species with circular mitochondrial genomes migrate as a diffuse population of linear molecules (termed polydisperse DNA) ranging from tens to several hundreds of kilobase pairs, containing only a small fraction of circular molecules (Fukuhara et al., 1993; Maleszka et al., 1991). In the case of C. subhashii mtDNA, we detected a strong distinct band with an estimated size of about 30 kb in samples prepared from yeast cells in agarose blocks. We observed that the appearance of the mtDNA band was dependent on the treatment of samples with proteinase K (Fig. 3a).
Fig. 3.
The termini of the linear mtDNA are bound by a protein. (a) DNA samples were prepared from C. subhashii (see Methods) and separated by PFGE in a 1.5 % agarose gel. The gel was stained with 0.5 μg ml−1 ethidium bromide (EtBr) and transferred onto a nylon membrane. The blot was hybridized with radioactively labelled mtDNA from C. subhashii. Lane 1, isolated mtDNA; lanes 2 and 3, total cellular DNA prepared in agarose blocks treated or untreated with proteinase K, respectively. (b) Approximately 1 μg of isolated mtDNA was treated with exonuclease III (ExoIII) (left panel) or BAL-31 nuclease (right panel), as indicated. The mtDNA was then extracted from reactions, digested with XbaI endonuclease and electrophoretically separated. Note that the terminal fragments are sensitive to ExoIII but apparently not to BAL-31, indicating the possibility that the linear molecules have their 5′ termini blocked. L and R, positions of the 1527 and 2803 bp terminal restriction enzyme fragments, respectively; C, position of the internal control (a 1040 bp long linear blunt-ended DNA fragment) mixed with mtDNA prior to digestion with BAL-31 nuclease. (c) The mtDNA–protein complexes were isolated as described in Methods, digested with restriction endonucleases ClaI or PvuI, and treated or not treated with proteinase K. The positions of terminal restriction enzyme fragments generated by PvuI (833 and 2339 bp) and ClaI (547 bp) are indicated as L, R and L+R, respectively. Note that both terminal ClaI fragments have identical sizes, as this enzyme digests the C. subhashii mtDNA within TIRs.
Finally, we assayed the presumed termini of linear mtDNA molecules for their sensitivity to exonuclease III and BAL-31 nuclease. The former enzyme is able to degrade 3′ ends of linear dsDNA molecules, strongly preferring blunt or 3′ recessed ends, while the exonuclease activity of BAL-31 progressively shortens linear DNA duplexes from both termini. We observed that the treatment of C. subhashii mtDNA with exonuclease III selectively affected both terminal XbaI fragments (1.5 and 2.8 kb) (Fig. 3b, left panel). In contrast, we did not observe any preferential degradation of mtDNA treated with BAL-31. Therefore, we included a linear blunt-ended duplex DNA as an internal control in the mtDNA sample, which gradually disappeared, thus confirming that BAL-31 was active in the reaction (Fig. 3b, right panel). Moreover, we found that the 5′ ends appeared to be refractory to T4 polynucleotide kinase (data not shown). These results indicate that the linear mtDNA molecules have accessible 3′ ends, but that the 5′ ends are blocked either by a special arrangement of DNA or by a protein or peptide covalently linked to mtDNA termini. Therefore, we tested the possibility that a protein blocked the mtDNA termini. Presumed mtDNA–protein complexes were isolated from mitochondrial lysates incubated with 1 % SDS at 65 °C, which denatures most proteins and allows dissociation of non-covalently bound proteins from DNA molecules. The mtDNA samples were then digested with the restriction endonucleases ClaI or PvuI. We observed that migration of terminal restriction enzyme fragments was selectively affected by proteinase K treatment (Fig. 3c). This experiment clearly shows that the electrophoretic mobilities of the terminal ClaI (547 bp) and PvuI (833 and 2339 bp) restriction fragments were affected by the protease activity, and suggests that most if not all mtDNA molecules possess a protein covalently linked to both termini.
DISCUSSION
In yeasts, two types of linear mitochondrial genomes have been described (Fukuhara et al., 1993; Nosek et al., 1995). The type I genomes terminate with covalently closed ssDNA hairpins (t-hairpins), and their replication strategy involves monomeric and dimeric circular molecules as the intermediates (Dinouel et al., 1993). In contrast, the type II linear genomes possess telomeres consisting of tandem repeat arrays (t-arrays), and mitochondria do not contain any circular genome forms (Nosek et al., 1995). The maintenance of the terminal sequences relies on the rolling-circle amplification of telomeric circles (t-circles), extragenomic circular molecules derived exclusively from the telomeric sequence (Nosek et al., 2005; Tomaska et al., 2000, 2009). In this study, we describe a novel type of linear mitochondrial genome architecture in yeasts. Using direct mtDNA sequencing, PFGE analysis and mapping of termini with exonucleases, we demonstrated that mitochondria of C. subhashii contain linear dsDNA molecules with TIRs. Moreover, our results indicate that the mtDNA molecules may have a protein covalently bound to the 5′ ends. We will refer to the C. subhashii mtDNA as the type III linear mitochondrial genome. These telomeres are similar to invertron-like elements, such as linear mitochondrial plasmids [e.g. the pPK2 plasmid from mitochondria of the yeast Pichia kluyveri (Blaisonneau et al., 1999)]. Although we did not identify any linear plasmid in C. subhashii and no plasmids have been reported in mitochondria of related Candida species, the presence of TIRs and ORFs for putative DNA polymerases supports the idea that a linear plasmid was involved in the formation of this linear genome. This is in line with the hypothesis that linear mitochondrial genomes have emerged as a result of the invasion of selfish genetic elements followed by linearization of an ancestral circular-mapping genome, and that subsequently these elements have become essential for the maintenance of the linear genome form (Nosek & Tomaska, 2003). In contrast to the plasmid-driven linearization of mitochondrial genomes observed in maize and slime moulds (Schardl et al., 1984; Takano et al., 1996), the linear genome present in C. subhashii mitochondria has plasmid-derived telomeres, although the plasmid-related genes, such as dpoBa, dpoBb and possibly also orf756, are integrated within internal regions of the genome. This indicates that the plasmid integration was not a recent event and that the linearization of an ancestral genome was followed by its rearrangement. Importantly, linear mitochondrial plasmids usually carry only a single gene for DNA polymerase, but C. subhashii mtDNA contains two related but non-identical dpoB genes. The dpoB copies could have emerged either by gene duplication or by repeated recombination with linear plasmid(s).
Linear mtDNA molecules may contain TPs covalently linked to their 5′ ends
The results of our PFGE analysis (Fig. 3a) not only confirmed that the genome is linear but also strongly suggested that the mtDNA interacts with a protein, which can be eliminated by treatment with proteinase K. The shift in electrophoretic mobility of terminal restriction enzyme fragments after protease digestion indicated that the protein interacts with the termini of mtDNA (Fig. 3c). Because this interaction survived incubation in 1 % SDS at 65 °C and we observed that the 5′ ends of the linear DNA molecules are blocked (Fig. 3b), we suggest that the protein is covalently linked to the 5′ termini, analogous to the TPs present at the ends of invertrons.
DNA polymerases and the nature of the TP
In adenoviral and bacteriophage genomes, separate ORFs encode the TP and DNA polymerase. However, in fungal cytoplasmic and mitochondrial linear plasmids, the TP is generated from the N-terminal domain of the plasmid-encoded DNA polymerase (Kim et al., 2000; Takeda et al., 1996), or an entire DNA polymerase molecule may serve as a TP (Chan et al., 1991; Vierula et al., 1990). At present, we cannot rule out the possibility that in C. subhashii mitochondria, the TP is encoded by an ORF of unknown function, such as orf756. On the other hand, DNA polymerase homologues encoded by C. subhashii mtDNA contain cryptic N-terminal domains exhibiting weak similarity to the corresponding region of the pPK2-encoded DpoB (Fig. 1d), indicating that these regions may represent TP precursor(s). Moreover, DpoBa and DpoBb contain an insertion between exonuclease (ExoIII) and polymerase (PolI) domains, reminiscent of the linker domain of the phi29 DNA polymerase important for DNA and TP binding (Truniger et al., 2005). This supports the idea that DpoBa and/or DpoBb may be involved in replication initiation by a protein-priming mechanism. However, we noticed that the sequence corresponding to the highly conserved PolI motif diverged in DpoBa, which questions its role in mtDNA replication and suggests that protein priming of mtDNA replication might rely solely on the activity of DpoBb.
Base composition bias
Unusual base composition is an intriguing feature of the C. subhashii mtDNA. The genome contains a high proportion of G+C residues, which preferentially localize at neutral positions of the protein-coding regions (Supplementary Table S3, Fig. 1c). This contrasts with most mtDNAs analysed so far, which have a high A+T content. Such a composition of an organellar genome is commonly explained by mutational (GC→AT) bias (e.g. cytosine deamination and guanine oxidation) and inefficient mtDNA repair. In the hemiascomycete mtDNAs, the G+C content varies from 10.9 % (Nakaseomyces bacillisporus) to 37.3 % (C. tropicalis), and apparently decreases with the genome length (Fig. 1b, Supplementary Table S1). Moreover, G+C residues are unevenly distributed in most species. For example, the 85.8 kb mtDNA of S. cerevisiae containing 82.9 % A+T has extensive intergenic spacers composed mostly of A+T residues, while a higher proportion of G+C bases occurs in genes, introns and several GC clusters, such as the ori/rep sequences (Foury et al., 1998). Importantly, GC clusters play a key role in genome dynamics because they represent preferential sites of mtDNA recombination (Dieckmann & Gandy, 1987).
Extremely G+C-rich mitochondrial genomes have recently been reported in the colourless green alga Polytomella capuana (57 %) and the lycophyte Selaginella moellendorffii (∼67.8 %), in which the unusual base composition seems to be associated with GC-biased gene conversion (gBGC) resulting from a high level of recombination and extensive C-to-U RNA editing, respectively (Smith, 2009; Smith & Lee, 2008).
The phylogenetic position of C. subhashii among hemiascomycetes (Fig. 2a) indicates that the base composition shift in the mitochondrial genome is a relatively recent event, although its molecular basis remains unknown. Possible explanations may include a recombination-driven process, such as gBGC and/or RNA editing mentioned above, although the latter mechanism has not yet been demonstrated in yeast mitochondria. In addition, we propose a hypothesis that takes into account a plasmid invasion followed by genome linearization. Our results suggest that the protein-priming replication acquired from the integrated plasmid could contribute to the unusual base composition of the C. subhashii mtDNA. During the synthesis of DNA, a strand asymmetry in base composition is generated that leads to the enrichment of keto (G+T) and amino (C+A) bases in leading and lagging strands, respectively (Lobry, 1999). Cumulative GC skew analysis thus allows identification of origins and termini of DNA replication in prokaryotic genomes (Grigoriev, 1998). In C. subhashii mtDNA, the analysis revealed a peak in the region 18 501–21 501, with a maximum at position 21 101, suggesting that a switch between leading and lagging strand synthesis may occur around this region (Fig. 1c). If mtDNA replication begins at the termini by a protein-priming mechanism, the two replication forks approaching from the termini may meet at this region. Distribution of G+T and C+A residues in the genome segments divided by the peak in cumulative GC skew supports this idea (Supplementary Fig. S1). Although linear mitochondrial plasmids usually have relatively low G+C content (e.g. the pPK2 sequence contains only 23.2 % G+C), diverged motifs within the 3′–5′ proofreading domain of plasmid-derived DNA polymerases (Fig. 1d) suggest that it might exhibit a mutator phenotype, eventually contributing to the increased G+C content. The resulting shift in base composition would then be subjected to purifying selection for maintaining the mitochondrial functions.
Besides the hypotheses mentioned above, it remains unknown how the base composition shift influences mtDNA maintenance and the expression of mitochondrial genes. We propose that one outcome of the increased G+C content is the absence of intronic sequences. The sequence changes could erase recognition sites for intron insertions, thus preventing transmission of these mobile elements into C. subhashii mtDNA.
Implications for clinical microbiology
C. subhashii is represented by a single clinical isolate from a patient with fungal peritonitis (Adam et al., 2009). At present, neither the distribution nor the epidemiological importance of this species is known. The unique features of its mtDNA sequence described in our article provide an opportunity to design specific molecular probes suitable for simple screening of unidentified Candida spp. among clinical isolates as well as in yeast culture collections.
Acknowledgments
The authors wish to thank Ladislav Kovac (Comenius University, Bratislava) for inspiration and continuous support, Heather Adam (University of Toronto, Canada) and Subhash Mohan (Mount Sinai Hospital, Toronto, Canada) for their generous gift of the C. subhashii type strain, and the members of our laboratories for comments and discussions. We also thank the reviewers for their helpful comments and suggestions. This work was supported by grants from the Howard Hughes Medical Institute (55005622), the Fogarty International Research Collaboration Award (2-R03-TW005654-04A1), and the Slovak grant agencies VEGA (1/0219/08, 1/0132/09) and APVV (0024-07, 20-001604, LPP-0164-06).
Abbreviations
gBGC, GC-biased gene conversion
ML, maximum-likelihood
mtDNA, mitochondrial DNA
TIR, terminal inverted repeat
TP, terminal protein
Footnotes
The GenBank/EMBL/DDBJ accession number for the mitochondrial genome sequence of C. subhashii is GU126492.
A supplementary figure, showing the distribution of G+T and C+A bases in C. subhashii mtDNA, and three supplementary tables, showing the G+C contents in yeast mitochondrial genomes, the strong bias of C. subhashii ORFs toward G+C-rich codons, and the base composition of coding sequences, are available with the online version of this paper.
References
- Adam, H., Groenewald, M., Mohan, S., Richardson, S., Bunn, U., Gibas, C. F., Poutanen, S. & Sigler, L. (2009). Identification of a new species, Candida subhashii, as a cause of peritonitis. Med Mycol 47, 305–311. [DOI] [PubMed] [Google Scholar]
- Anderson, J. B., Wickens, C., Khan, M., Cowen, L. E., Federspiel, N., Jones, T. & Kohn, L. M. (2001). Infrequent genetic exchange and recombination in the mitochondrial genome of Candida albicans. J Bacteriol 183, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisonneau, J., Nosek, J. & Fukuhara, H. (1999). Linear DNA plasmid pPK2 of Pichia kluyveri: distinction between cytoplasmic and mitochondrial linear plasmids in yeasts. Yeast 15, 781–791. [DOI] [PubMed] [Google Scholar]
- Burger, G., Lang, B. F. & Gray, M. W. (2000). Phylogenetic relationships of stramenopile algae, based on complete mitochondrial genome sequences. Genbank Acc.no. AF287134.
- Chan, B. S., Court, D. A., Vierula, P. J. & Bertrand, H. (1991). The kalilo linear senescence-inducing plasmid of Neurospora is an invertron and encodes DNA and RNA polymerases. Curr Genet 20, 225–237. [DOI] [PubMed] [Google Scholar]
- Coleman, A. W., Thompson, W. & Goff, L. J. (1991). Identification of the mitochondrial genome in the chrysophyte alga Ochromonas danica. J Eukaryot Microbiol 38, 129–135. [Google Scholar]
- Dieckmann, C. L. & Gandy, B. (1987). Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J 6, 4197–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinouel, N., Drissi, R., Miyakawa, I., Sor, F., Rousset, S. & Fukuhara, H. (1993). Linear mitochondrial DNAs of yeasts: closed-loop structure of the termini and possible linear-circular conversion mechanisms. Mol Cell Biol 13, 2315–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J. & Ashton, B. M. C., Heled, J., Kearse, M., Moir, R., Stones-Havas, S., Thierer, T. & Wilson, A. (2009). Geneious v4.8; http://www.geneious.com/.
- Edgar, R. C. (2004). muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickett, J. W. (1982). Recognition of protein coding regions in DNA sequences. Nucleic Acids Res 10, 5303–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, D. A., Logue, M. E., Stajich, J. E. & Butler, G. (2006). A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget, L., Ustinova, J., Wang, Z., Huss, V. A. & Lang, B. F. (2002). Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol Biol Evol 19, 310–319. [DOI] [PubMed] [Google Scholar]
- Foury, F., Roganti, T., Lecrenier, N. & Purnelle, B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440, 325–331. [DOI] [PubMed] [Google Scholar]
- Fukuhara, H., Sor, F., Drissi, R., Dinouel, N., Miyakawa, I., Rousset, S. & Viola, A. M. (1993). Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features. Mol Cell Biol 13, 2309–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev, A. (1998). Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res 26, 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Handa, H. (2008). Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion 8, 15–25. [DOI] [PubMed] [Google Scholar]
- Hermanns, J. & Osiewacz, H. D. (1992). The linear mitochondrial plasmid pAL2-1 of a long-lived Podospora anserina mutant is an invertron encoding a DNA and RNA polymerase. Curr Genet 22, 491–500. [DOI] [PubMed] [Google Scholar]
- Kayal, E. & Lavrov, D. V. (2008). The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene 410, 177–186. [DOI] [PubMed] [Google Scholar]
- Kim, E. K., Jeong, J. H., Youn, H. S., Koo, Y. B. & Roe, J. H. (2000). The terminal protein of a linear mitochondrial plasmid is encoded in the N-terminus of the DNA polymerase gene in white-rot fungus Pleurotus ostreatus. Curr Genet 38, 283–290. [DOI] [PubMed] [Google Scholar]
- Klassen, R. & Meinhardt, F. (2007). Linear protein-primed replicating plasmids in eukaryotic microbes. In Microbial Linear Plasmids, pp. 187–226. Edited by F. Meinhardt & R. Klassen. Berlin: Springer-Verlag.
- Lobry, J. R. (1999). Genomic landscapes. Microbiology Today 26, 164–165. [Google Scholar]
- Longas, E., de Vega, M., Lazaro, J. M. & Salas, M. (2006). Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res 34, 6051–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T. M. & Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleszka, R., Skelly, P. J. & Clark-Walker, G. D. (1991). Rolling circle replication of DNA in yeast mitochondria. EMBO J 10, 3923–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt, F. & Klassen, R. (2007). Microbial Linear Plasmids. Berlin: Springer-Verlag.
- Nicholas, K. B., Nicholas, H. B. J. & Deerfield, D. W. I. (1997). GeneDoc: analysis and visualization of genetic variation. EMBnet News 4, 14 [Google Scholar]
- Nosek, J. & Fukuhara, H. (1994). NADH dehydrogenase subunit genes in the mitochondrial DNA of yeasts. J Bacteriol 176, 5622–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek, J. & Tomaska, L. (2002). Mitochondrial telomeres: alternative solutions to the end-replication problem. In Telomeres, Telomerases and Cancer, pp. 396–417. Edited by G. Krupp & R. Parwaresch. New York: Kluwer Academic/Plenum Publishers.
- Nosek, J. & Tomaska, L. (2003). Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr Genet 44, 73–84. [DOI] [PubMed] [Google Scholar]
- Nosek, J. & Tomaska, L. (2008). Mitochondrial telomeres: an evolutionary paradigm for the emergence of telomeric structures and their replication strategies. In Origin and Evolution of Telomeres, pp. 163–171. Edited by J. Nosek & L. Tomaska. New York: Landes Bioscience.
- Nosek, J., Dinouel, N., Kovac, L. & Fukuhara, H. (1995). Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol Gen Genet 247, 61–72. [DOI] [PubMed] [Google Scholar]
- Nosek, J., Tomaska, L., Fukuhara, H., Suyama, Y. & Kovac, L. (1998). Linear mitochondrial genomes: 30 years down the line. Trends Genet 14, 184–188. [DOI] [PubMed] [Google Scholar]
- Nosek, J., Novotna, M., Hlavatovicova, Z., Ussery, D. W., Fajkus, J. & Tomaska, L. (2004). Complete DNA sequence of the linear mitochondrial genome of the pathogenic yeast Candida parapsilosis. Mol Genet Genomics 272, 173–180. [DOI] [PubMed] [Google Scholar]
- Nosek, J., Rycovska, A., Makhov, A. M., Griffith, J. D. & Tomaska, L. (2005). Amplification of telomeric arrays via rolling-circle mechanism. J Biol Chem 280, 10840–10845. [DOI] [PubMed] [Google Scholar]
- Olovnikov, A. M. (1973). A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41, 181–190. [DOI] [PubMed] [Google Scholar]
- Rodriguez, I., Lazaro, J. M., Salas, M. & De Vega, M. (2004). phi29 DNA polymerase–terminal protein interaction. Involvement of residues specifically conserved among protein-primed DNA polymerases. J Mol Biol 337, 829–841. [DOI] [PubMed] [Google Scholar]
- Sakaguchi, K. (1990). Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol Rev 54, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas, M. (1991). Protein-priming of DNA replication. Annu Rev Biochem 60, 39–71. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. (2001). Molecular Cloning: a Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schardl, C. L., Lonsdale, D. M., Pring, D. R. & Rose, K. R. (1984). Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature 310, 292–296. [Google Scholar]
- Shao, Z., Graf, S., Chaga, O. Y. & Lavrov, D. V. (2006). Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381, 92–101. [DOI] [PubMed] [Google Scholar]
- Smith, D. R. (2009). Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol 71, 627–639. [DOI] [PubMed] [Google Scholar]
- Smith, D. R. & Lee, R. W. (2008). Mitochondrial genome of the colorless green alga Polytomella capuana: a linear molecule with an unprecedented GC content. Mol Biol Evol 25, 487–496. [DOI] [PubMed] [Google Scholar]
- Takano, H., Kawano, S. & Kuroiwa, T. (1994). Genetic organization of a linear mitochondrial plasmid (mF) that promotes mitochondrial fusion in Physarum polycephalum. Curr Genet 26, 506–511. [DOI] [PubMed] [Google Scholar]
- Takano, H., Mori, K., Kawano, S. & Kuroiwa, T. (1996). Rearrangements of mitochondrial DNA and the mitochondrial fusion-promoting plasmid (mF) are associated with defective mitochondrial fusion in Physarum polycephalum. Curr Genet 29, 257–264. [DOI] [PubMed] [Google Scholar]
- Takeda, M., Hiraishi, H., Takesako, T., Tanase, S. & Gunge, N. (1996). The terminal protein of the linear DNA plasmid pGKL2 shares an N-terminal domain of the plasmid-encoded DNA polymerase. Yeast 12, 241–246. [DOI] [PubMed] [Google Scholar]
- Tomaska, L., Nosek, J., Makhov, A. M., Pastorakova, A. & Griffith, J. D. (2000). Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res 28, 4479–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaska, L., Nosek, J., Kramara, J. & Griffith, J. D. (2009). Telomeric circles: universal players in telomere maintenance? Nat Struct Mol Biol 16, 1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger, V., Bonnin, A., Lazaro, J. M., de Vega, M. & Salas, M. (2005). Involvement of the “linker” region between the exonuclease and polymerization domains of phi29 DNA polymerase in DNA and TP binding. Gene 348, 89–99. [DOI] [PubMed] [Google Scholar]
- Valach, M., Tomaska, L. & Nosek, J. (2008). Preparation of yeast mitochondrial DNA for direct sequence analysis. Curr Genet 54, 105–109. [DOI] [PubMed] [Google Scholar]
- Vierula, P. J., Cheng, C. K., Court, D. A., Humphrey, R. W., Thomas, D. Y. & Bertrand, H. (1990). The kalilo senescence plasmid of Neurospora intermedia has covalently-linked 5′ terminal proteins. Curr Genet 17, 195–201. [Google Scholar]
- Voigt, O., Erpenbeck, D. & Worheide, G. (2008). A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics 9, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, J. D. (1972). Origin of concatemeric T7 DNA. Nat New Biol 239, 197–201. [DOI] [PubMed] [Google Scholar]