Abstract
The Porphyromonas gingivalis vimA gene has been previously shown to play a significant role in the biogenesis of gingipains. Further, in P. gingivalis FLL92, a vimA-defective mutant, there was increased auto-aggregation, suggesting alteration in membrane surface proteins. In order to determine the role of the VimA protein in cell surface biogenesis, the surface morphology of P. gingivalis FLL92 was further characterized. Transmission electron microscopy demonstrated abundant fimbrial appendages and a less well defined and irregular capsule in FLL92 compared with the wild-type. In addition, atomic force microscopy showed that the wild-type had a smoother surface compared with FLL92. Western blot analysis using anti-FimA antibodies showed a 41 kDa immunoreactive protein band in P. gingivalis FLL92 which was missing in the wild-type P. gingivalis W83 strain. There was increased sensitivity to globomycin and vancomycin in FLL92 compared with the wild-type. Outer membrane fractions from FLL92 had a modified lectin-binding profile. Furthermore, in contrast with the wild-type strain, nine proteins were missing from the outer membrane fraction of FLL92, while 20 proteins present in that fraction from FLL92 were missing in the wild-type strain. Taken together, these results suggest that the VimA protein affects capsular synthesis and fimbrial phenotypic expression, and plays a role in the glycosylation and anchorage of several surface proteins.
INTRODUCTION
A variety of bacterial cell surface components, including capsule, polysaccharides, proteases (gingipains), haemagglutinin, lipopolysacharides (LPSs), major outer-membrane proteins and fimbriae contribute to the virulence of Porphyromonas gingivalis (reviewed by Lamont & Jenkinson, 1998; Yoshimura et al., 2009). This organism, a black-pigmented, Gram-negative anaerobe, is an important aetiological agent of periodontal disease and is associated with other systemic illnesses including cardiovascular disease (Dave & Van, 2008; Demmer & Desvarieux, 2006; Ford et al., 2007; Nakano et al., 2009). Cell surface components like the fimbriae (Amano et al., 1999; Murakami et al., 2004; Wang et al., 2009), lectin-like adhesins (Cutler et al., 1995) and gingipains (McAlister et al., 2009; Pavloff et al., 1997; Pike et al., 1996) are integrally involved in the adhesion, invasion and colonization of periodontal tissue. In addition, the gingipains are involved in several cellular processes (Kuramitsu, 1998) including growth, inactivation of cytokines and their receptors, platelet aggregation, attenuation of neutrophil antibacterial activities, increasing vascular permeability, and apoptosis of gingival epithelial cells (Stathopoulou et al., 2009). Gingipains consist of a lysine-specific protease (Kgp) and an arginine-specific protease (Rgp), which is further subdivided into RgpA and RgpB; these proteases cleave proteins after arginine residues, and are encoded by the rgpA and rgpB genes, respectively. The mature form of RgpA contains a catalytic domain and a haemagglutinin domain, while RgpB possesses only a catalytic domain. Lysine-specific protease (Kgp) (Imamura, 2003; Mikolajczyk-Pawlinska et al., 1998; Pavloff et al., 1995) cleaves proteins after lysine residues.
Post-translational modifications of surface components such as gingipains, fimbriae and LPS play a key functional role in regulating the virulence of the organism. The maturation pathways of the gingipains are linked to the biosynthesis of surface carbohydrates (Paramonov et al., 2005; Shoji et al., 2002) and several other proteins, including PorR (Shoji et al., 2002), PorT (Nguyen et al., 2009; Sato et al., 2005), Sov (Sato et al., 2005) and VimA (Vanterpool et al., 2004, 2005b, 2006). Arg-X also plays a role in processing fimbriae protein precursors, serving as a proteinase for fimbrillin maturation (Kato et al., 2007; Nakayama et al., 1996). The LPS of P. gingivalis, which has recently been shown to contain two macromolecules [O-LPS (O antigen attached to a lipid A core) and A-LPS (phosphorylated branched mannan repeating unit attached to a lipid A core)], undergoes several stages of assembly, including catalysis by a relaxed-specificity ligase which is able to attach either a branched phosphorylated mannan or a tetrasaccharide repeating unit to the Lipid A core (Rangarajan et al., 2008). Lipid A is also present in several structural forms (Kumada et al., 1995) and probably functions in response to strain differences as well as environmental cues.
The P. gingivalis vimA gene, which is part of the recA operon and codes for a 39 kDa protein (putative acyl-CoA N-acyltransferase) has been previously shown to be part of the maturation pathway for gingipains (Abaibou et al., 2001; Vanterpool et al., 2006). Though the mechanism for gingipain regulation has not been thoroughly elucidated, we have demonstrated that vimA modulates the phenotypic expression of the gingipains. Additionally, inactivation of this gene resulted in a non-black pigmented strain designated P. gingivalis FLL92, which showed reduced levels of proteolytic, haemagglutinating and haemolytic activities (Abaibou et al., 2001; Olango et al., 2003). Using a mouse model, the virulence of P. gingivalis FLL92 was shown to be drastically reduced when compared with wild-type W83 strain. The reduced proteolytic activity observed in this vimA-defective mutant is as a result of a defect in the post-translational regulation of the gingipain genes. This is further supported by the presence of the pro-enzyme form of the gingipains in addition to their unaltered transcription in the vimA-defective mutant (Olango et al., 2003). There is also a late onset of gingipain activity in FLL92, which is mostly soluble, with little or no cell-associated activity (Olango et al., 2003; Vanterpool et al., 2005b). Other studies have shown that the recombinant VimA can interact with the gingipains in addition to several proteins, including HtrA, β-lactamase and sialidase, which are known in other bacterial systems to be involved in post-translational regulation (Vanterpool et al., 2006). It still remains unclear whether the vimA gene product is part of a central mechanism that may be involved in the maturation or post-translational regulation of other cell surface proteins in P. gingivalis.
In the present study, we further characterized the cell surface of the vimA-defective mutant of P. gingivalis. We investigated the role of VimA on capsule and fimbriae formation and outer-membrane protein expression. In addition, the effect of cell surface changes on antibiotic resistance was also evaluated.
METHODS
Bacterial strains and growth conditions.
P. gingivalis strains (W83, ATCC 33277 and FLL92) were grown in either brain–heart infusion (BHI) broth (Difco) supplemented with cysteine (0.1 %), vitamin K (0.5 μg ml−1) and haemin (5 μg ml−1) (BHI-HK) or trypticase soy broth containing menadione and haemin (TSBKH). Solid medium was prepared by supplementing with 1.5 % agar and 5 % defibrinated sheep blood (Haemostat laboratories). All cultures were incubated at 37 °C in an anaerobic chamber (Coy Manufacturing) in 80 % N2, 10 % H2 and 10 % CO2. Growth rates were determined spectrophotometrically at 600 nm (OD).
Biofilm formation assay.
Biofilm formation was assayed in 96-well microtitre dishes (R. J. Lamont, personal communication). Briefly, P. gingivalis was cultured overnight in TSBKH media. One millilitre of the overnight culture (mid-exponential phase) was centrifuged at 12 851 r.c.f. for 2 min at room temperature. The pellet was collected and washed twice with 1 ml PBS, then resuspended in a 1 ml mixture of TSBKH and PBS (1 : 2). Aliquots of the sample were added to each well. Plates were then incubated with a slight rotation for 24 h in an anaerobic chamber, to allow for the development of biofilm. After removal of the culture supernatant, the plates were washed twice by immersion in distilled water, then allowed to air dry for 1 h. The biofilm was stained with 0.5 % safranin for 15 min (100 μl per well), then washed twice with distilled water. Ethanol (95 %) was added to solubilize the safranin which was transferred to a new microtitre dish. Biofilm formation was obtained by determining the absorbance with a plate reader at 490 nm (Davey & Duncan, 2006).
Analysis of P. gingivalis strains by RT-PCR.
Total RNA was extracted from P. gingivalis at the early (OD600 1.0) phase of the growth cycle, using the SV total RNA isolation system (Promega). RT-PCR was performed using primers specific for the FimA (forward, 5′-GGCAGAACCCGTTGTAGAAA; reverse, 5′-GACCAAAGAATTGCCGAAAA) and 16S ribosomal (Bogen & Slots, 1999) gene product and the one-step RT-PCR kit (Invitrogen), following the manufacturer's instructions, with 1 μg template RNA in 50 μl RT-PCR mixture. RT-PCR in the absence of reverse transcriptase served as the negative control.
Protein electrophoresis and blotting.
SDS-PAGE was performed according to the manufacturer's instructions (Invitrogen) on 10 % Bis/Tris separating gel in morpholinepropanesulfonic acid (MOPS) SDS running buffer. Proteins separated by SDS-PAGE were electrophoretically transferred to a nitrocellulose membrane (Whatman Optitran BA-S 85) and the blotted membranes were immunostained with polyclonal anti-FimA antibody (Lin et al., 2006). Studies using lectin from Agaricus bisporus [ABA; specific for galactose (β 1,3) N-acetylgalactosamine], Concanavalin A (ConA; specific for α-mannose and α-glucose), Dolichos biflorus (DBA; specific for N-acetyl-α-d-galactosamine), Erythrina crista-galli [ECA; specific for galactose (β 1,4) N-acetylglucosamine], Glycine max [SBA; specific for N-acetyl-d-galactosamine], Limulus polyphemus [LPA; specific for sialic acid (N-acetyl neuraminic acid)] and Triticum vulgaris [WGA; specific for N-acetyl-β-d-glucosamine] were performed according to the manufacturer's recommendations (Sigma Aldrich). In brief, blots were blocked in PBS containing 2 % Tween for 2 min at 20 °C. Blots were then rinsed twice in PBS and incubated with 1 μg lectin-peroxidase ml−1 in PBS containing 0.05 % Tween, with 1 mM CaCl2, 1 mM MnCl2 and 1 mM MgCl2, for 16 h at 20 °C. Surplus lectins were removed by rinsing with PBS; the blots were developed using the DAB staining kit with nickel enhancement (Vector Laboratories) (Linton et al., 2002).
Microscopic examination.
Atomic force microscopy (AFM) and transmission electron microscopy (TEM) were used to provide detailed visualizations of the surface morphology. AFM was carried out using W83 and FLL92 stains that were grown overnight to OD600 1.2, washed twice in 0.1 M PBS buffer pH 7.4 and mounted on a cover slide. AFM was performed using the Innova Scanning Probe Microscope (Veeco Instruments) in air, using the tapping mode. Images were captured in the height mode with a resolution of 256×256 (Pratto et al., 2009). TEM was performed using the Philips Tecnai 12 TEM as per the method described by Harris (1991). Briefly, Formvar-carbon coated grids were prepared; the Formvar support was removed by placing the grids in an atmosphere of solvent vapour. Grids were then placed on a wire mesh in a glass Petri dish with carbon tetrachloride below the wire mesh. One microlitre of serially diluted sample was placed under the carbon side of a 4×5 mm square of mica (approx. twice the size of an electron microscope grid). The grid was washed in 0.5 % acetic acid then acetone. The carbon film was broken to free the specimen grid, after which the grid was placed in stain solution (neutral 1 % aqueous phosphotungstic acid) for 30 s. After blotting dry, the grid was examined using the Tecnai TEM (FEI).
Localization of FimA in P. gingivalis by immunogold staining.
Anti-FimA antibodies (Lin et al., 2006) were purified and used at a working concentration of 1 : 100. The colloidal gold conjugate (10 nm; Aurion) was diluted using incubation buffer (20 mM phosphate buffer, 150 mM NaCl pH 7.4, 0.2 % BSA, 15 mM NaN3) to 1 : 1000. A checker board titration of primary antibody to immunogold conjugate was made to identify the optimal working concentration. Processed nickel grids were subjected to charging of the processed P. gingivalis strains for 1 h, then washed 20 times in 0.025 M Tris buffer (pH 7.4) and lightly blotted. Grids were subsequently blocked with 5 % BSA in 0.025 M Tris buffer for 15 min at room temperature, then incubated in diluted antibody–gold complex for 4 h at 37 °C. Staining was done using uranyl acetate (0.8 g uranyl acetate dissolved in 10 ml absolute ethanol) and freshly prepared lead citrate (10–40 mg lead citrate, 10 ml filtered water, 100 μl 10 M NaOH). Grids were immersed in uranyl acetate for 7 min at room temperature and then washed, once by immersion in 25 % ethanol and twice by immersion in water. Grids were dried on filter paper for 10 min at room temperature and then floated on lead citrate drops for 5 min. Following two washes by immersion in 0.02 M NaOH and drying, grids were visualized using the Philips Tecnai 12 TEM.
Hydrophobicity assay.
Hydrophobicity assays of P. gingivalis strains were based on adhesion to hexadecane, as described by Grivet et al. (2000). Bacteria were harvested during exponential growth phase by centrifugation at 4 °C for 15 min, then they were washed with 0.1 M PBS (pH 7) and resuspended in PBS. The OD600 of the samples was adjusted to 0.85. Bacterial suspension (3 ml) was placed in a polystyrene tube to which 400 μl hexadecane was added. No hexadecane was added to the control suspensions. Suspensions were equilibrated in a water bath at 37 °C, vortexed for two 30 s periods with 5 s intervals and these were allowed to stand until the phases separated. The lower aqueous phase was carefully removed and its OD600 was determined. The values obtained were expressed as the percentage of bacteria remaining in the aqueous phase compared with the control suspension.
Determination of minimum inhibitory concentrations (MIC).
Serial dilutions of antibiotics were used to determine MIC values in broth. These experiments were done in duplicate. In brief, broth macro dilutions of W83 and FLL92 were carried out using tubes containing 9 ml BHI-HK broth plus 1 ml of antibiotic dilution. Tubes were examined for turbidity after incubation at 37 °C for 72 h in an anaerobic chamber (Wright et al., 1997). The MIC was taken as the concentration at which no visible growth was observed.
Labelling of P. gingivalis with [3H]-palmitic acid.
P. gingivalis cells were labelled according the protocol described by Shoji et al. (2004). Briefly, 5 ml cultures of W83 and FLL92 were incubated for 24 h in BHI-HK in the presence of 50 μCi palmitic acid [9,10(n)-3H] ml−1 (MP Biomedicals). Cells were then centrifuged, washed in PBS buffer and homogenized using glass beads to derive total proteins. Protein fractions were subsequently separated on an SDS-PAGE gel. Radiolabelled proteins were detected using autoradiography as described by Voytas & Ke (2001).
Bioinformatic analysis of VimA.
Protein modelling of VimA was conducted using I-Tasser (Zhang, 2008, 2009). The subcellular location of the protein was predicted from its amino acid sequence using SignalP 3.0 (Nielsen et al., 1997), Phobius (Kall et al., 2007) and PSORTb (Gardy et al., 2005). Protein motifs and domains of VimA were identified using PPSearch (Pearson & Lipman, 1988) and Superfamily (Gough et al., 2001).
Cell fractionation.
The Sarkosyl method was used to prepare outer membrane fractions of P. gingivalis (Potempa et al., 1995). In brief, P. gingivalis cells were harvested from a 500 ml culture during the exponential and stationary phase of the growth cycle (OD600 0.6–1.0 and 1.5–1.8, respectively). Cells were pelleted at 9000 g for 30 min at 4 °C then resuspended in 5 ml 20 mM Tris/HCl (pH 7.4), 10 mM EDTA, 10 mM N-α-p-tosyl-l-lysine chloromethyl ketone (TLCK) and 1 % sodium lauryl sarcosinate. Cells were disrupted by passage through a French pressure cell at 109 MPa. Whole cells were removed by further centrifugation at 5000 g for 20 min at 4 °C. The outer membrane fraction was separated by centrifugation (100 000 g for 30 min), and then the pellet was washed three times in 0.5 % sodium lauryl sarcosinate and resuspended in 20 mM Tris/HCl (pH 7.4). Extracellular fractions were prepared from cell-free culture fluid precipitated with 60 % acetone (−20 °C) (Vanterpool et al., 2006). The protein pellet was resuspended in 7 ml 100 mM Tris/HCl buffer (pH 7.4) in the presence of 1 mM TLCK, dialysed for 48 h against the same buffer, then stored at −20 °C. Total proteins were harvested from a 50 ml culture during the late exponential phase. Cells were pelleted at 8000 g for 30 min then resuspended in 1 ml 25 mM Tris/HCl (pH 8.0) with a protease inhibitor cocktail tablet (Complete EDTA-free, Roche), RNase and DNase (1 μg ml−1). Resuspended cells were then transferred to a 2 ml cryogenic tube containing 1 g of 0.1 mm glass beads. Cells were lysed in a Beadbeater homogenizer for 4 min with 30 s intervals, with 1 min cooling on ice. Samples were then centrifuged (10 000 g) for 12 min at 4 °C. The upper aqueous layer containing the total protein fraction was removed and stored on ice or at −20 °C.
Digestion of P. gingivalis wild-type and FLL92 proteins.
Late exponential and stationary phase membrane proteins were separated on a 10 % Bis/Tris gel (Invitrogen) in 1× MOPS running buffer for 1.5 cm, then visualized by staining with SimplyBlue safe stain (Invitrogen) (Henry et al., 2008). After destaining in water, the gel was cut into 1–2 mm slices. Gel slices were subsequently dehydrated in acetonitrile and dried in a vacuum centrifuge for 30 min. The gel slices were incubated for 1 h at 60 °C in a solution containing 20 μl of 20 mM DTT in 100 mM NH4HCO3 (enough to cover the gel pieces). The DTT solution was replaced with an alkylating solution (20 μl of 200 mM iodoacetamide in 100 mM NH4HCO3) after cooling the proteins to room temperature. Gel slices were further incubated at ambient temperature for 30 min in the dark, followed by two washes with 150 μl of 100 mM NH4HCO3; the slices were then finely minced with a flame-sealed polypropylene pipette tip, dehydrated by the addition of acetonitrile and vacuum dried. Following an overnight incubation of the gel pieces with 20 μl digestion buffer [1 μl of mass spectrometry (MS)-grade trypsin (www.promega.com) in 50 mM acetic acid with 1 μl of 100 mM NH4HCO3], the digestion reaction was stopped with 10 μl of 5 % formic acid. After transferring the digest solution (aqueous extraction) to a 0.65 ml siliconized tube, 30 μl of 50 % acetonitrile with 0.1 % formic acid was added, the mixture was vortexed for 3 min, centrifuged and then sonicated for 5 min. The process was repeated and both extractions were pooled and concentrated to 10 μl in a vacuum centrifuge. Peptide extraction was accomplished using standard C18 ZipTip technology following the manufacturer's directions (Millipore).
MS and data analysis.
An LCQ Deca XP Plus system (www.thermo.com) with nano-electrospray technology (www.newobjective.com) consisting of a reverse-phase C18 separation of peptides on a 10 cm by 75 μm capillary column using Microm Magic RP-18AQ resin (www.michrom.com) with direct electrospray injection was used to analyse the extracted peptides from each gel piece (Henry et al., 2008). A four part protocol was used for the MS and MS/MS analyses; this included one full MS analysis (from 450 to 1750 m/z) followed by three MS/MS events using data-dependent acquisition, where the most intense ion from a given full MS scan was subjected to collision-induced dissociation, followed by the second and third most intense ions. The nanoflow buffer gradient was extended over 45 min in conjunction with the cycle repeating itself every 2 s, using a 0–60 % acetonitrile gradient from buffer B (95 % acetonitrile with 0.1 % formic acid) developed against buffer A (2 % acetonitrile with 0.1 % formic acid) at a flow rate of 250–300 nl min−1, with a final 5 min 80 % bump of buffer B before re-equilibration. In order to move the 20 μl sample from the autosampler to the nanospray unit, flow stream splitting (1 : 1000) and a Scivex 10 port automated valve (Upchurch Scientific) together with a Michrom nanotrap column was used. The spray voltage and current were set at 2.2 kV and 5.0 μA, with a capillary voltage of 25 V in positive ion mode. The spray temperature for peptides was 160 °C. Data collection was achieved using the Xcalibur software (Thermo Electron), then screened with Bioworks 3.1. mascot software (www.matrixscience.com) was used for each analysis to produce unfiltered data and output files. Statistical validation of peptide and protein findings was achieved using X! Tandem (www.thegpm.org) and scaffold 2 meta analysis software (www.proteomesoftware.com). The presence of two different peptides at a probability of at least 95 % was required for consideration of positive identification. Confirmation of individual peptide matches was achieved using the blast database (www.oralgen.lanl.gov).
RESULTS
In silico analysis of VimA reveals close similarity to the Fem family of proteins
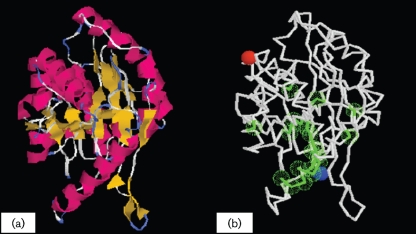
Using I-Tasser, the putative structure of the VimA was modelled; this structure has a confidence score of 0.54 (Fig. 1a). Several binding sites were also predicted (Fig. 1b). The predicted structure is similar to the FemA of Staphylococcus aureus (TM-score 0.81) and FemX of Weisella viridiscens (TM-score 0.89). The gene ontology is predicted to include a role in one or more of the following: peptidoglycan biosynthetic process, cellular component biogenesis, cell wall biogenesis, peptidoglycan-based cell wall biogenesis, metabolic process, biosynthetic process, carbohydrate biosynthetic process, primary metabolic process, carbohydrate metabolic process and peptidoglycan metabolic process. SignalP 3.0 and PSORTb predicted no signal peptide in the VimA sequence, suggesting that this protein is not secreted. Phobius scored the likelihood of VimA being a non-cytoplasmic protein at 0.83. Using EMBL protein interaction software v.8.1, VimA was predicted to interact directly with several proteins, including several uncharacterized putative proteins of unknown functional aetiology. The cysteine proteases (PG0506-RgpB) were associated with VimA with a score of 0.808. The bacterioferritin combinatory protein had an interaction score of 0.582, meaning that it is involved in storage of iron in the form of hydrated ferric oxide. Lys1 mediates lysine biosynthesis through the enzyme saccharopine dehydrogenase, while RecA has multiple activities, all related to DNA repair. Position-specific iterative protein classifier and Superfam 3.0 predicted that VimA belongs to the DUF482/CH1444 and/or Acyl-CoA N-acyltransferase superfamily.
Fig. 1.
(a) Model of the VimA protein (confidence score 0.54). The secondary structures are highlighted in red (α-helices) and yellow (β-strands). (b) Predicted binding site residues (green): Leu : 55 Gly : 56 Ser : 57, Phe : 71 Arg : 72 Ala : 73, Val : 77 His : 78 Met : 84, Tyr : 95 Ser : 96 Lys : 97, Tyr : 58 Ser : 59, Arg : 75 Arg : 76, His : 88 Phe : 91, and Gln : 108 Trp : 297. The N- and C-terminal residues are marked by blue and red spheres, respectively.
Surface morphology is affected by the vimA mutation
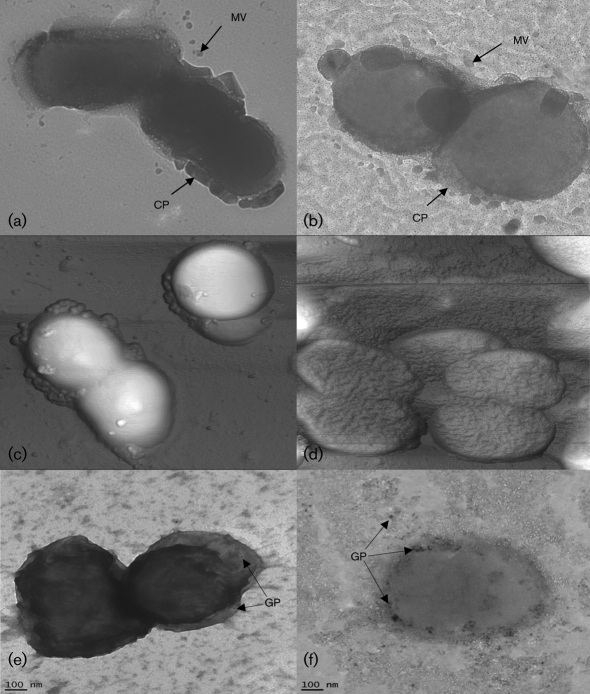
Auto-aggregation in P. gingivalis seems to be correlated with the absence of a capsule and the presence of fimbriae (Davey & Duncan, 2006). AFM and TEM were used to visualize potential differences between the mutant and the wild-type. Micrographs of W83 (Fig. 2a) and FLL92 (Fig. 2b) revealed differences in the capsular structure. The capsule of W83 was solid and well-defined in contrast with the capsule of FLL92, which was less defined and irregular. AFM micrographs of W83 (Fig. 2c) and FLL92 (Fig. 2d) showed that FLL92 had distinct fine structures, resulting in a corrugated appearance, in contrast with the smooth morphology of W83. FLL92 cells also tended to clump together. Immunogold staining of W83 (Fig. 2e) and FLL92 (Fig. 2f) showed adherence of gold particles to abundant fimbrial appendages in FLL92 in contrast with few gold particles adhering to W83.
Fig. 2.
TEM micrographs of W83 (a) and FLL92 (b) showing capsule (CP) with vesicles (MV). AFM micrographs of W83 (c) and FLL92 (d). Immunogold localization of P. gingivalis FimA in W83 (e) and FLL92 (f), visualized by TEM after incubation with anti-FimA serum conjugated to 10 nm gold particles (GP).
Expression of fimbrial genes in the vimA-defective mutant
The autoaggregation observed in FLL92 could have been as a result of changes to the fimbrial protein. It is also possible that VimA may also be involved in the post-translational regulation of fimbrial expression. To confirm the presence of the mRNA transcript for the fimA gene, total RNA was isolated from wild-type and FLL92 strains grown to stationary phase. Using reverse transcriptase with specific oligonucleotide primers for an intragenic region of the fimA gene, a predicted 0.6 kb fragment was amplified for both the wild-type and FLL92 isogenic strain. Immunoblotting of outer membrane and total protein fractions of FLL92 and W83 with the FimA antibody revealed strong immunoreactivity with a 41–43 kDa band in both fractions of FLL92 (Fig. 3). This band, which corresponds with the expected size of FimA, was absent in both W83 fractions.
Fig. 3.
Immunoblot analysis using anti-FimA antibody with outer membrane (OM) and total protein (TP) fractions of P. gingivalis ATCC 33277, W83 and FLL92. A 43 and 41 kDa protein corresponding with the expected size of FimA was observed in 33277 and in the vimA mutant, respectively (arrows).
There is no change in the hydrophobicity or ability to form biofilm in FLL92
Adhesion to hexadecane was used to determine the hydrophobicity of W83 and FLL92; ATCC 33277 served as a control (known to form biofilm and to auto-aggregate). There was no change in the hydrophobicity profile of FLL92 compared with W83. Over half (55 %) of ATCC 33277 cells adhered to hexadecane; this reflects their hydrophilic surface properties. The ability of the vimA-defective mutant FLL92 to form biofilm, as quantified by adherence to the surface of 96-well plates, was unchanged when compared with W83 (data not shown).
Bacterial cell surface/membrane modifications can alter antimicrobial sensitivity
Several antibiotics with varying targets were employed in antibiotic sensitivity testing of FLL92. FLL92 showed increased sensitivity to globomycin (10 μg ml−1 compared with 25 μg ml−1 in W83) and vancomycin (5 μg ml−1 compared with 10 μg ml−1 in W83).
VimA affects outer-membrane proteins glycosylated with specific carbohydrate moieties
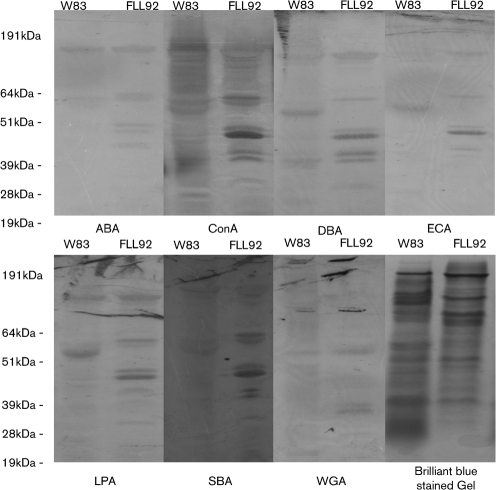
Previous reports from our laboratory have documented the changes in the glycosylation profile of FLL92 compared with wild-type W83 (Vanterpool et al., 2005b). To further confirm the role of VimA in post-translational glycosylation of outer-membrane proteins, lectins were used. Differential lectin-bound protein profiles were observed using ConA, DBA, ECA, LPA and SBA (Fig. 4). No discernible differences were observed using WGA or ABA.
Fig. 4.
Outer-membrane preparations from P. gingivalis W83 and FLL92 were separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked in PBS then incubated with lectin-peroxidase overnight. Peroxidase activity was detected using the DAB peroxidase substrate kit. Differential lectin binding was observed using ABA, ConA, DBA, ECA, LPA, SBA and SGA.
VimA affects the anchorage of several membrane-bound proteins
MS analysis of membrane fractions of W83 and FLL92 identified 20 proteins that were absent from the wild-type membrane fraction but present in the FLL92 fraction (Table 1). While 13 of these proteins are hypothetical, PG0083, PG0581, PG0602, PG0703 and PG1496 are predicted to be involved in transport (www.oralgen.lanl.gov). Nine proteins were identified as missing from the FLL92 fraction compared with the wild-type (Table 2). These included three proteases – RgpA (PG1768), Kgp (PG1605) and carboxypeptidase D (PG0212) – in addition to PG0554 and PG1136, which are predicted to be involved in amino acid biosynthesis (www.oralgen.lanl.gov). PG1857, which is predicted to be involved glycolysis or gluconeogenesis (www.oralgen.lanl.gov), was also missing. Twenty-one membrane proteins were identified as having variable spectral counts (Table 3), corresponding with a fold change of 1.4 or greater. Ten of these proteins showed a positive fold change in the vimA mutant, while 11 showed a negative fold change. The majority of these proteins were predicted to be involved in cell envelope biogenesis or transport (www.oralgen.lanl.gov).
Table 1.
MS analysis of aberrantly expressed proteins in FLL92
The spectral count value (SCV) for W83 was 0 for all proteins.
| Identified protein | GenBank accession no. | Putative role/function | Molecular weight (kDa) | FLL92 SCV |
|---|---|---|---|---|
| Conserved hypothetical protein (possible alkaline secretion protease) | PG0083 | Transport and binding | 56 | 4 |
| Conserved hypothetical protein (possible lipoprotein) | PG0219 | Unknown | 32 | 82 |
| Hypothetical protein | PG0302 | Unknown | 47 | 7 |
| Conserved hypothetical protein | PG0332 | Unknown | 22 | 1 |
| Hypothetical protein | PG0334 | Unknown | 82 | 2 |
| Conserved hypothetical protein | PG0373 | Unknown | 30 | 16 |
| Conserved hypothetical protein | PG0410 | Unknown | 48 | 4 |
| Periplasmic serine protease | PG0535 | Proteolysis | 53 | 1 |
| TonB receptor tlr (C-terminal part) | PG0581 | Transport | 44 | 4 |
| Haem-binding protein/peripheral outer membrane chelatase | PG0602 | Transport and binding | 33 | 4 |
| Probable biopolymer transport (TolQ) protein | PG0703 | Transport | 29 | 3 |
| Hypothetical protein | PG0809 | Unknown | 16 | 2 |
| Conserved hypothetical protein | PG0922 | Unknown | 55 | 7 |
| Probable outer membrane lipoprotein | PG1177 | Cell wall biogenesis | 21 | 18 |
| Conserved hypothetical protein | PG1308 | Unknown | 100 | 24 |
| Conserved hypothetical protein | PG1496 | Transport | 99 | 19 |
| Conserved hypothetical protein | PG1772 | Proteolysis | 98 | 4 |
| Hypothetical protein | PG1790 | Unknown | 10 | 1 |
| Outer-membrane protein (immunogenic 23 kDa lipoprotein) | PG1793 | Cell wall biogenesis | 23 | 44 |
| Probable integral outer-membrane protein | PG1840 | Cell wall biogenesis | 24 | 24 |
Table 2.
MS analysis of missing proteins in FLL92
The spectral count value (SCV) for FLL92 was 0 for all proteins.
| Identified protein | GenBank accession no. | Putative role/function | Molecular weight (kDa) | W83 SCV |
|---|---|---|---|---|
| Carboxypeptidase D/immunoreactive 92 kDa antigen | PG0212 | Proteolysis | 92 | 3 |
| Hypothetical protein | PG0554 | Cell redox homeostasis | 38 | 95 |
| Conserved hypothetical protein | PG0981 | Unknown | 107 | 3 |
| Branched-chain amino acid aminotransferase | PG1136 | Amino acid biosynthesis | 38 | 11 |
| Peptidylarginine deiminase | PG1249 | Arginine and proline metabolism | 62 | 100 |
| Porphypain polyprotein; lys-X proteinase/haemagglutinin | PG1605 | Proteolysis/pathogenesis/DNA replication and repair | 188 | 60 |
| Hypothetical protein | PG1722.1 | Unknown | 24 | 5 |
| Arginine-specific cysteine proteinase; gingipain | PG1768 | Proteolysis/pathogenesis/DNA replication and repair | 186 | 92 |
| Glyceraldehyde 3-phosphate dehydrogenase | PG1857 | Glycolysis/gluconeogenesis | 36 | 60 |
Table 3.
MS analysis of proteins with different spectral count values in W83 and FLL92
| Identified protein | GenBank accession no. | Putative role/involvement | Molecular weight (kDa) | Fold change | W83 SCV | FLL92 SCV |
|---|---|---|---|---|---|---|
| Probable outer membrane lipoprotein | PG0916 | Cell envelope | 63 | 7.1 | 26 | 185 |
| Possible outer membrane-associated protein | PG1893 | Cell envelope | 54 | 4.0 | 40 | 10 |
| TonB-linked outer membrane receptor | PG0601 | Transport and binding | 85 | 3.6 | 5 | 18 |
| Possible outer membrane-associated protein | PG1424 | Cell envelope | 61 | 3.2 | 17 | 54 |
| Outer-membrane protein | PG0626 | Cell envelope | 42 | 2.6 | 156 | 408 |
| Possible lipoprotein | PG1600 | Unknown | 51 | 2.2 | 5 | 11 |
| Outer-membrane protein | PG0627 | Cell envelope | 43 | 2.0 | 129 | 256 |
| Probable integral outer-membrane protein | PG1592 | Cell envelope | 24 | 1.9 | 29 | 56 |
| Probable integral outer-membrane protein | PG1562 | Cell envelope | 32 | 1.6 | 11 | 17 |
| TonB-dependent outer membrane receptor | PG0170 | Transport | 112 | 1.5 | 452 | 674 |
| Receptor antigen B | PG0171 | Cell envelope | 56 | −1.4 | 455 | 316 |
| TonB-linked receptor tlr | PG0582 | Transport | 51 | −1.4 | 14 | 10 |
| TonB-linked outer membrane receptor | PG1752 | Transport | 93 | −1.5 | 15 | 10 |
| Hypothetical protein | PG0197 | Unknown | 33 | −1.8 | 9 | 5 |
| Conserved hypothetical protein; possible TonB-dependent receptor | PG0899 | Transport | 100 | −2.0 | 8 | 4 |
| Outer-membrane protein, TonB-dependent receptor | PG0637 | Transport | 94 | −2.9 | 52 | 18 |
| Phenylalanine/histidine ammonia-lyase | PG0299 | Histidine catabolysis | 55 | −3.3 | 23 | 7 |
| Outer-membrane protein | PG1901 | Cell envelope | 32 | −3.6 | 29 | 8 |
| β-Galactosidase | PG0598 | Carbohydrate metabolism | 127 | −3.8 | 15 | 4 |
| Hypothetical protein | PG1894 | Unknown | 21 | −10.8 | 43 | 4 |
| TonB-linked outer membrane receptor | PG1242 | Transport | 96 | −16.0 | 80 | 5 |
Analysis of missing membrane proteins reveals several common motifs
Using PPSearch (www.ebi.ac.uk/Tools/ppsearch), several common domains were identified in the missing proteins identified by MS analysis (Table 4). In PG1768 (RgpA), PG1605 (Kgp), PG0212 (carboxypeptidase D) and PG0981 (hypothetical), the most abundant motifs were myristoylation sites (13, 11, 49 and 37, respectively), casein kinase II phosphorylation motifs (19, 14, 29 and 35, respectively) and protein kinase C motifs (15, 16, 27 and 20, respectively).
Table 4.
Number of predicted motifs present in missing FLL92 membrane proteins
| PG0212* | PG0554 | PG0981 | PG1136 | PG1249 | PG1605 | PG1722.1 | PG1768 | PG1857 | |
|---|---|---|---|---|---|---|---|---|---|
| MYR† | 13 | 3 | 11 | 6 | 8 | 49 | 7 | 37 | 7 |
| TYR_PHOS‡ | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| CK2_PHOS§ | 19 | 5 | 14 | 4 | 5 | 29 | 3 | 25 | 8 |
| PKC_PHOS|| | 15 | 5 | 16 | 3 | 6 | 27 | 5 | 20 | 5 |
| CAMP_PHOS¶ | 1 | 0 | 1 | 0 | 1 | 2 | 1 | 2 | 1 |
| ASN_GLYC# | 8 | 4 | 0 | 3 | 5 | 15 | 1 | 17 | 3 |
| Amidation** | 1 | 0 | 3 | 0 | 1 | 4 | 0 | 4 | 0 |
| Unique | Zinc_Car†† | None | None | None | None | DNA-Lig‡‡ | None | DNA-Lig | GAPDH§§ |
*Each protein's GenBank accession number is given in the table; see Table 2 for details of the proteins.
†N-Myristoylation site.
‡Tyrosine kinase phosphorylation site.
§Casein kinase II phosphorylation site.
||Protein kinase C phosphorylation site.
¶cAMP- and cGMP-dependent protein kinase phosphorylation site.
#N-Glycosylation site.
**Amidation site.
††Zinc carboxypeptidase.
‡‡ATP-dependent DNA ligase AMP-binding site.
§§GAPDH active site.
Palmitic acid assay shows that VimA is involved in the acylation of proteins
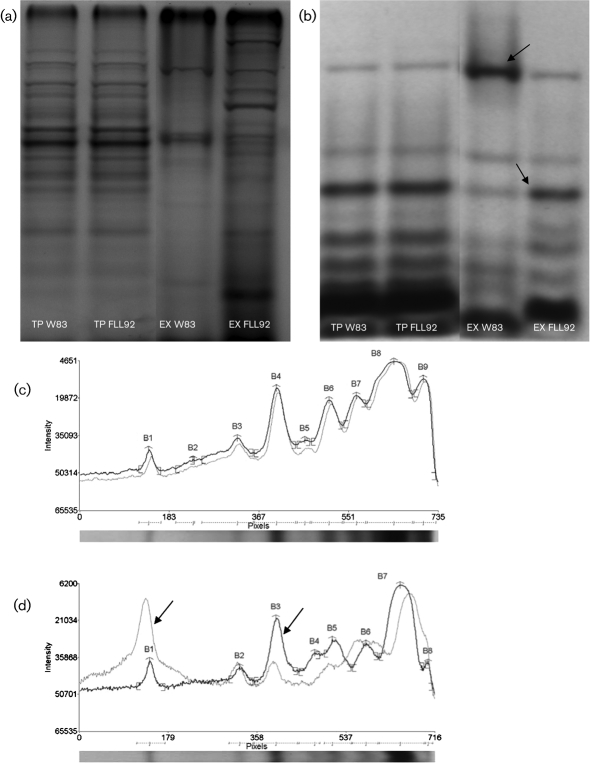
Several outer-membrane proteins are lipid-modified in P. gingivalis (Yoshimura et al., 2009). To determine whether VimA could play a role in lipid modification of proteins, P. gingivalis was grown in the presence of 3H-labelled palmitic acid. No change in the acylation profile was observed in the total protein fractions of the wild-type or FLL92 grown to exponential phase (Fig. 5b). In the extracellular fraction from FLL92 grown to exponential phase (Fig. 5b), a 54 kDa protein was observed in W83 which had three times the intensity of that observed in FLL92; while a 27 kDa protein was observed in FLL92 with twice the intensity of W83 (Fig. 5c).
Fig. 5.
Autoradiography of total protein (TP) and extracellular fractions (EX) of W83 and FLL92 grown overnight in the presence of 3H-labelled palmitic acid. (a, b) Brilliant blue-stained SDS-PAGE gel (a) and 3H-labelled palmitic acid nitrocellulose membrane (b) with W83 and FLL92 TP and EX fractions. (c, d) Densitometric analysis of 3H lipid-labelled proteins of W83 (grey) and FLL92 (black) total protein (c) and extracellular (d) fractions. A 54 kDa protein (B1) was three times as abundant in W83 compared with FLL92 and the 27 kDa protein (B3) was twice as abundant in FLL92 compared with W83 (see arrows).
DISCUSSION
Membrane integrity is critical to the survival, adaptability and pathogenicity of bacteria (Duban et al., 1993; Frirdich & Whitfield, 2005; Zaas et al., 2005). In this study, we examined the role of VimA on membrane proteins and structures, with a view to further elucidate its role in modulating virulence in P. gingivalis. The role of VimA in gingipain maturation, auto-aggregation, haemolysis, haemagglutination and LPS synthesis has been alluded to in previous reports (Vanterpool et al., 2004, 2005a, b, 2006); however, its role in membrane biogenesis has not been explored. The unique phenotype of FLL92 led us to hypothesize that VimA plays a significant role in membrane biogenesis of P. gingivalis and is integral for anchorage and maturation of membrane proteins.
In silico analysis of the VimA protein predicted a structure that showed similarity to the Fem family of proteins. These proteins are found in Gram-positive bacteria and are involved in cell envelope biogenesis, particularly in peptidoglycan formation (Brakstad & Maeland, 1997; Mainardi et al., 2008; Stapleton & Taylor, 2002). Interestingly, this protein is not predicted to be involved in secretion or cytoplasmic localization, suggesting that it is likely a transmembrane or periplasmic protein. Attempts to localize this protein with antibodies raised against the overexpressed protein in Escherichia coli have so far proven unsuccessful. The VimA protein could be classified in the DUF482/CH1444 and/or acyl-CoA N-acyltransferase superfamily. DUF482/CH1444 is part of the PEP-CTERM system that has sortase-like function and is involved in bacterial exopolysaccharide biosynthesis (Haft et al., 2006). Proteins exported in this system have conserved C-terminal sorting signals that are recognized by a sortase in Gram-positive bacteria (Desvaux et al., 2006; Dramsi et al., 2005; Marraffini et al., 2006; Ton-That et al., 2004; Tsukiji & Nagamune, 2009) or an analogous system termed exosortase in Gram-negative bacteria (Salinero et al., 2009). The second predicted family (acyl-CoA N-acyltransferase) is a broad family which includes N-acetyltransferase, N-myristoyl transferase, autoinducer synthase, FemXAB nonribosomal peptidyltransferases, ornithine decarboxylase antizyme-like, AstA-like, DUF1122 and EF1021-like subfamilies. VimA is predicted to belong to the FemXAB nonribosomal peptidyltransferase subfamily. Notably, VimA is predicted to interact with RgpB (score 0.81) in addition to several other proteins; this is consistent with previous reports which demonstrate that VimA is able to interact with RgpB. Interestingly, several proteins, including those coded for upstream and downstream of vimA, are predicted to interact with VimA, and it is noteworthy that several proteins, including PG1605 (Kgp), PG0010 (β-lactamase), PG1101 (alanyl-tRNA synthase), PG0535 (HtrA), PG1833 (RegT), PG0324 (putative sialidase) and PG1768 (RgpA), that have been previously shown to interact (Vanterpool et al., 2006) were not predicted for interaction with Vim A. These data suggest that VimA is likely to be a multifunctional protein, possibly forming a complex with those proteins encoded at the recA locus, and may modulate several virulence factors in addition to being involved in the biosynthesis of membrane proteins and structures.
We have demonstrated in previous studies that compared with the wild-type, FLL92 autoaggregates (Olango et al., 2003). Several reports have suggested that the expression of fimbriae and the absence of a capsule may contribute to auto-aggregation, which in turn increases the organism's ability to form biofilm (Davey & Duncan, 2006). Visible differences were observed between FLL92 and W83. These differences related to capsular formation and surface morphology. The capsule of FLL92 was irregular and fuzzy in contrast with the solid defined capsule of W83. FLL92 also had a corrugated surface morphology compared with the smooth morphology present in W83. Abundant immunogold particles were observed in the appendage-like structures of FLL92; however, few gold particles were incorporated in the few surface-related appendages of W83. Interestingly, RT-PCR demonstrated that the fimA gene was similarly expressed in both the wild-type and vimA-defective isogenic mutant.
The presence of major fimbriae in wild-type W83 was previously disputed (Nishiyama et al., 2007), as screening by immunological means such as bacterial agglutination, ouchterlony immunodiffusion and Western blotting were all negative; however, electron microscopy has been used successfully to establish that W83 is poorly fimbriated (Handley & Tipler, 1986; Suzuki et al., 1988). This is in contrast with highly fimbriated strains such as 33277 and 381 which show positive immunological fimbrial results and positive visualization by electron microscopy. In P. gingivalis, there are six classifications of the fimA gene (I, Ib, II, III, IV and V) (Amano et al., 2004); the large majority of patients carry either type II or IV fimA. Interestingly, these genotypic differences do not correlate with morphological differences in fimbriae (Amano et al., 2000). W83 is type IV, whereas 33277 and 381 are type I (Nishiyama et al., 2007). The relationship between capsular and fimbrial synthesis is at present unknown in P. gingivalis; however, as has been demonstrated in Klebsiella pneumoniae (Matatov et al., 1999; Sahly et al., 2000), fimbrial function can be impeded by expression of the polysaccharide capsule and can be restored by inhibiting capsular synthesis. Capsular formation may also impede assembly of preformed fimbrial subunits on the bacterial surface (Sahly et al., 2000). This effect in K. pneumoniae is due to direct physical interaction as opposed to transcriptional or translational changes, and is in keeping with our own observations which show that the fimA transcript is made in the wild-type and the mutant. Therefore, in light of the effect of the vimA mutation on capsular synthesis, fimbrial function and synthesis may no longer be impeded, resulting in the assembly of fimbriae on the surface. The presence of the fimA message coupled with undetected FimA via immunoblotting, may also indicate a role for vimA in protein synthesis and turnover. Interestingly, these alterations in membrane structure did not affect the organism's ability to form biofilm. Taken together, these findings indicate a critical role for VimA in capsular biogenesis and probably an indirect role in fimbrial synthesis. Further studies are actively being pursued in our laboratory to elucidate the role of VimA in synthesis and regulation of the capsule and fimbriae.
The corrugated surface morphology of FLL92 coupled with previous reports which demonstrated that although there are more proteins present in the membrane fraction, other proteins such as RgpA and Kgp are missing (Vanterpool et al., 2005b, 2006), could suggest that this morphology was likely due to the aberrant anchorage and/or expression of proteins on the cell membrane. Our findings revealed 20 proteins that were aberrantly anchored to the cell wall of FLL92, 9 proteins that were missing and 21 proteins which had variable spectral count values. These normalized spectral count values correlate to the relative abundance of these proteins. Of the nine missing proteins; three are involved in proteolysis; these include carboxypeptidase D (PG0212), RgpA (1768) and Kgp (1605). GAPDH (PG1857) and peptidyl–arginine deiminase (PG1249) were also missing.
RgpA and Kgp proteinases and adhesins are C-terminally processed by carboxypeptidase D, otherwise known as carboxypeptidase CPG70. This protein shares C-terminal sequence similarity to cysteine proteinases, indicating a common mechanism for cell surface attachment and secretion (Chen et al., 2002; Veith et al., 2004). The absence of this carboxypeptidase on the surface of FLL92 likely explains why RgpA and Kgp are incorrectly modified and inactive, existing in the extracellular milieu, but not anchored to the surface. This is in contrast with the surface-bound yet inactive RgpB. GAPDH is a metabolic enzyme of the glycolytic pathway which phosphorylates glyceraldehyde-3-phosphate to generate 1,3 biphosphoglycerate (Lama et al., 2009). GAPDH has also been shown to exist on the surface of bacterial pathogens, functioning as a novel virulence factor in several bacterial species by binding host proteins (Maeda et al., 2004; Pancholi & Fischetti, 1992). In Trichomonas vaginalis, this enzyme binds fibronectin, plasminogen and collagen (Lama et al., 2009), while in enterohaemorrhagic E. coli and enteropathogenic E. coli, it binds human plasminogen and fibrinogen (Egea et al., 2007). In Candida albicans, this surface-associated enzyme binds to laminin and fibronectin (Gozalbo et al., 1998). Surface-bound GADPH of Streptococcus oralis has been shown to play a role in P. gingivalis colonization. FimA of P. gingivalis is able to bind to GADPH present on the streptococcal surface (Nagata et al., 2009); this interaction is further used to bind to human oral epithelial cells (Kinoshita et al., 2008). The function of surface-bound GADPH in P. gingivalis has not been elucidated; however, we speculate that it is probably serving to bind host extracellular matrix proteins present within the periodontal pocket for proteolytic degradation by the gingipains. It is also possible that this protein may itself contain a binding receptor for other oral bacteria so as to allow for biofilm formation and colonization of the oral cavity. Peptidyl arginine deiminase (PAD), which catalyses the deimination of the guanidino group from carboxy-terminal Arg residues to produce ammonia, is released by P. gingivalis, which leads to the progression and development of adult-onset periodontitis. By producing ammonia, PAD provides a protective effect to the bacterium during the acid cleansing cycles of the mouth (Shirai et al., 2001). The activity of PAD among cells, supernatant and vesicles is growth dependent. It has also been suggested that the interaction between PAD and Arg-X may arise from Arg-X cleaving anti-adhesive humoral defence peptides containing internal arginyl residues and other arginine carboxyl terminal defence peptides, which are then inactivated by PAD (McGraw et al., 1999; Rodriguez et al., 2009). It is likely that anchorage of PAD may be dependent on the carboxypeptidase D processing. The reported interaction between Arg-X and PAD leads us to hypothesize that the enzymic capabilities of PAD may be compromised as Arg-X is inactive, thus reducing its efficacy. Further studies are needed to clarify this hypothesis.
When missing proteins were examined for protein motifs using PPsearch, a protein motif identification software, myristoylation and phosphorylation sites were in great abundance in all of the missing proteins. Of the 20 proteins, 13 were hypothetical proteins, 9 of which had an unknown function. Further, PG0535 (serine periplasmic protein), which has been shown to interact with VimA, is aberrantly expressed on the membrane of FLL92. This study also confirmed the missing membrane-associated RgpA and Kgp in FLL92, as previously reported (Vanterpool et al., 2005b, 2006). These findings potentially explain the auto-aggregation that is observed in FLL92 and support a role for VimA in anchorage and maturation of membrane proteins, possibly through myristoylation or phosphorylation. The specific interaction between VimA and missing or aberrantly expressed proteins is being investigated.
In the vimA-defective mutant, there is aberrant protein glycosylation (Vanterpool et al., 2005b). It is not known, however, whether this aberrant glycosylation is specific to certain carbohydrate moieties. To clarify the effect of the vimA mutation on glycosylation of outer-membrane proteins, lectins were used. Outer-membrane proteins glycosylated with galactose (β 1,3) N-acetylgalactosamine, N-acetyl-α-d-galactosamine, galactose (β 1,4) N-acetylglucosamine, N-acetyl-d-galactosamine and sialic acid (N-acetyl neuramic acid) were affected by the vimA mutation. We are still actively exploring whether this is due to the glycosylation of aberrantly expressed proteins (i.e. proteins that are not present on the wild-type membrane but are present on the membrane of FLL92) or proteins that are incorrectly processed, as is the case with RgpB. These findings suggest that VimA plays a role in the correct glycosylation of several membrane proteins.
Since several outer-membrane proteins are lipid-modified in P. gingivalis (Slakeski et al., 2002), and in keeping with the likely role of the VimA protein as a possible acyl transferase, we incubated W83 and FLL92 with 3H-labelled palmitic acid (lipid donor). The expression profile of lipid-modified proteins in the wild-type and mutant was unchanged. This suggests either that VimA is not involved in the acylation of outer-membrane proteins or that there is a redundant mechanism of protein acylation. To this end, PG1355 (putative acyltransferase) was upregulated (3.5-fold change) in FLL92 (unpublished microarray data) compared with the mutant. Interestingly, densitometric analysis of the extracellular fractions of W83 and FLL92 for lipid-modified proteins revealed a 54 kDa protein that was three times more abundant in W83 than was observed in FLL92. We also observed a 27 kDa protein that was twice as abundant in FLL92 as in W83. The above findings may indicate a likely role for VimA in the proper acylation of a 54 kDa extracellular protein; however, we have not ruled out the possibility that this protein is a dimer of the 27 kDa protein observed in the extracellular fraction of FLL92. Several reports have demonstrated that acylation is important for protein folding and protein–protein interaction, as well as ligand-induced conformational changes (Athavankar & Peterson, 2003; Herlax et al., 2009; Olsen & Kaarsholm, 2000). It is possible that correct acylation of the 27 kDa extracellular protein leads to dimerization. In the vimA mutant, aberrant acylation of the 27 kDa extracellular protein may occur, resulting in higher levels of monomeric protein form. Further studies in our laboratory are seeking to clarify this position.
The expression of additional proteins attributed to membrane biosynthesis and transport in the VimA mutant suggest that several post-translational modification pathways may be affected by this protein. These modifications could include myristoylation and glycosylation. Co-translational modifications of proteins could be attributed to their involvement in signal transduction pathways as molecular switches associated with glycine-rich protein motifs. Myristoylation of proteins is reported to be involved in the type III secretory system in prokaryotes (Nimchuk et al., 2000)
The missing and aberrantly expressed proteins in FLL92 suggest that VimA could have sorting/transport functions. VimA could also function as an acyl-CoA N-acyltransferase enzyme involved in protein modification or anchorage of membrane proteins. In conclusion, our results highlight a putative central role for VimA in protein modification and transport, possibly triggering cascade reactions that may modulate the virulence potential of P. gingivalis. The putative multiple functions of VimA need further clarification.
Acknowledgments
This work was supported by Loma Linda University and Public Health Grant DE13664 and DE019730 from NIDCR (to H. M. F.). We thank Professor Masatoshi Inukai for the globomycin antibiotic, Dr Hua Xie for the FimA antibody and Dr Danielo Boskovic for helpful comments concerning the acylation studies.
Abbreviations
ABA, Agaricus bisporus lectin
AFM, atomic force microscopy
BHI, brain–heart infusion
ConA, Concanavalin A
DBA, Dolichos biflorus lectin
ECA, Erythrina cristagalli lectin
LPA, Limulus polyphemus lectin
LPS, lipopolysaccharide
MS, mass spectrometry
PAD, peptidyl arginine deiminase
SBA, Glycine max lectin
TEM, transmission electron microscopy
TLCK, N-α-p-tosyl-l-lysine chloromethyl ketone
TSBKH, trypticase soy broth containing menadione and haemin
WTA, Triticum vulgaris lectin
References
- Abaibou, H., Chen, Z., Olango, G. J., Liu, Y., Edwards, J. & Fletcher, H. M. (2001). vimA gene downstream of recA is involved in virulence modulation in Porphyromonas gingivalis W83. Infect Immun 69, 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, A., Nakagawa, I., Kataoka, K., Morisaki, I. & Hamada, S. (1999). Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol 37, 1426–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano, A., Kuboniwa, M., Nakagawa, I., Akiyama, S., Morisaki, I. & Hamada, S. (2000). Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res 79, 1664–1668. [DOI] [PubMed] [Google Scholar]
- Amano, A., Nakagawa, I., Okahashi, N. & Hamada, N. (2004). Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res 39, 136–142. [DOI] [PubMed] [Google Scholar]
- Athavankar, S. & Peterson, B. R. (2003). Control of gene expression with small molecules: biotin-mediated acylation of targeted lysine residues in recombinant yeast. Chem Biol 10, 1245–1253. [DOI] [PubMed] [Google Scholar]
- Bogen, G. & Slots, J. (1999). Black-pigmented anaerobic rods in closed periapical lesions. Int Endod J 32, 204–210. [DOI] [PubMed] [Google Scholar]
- Brakstad, O. G. & Maeland, J. A. (1997). Mechanisms of methicillin resistance in staphylococci. APMIS 105, 264–276. [DOI] [PubMed] [Google Scholar]
- Chen, Y. Y., Cross, K. J., Paolini, R. A., Fielding, J. E., Slakeski, N. & Reynolds, E. C. (2002). CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J Biol Chem 277, 23433–23440. [DOI] [PubMed] [Google Scholar]
- Cutler, C. W., Kalmar, J. R. & Genco, C. A. (1995). Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol 3, 45–51. [DOI] [PubMed] [Google Scholar]
- Dave, S. & Van, D. T. (2008). The link between periodontal disease and cardiovascular disease is probably inflammation. Oral Dis 14, 95–101. [DOI] [PubMed] [Google Scholar]
- Davey, M. E. & Duncan, M. J. (2006). Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol 188, 5510–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmer, R. T. & Desvarieux, M. (2006). Periodontal infections and cardiovascular disease: the heart of the matter. J Am Dent Assoc 137 (Suppl.), 14S–20S. [DOI] [PubMed] [Google Scholar]
- Desvaux, M., Dumas, E., Chafsey, I. & Hebraud, M. (2006). Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol Lett 256, 1–15. [DOI] [PubMed] [Google Scholar]
- Dramsi, S., Trieu-Cuot, P. & Bierne, H. (2005). Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol 156, 289–297. [DOI] [PubMed] [Google Scholar]
- Duban, M. E., Lee, K. & Lynn, D. G. (1993). Strategies in pathogenesis: mechanistic specificity in the detection of generic signals. Mol Microbiol 7, 637–645. [DOI] [PubMed] [Google Scholar]
- Egea, L., Aguilera, L., Gimenez, R., Sorolla, M. A., Aguilar, J., Badia, J. & Baldoma, L. (2007). Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int J Biochem Cell Biol 39, 1190–1203. [DOI] [PubMed] [Google Scholar]
- Ford, P. J., Yamazaki, K. & Seymour, G. J. (2007). Cardiovascular and oral disease interactions: what is the evidence? Prim Dent Care 14, 59–66. [DOI] [PubMed] [Google Scholar]
- Frirdich, E. & Whitfield, C. (2005). Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J Endotoxin Res 11, 133–144. [DOI] [PubMed] [Google Scholar]
- Gardy, J. L., Laird, M. R., Chen, F., Rey, S., Walsh, C. J., Ester, M. & Brinkman, F. S. (2005). PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21, 617–623. [DOI] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R. & Chothia, C. (2001). Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313, 903–919. [DOI] [PubMed] [Google Scholar]
- Gozalbo, D., Gil-Navarro, I., Azorin, I., Renau-Piqueras, J., Martinez, J. P. & Gil, M. L. (1998). The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun 66, 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet, M., Morrier, J. J., Benay, G. & Barsotti, O. (2000). Effect of hydrophobicity on in vitro streptococcal adhesion to dental alloys. J Mater Sci Mater Med 11, 637–642. [DOI] [PubMed] [Google Scholar]
- Haft, D. H., Paulsen, I. T., Ward, N. & Selengut, J. D. (2006). Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley, P. S. & Tipler, L. S. (1986). An electron microscope survey of the surface structures and hydrophobicity of oral and non-oral species of the bacterial genus Bacteroides. Arch Oral Biol 31, 325–335. [DOI] [PubMed] [Google Scholar]
- Harris, J. R. (1991). Electron Microscopy in Biology: a Practical Approach. Oxford, UK: IRL Press.
- Henry, L. G., Sandberg, L., Zhang, K. & Fletcher, H. M. (2008). DNA repair of 8-oxo-7,8-dihydroguanine lesions in Porphyromonas gingivalis. J Bacteriol 190, 7985–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlax, V., Mate, S., Rimoldi, O. & Bakas, L. (2009). Relevance of fatty acid covalently bound to Escherichia coli α-hemolysin and membrane microdomains in the oligomerization process. J Biol Chem 284, 25199–25210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, T. (2003). The role of gingipains in the pathogenesis of periodontal disease. J Periodontol 74, 111–118. [DOI] [PubMed] [Google Scholar]
- Kall, L., Krogh, A. & Sonnhammer, E. L. (2007). Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucleic Acids Res 35, W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T., Tsuda, T., Omori, H., Kato, T., Yoshimori, T. & Amano, A. (2007). Maturation of fimbria precursor protein by exogenous gingipains in Porphyromonas gingivalis gingipain-null mutant. FEMS Microbiol Lett 273, 96–102. [DOI] [PubMed] [Google Scholar]
- Kinoshita, H., Wakahara, N., Watanabe, M., Kawasaki, T., Matsuo, H., Kawai, Y., Kitazawa, H., Ohnuma, S., Miura, K. & other authors (2008). Cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Lactobacillus plantarum LA 318 recognizes human A and B blood group antigens. Res Microbiol 159, 685–691. [DOI] [PubMed] [Google Scholar]
- Kumada, H., Haishima, Y., Umemoto, T. & Tanamoto, K. (1995). Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J Bacteriol 177, 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu, H. K. (1998). Proteases of Porphyromonas gingivalis: what don't they do? Oral Microbiol Immunol 13, 263–270. [DOI] [PubMed] [Google Scholar]
- Lama, A., Kucknoor, A., Mundodi, V. & Alderete, J. F. (2009). Glyceraldehyde-3-phosphate dehydrogenase is a surface-associated, fibronectin-binding protein of Trichomonas vaginalis. Infect Immun 77, 2703–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont, R. J. & Jenkinson, H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62, 1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., Wu, J. & Xie, H. (2006). Porphyromonas gingivalis minor fimbriae are required for cell–cell interactions. Infect Immun 74, 6011–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton, D., Allan, E., Karlyshev, A. V., Cronshaw, A. D. & Wren, B. W. (2002). Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol Microbiol 43, 497–508. [DOI] [PubMed] [Google Scholar]
- Maeda, K., Nagata, H., Kuboniwa, M., Kataoka, K., Nishida, N., Tanaka, M. & Shizukuishi, S. (2004). Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect Immun 72, 5475–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainardi, J. L., Villet, R., Bugg, T. D., Mayer, C. & Arthur, M. (2008). Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol Rev 32, 386–408. [DOI] [PubMed] [Google Scholar]
- Marraffini, L. A., Dedent, A. C. & Schneewind, O. (2006). Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol Mol Biol Rev 70, 192–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatov, R., Goldhar, J., Skutelsky, E., Sechter, I., Perry, R., Podschun, R., Sahly, H., Thankavel, K., Abraham, S. N. & other authors (1999). Inability of encapsulated Klebsiella pneumoniae to assemble functional type 1 fimbriae on their surface. FEMS Microbiol Lett 179, 123–130. [DOI] [PubMed] [Google Scholar]
- McAlister, A. D., Sroka, A., Fitzpatrick, R. E., Quinsey, N. S., Travis, J., Potempa, J. & Pike, R. N. (2009). Gingipain enzymes from Porphyromonas gingivalis preferentially bind immobilized extracellular proteins: a mechanism favouring colonization? J Periodontal Res 44, 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw, W. T., Potempa, J., Farley, D. & Travis, J. (1999). Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun 67, 3248–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk-Pawlinska, J., Kordula, T., Pavloff, N., Pemberton, P. A., Chen, W. C., Travis, J. & Potempa, J. (1998). Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol Chem 379, 205–211. [DOI] [PubMed] [Google Scholar]
- Murakami, Y., Masuda, T., Imai, M., Iwami, J., Nakamura, H., Noguchi, T. & Yoshimura, F. (2004). Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol Immunol 48, 561–569. [DOI] [PubMed] [Google Scholar]
- Nagata, H., Iwasaki, M., Maeda, K., Kuboniwa, M., Hashino, E., Toe, M., Minamino, N., Kuwahara, H. & Shizukuishi, S. (2009). Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun 77, 5130–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, K., Nemoto, H., Nomura, R., Inaba, H., Yoshioka, H., Taniguchi, K., Amano, A. & Ooshima, T. (2009). Detection of oral bacteria in cardiovascular specimens. Oral Microbiol Immunol 24, 64–68. [DOI] [PubMed] [Google Scholar]
- Nakayama, K., Yoshimura, F., Kadowaki, T. & Yamamoto, K. (1996). Involvement of arginine-specific cysteine proteinase (Arg–gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol 178, 2818–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, K. A., Zylicz, J., Szczesny, P., Sroka, A., Hunter, N. & Potempa, J. (2009). Verification of a topology model of PorT as an integral outer-membrane protein in Porphyromonas gingivalis. Microbiology 155, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Marois, E., Kjemtrup, S., Leister, R. T., Katagiri, F. & Dangl, J. L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101, 353–363. [DOI] [PubMed] [Google Scholar]
- Nishiyama, S., Murakami, Y., Nagata, H., Shizukuishi, S., Kawagishi, I. & Yoshimura, F. (2007). Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153, 1916–1925. [DOI] [PubMed] [Google Scholar]
- Olango, G. J., Roy, F., Sheets, S. M., Young, M. K. & Fletcher, H. M. (2003). Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92. Infect Immun 71, 3740–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, H. B. & Kaarsholm, N. C. (2000). Structural effects of protein lipidation as revealed by LysB29-myristoyl, des(B30) insulin. Biochemistry 39, 11893–11900. [DOI] [PubMed] [Google Scholar]
- Pancholi, V. & Fischetti, V. A. (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med 176, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov, N., Rangarajan, M., Hashim, A., Gallagher, A., Aduse-Opoku, J., Slaney, J. M., Hounsell, E. & Curtis, M. A. (2005). Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol Microbiol 58, 847–863. [DOI] [PubMed] [Google Scholar]
- Pavloff, N., Potempa, J., Pike, R. N., Prochazka, V., Kiefer, M. C., Travis, J. & Barr, P. J. (1995). Molecular cloning and structural characterization of the Arg–gingipain proteinase of Porphyromonas gingivalis. Biosynthesis as a proteinase–adhesin polyprotein. J Biol Chem 270, 1007–1010. [DOI] [PubMed] [Google Scholar]
- Pavloff, N., Pemberton, P. A., Potempa, J., Chen, W. C., Pike, R. N., Prochazka, V., Kiefer, M. C., Travis, J. & Barr, P. J. (1997). Molecular cloning and characterization of Porphyromonas gingivalis lysine-specific gingipain. A new member of an emerging family of pathogenic bacterial cysteine proteinases. J Biol Chem 272, 1595–1600. [DOI] [PubMed] [Google Scholar]
- Pearson, W. R. & Lipman, D. J. (1988). Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike, R. N., Potempa, J., McGraw, W., Coetzer, T. H. & Travis, J. (1996). Characterization of the binding activities of proteinase–adhesin complexes from Porphyromonas gingivalis. J Bacteriol 178, 2876–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa, J., Pike, R. & Travis, J. (1995). The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg–gingipain or Lys–gingipain. Infect Immun 63, 1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto, F., Suzuki, Y., Takeyasu, K. & Alonso, J. C. (2009). Single-molecule analysis of protein–DNA complexes formed during partition of newly replicated plasmid molecules in Streptococcus pyogenes. J Biol Chem 284, 30298–30306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan, M., Aduse-Opoku, J., Paramonov, N., Hashim, A., Bostanci, N., Fraser, O. P., Tarelli, E. & Curtis, M. A. (2008). Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J Bacteriol 190, 2920–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, S. B., Stitt, B. L. & Ash, D. E. (2009). Expression of peptidylarginine deiminase from Porphyromonas gingivalis in Escherichia coli: enzyme purification and characterization. Arch Biochem Biophys 488, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahly, H., Podschun, R., Oelschlaeger, T. A., Greiwe, M., Parolis, H., Hasty, D., Kekow, J., Ullmann, U., Ofek, I. & other authors (2000). Capsule impedes adhesion to and invasion of epithelial cells by Klebsiella pneumoniae. Infect Immun 68, 6744–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinero, K. K., Keller, K., Feil, W. S., Feil, H., Trong, S., Di, B. G. & Lapidus, A. (2009). Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genomics 10, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., Sakai, E., Veith, P. D., Shoji, M., Kikuchi, Y., Yukitake, H., Ohara, N., Naito, M., Okamoto, K. & other authors (2005). Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J Biol Chem 280, 8668–8677. [DOI] [PubMed] [Google Scholar]
- Shirai, H., Blundell, T. L. & Mizuguchi, K. (2001). A novel superfamily of enzymes that catalyze the modification of guanidino groups. Trends Biochem Sci 26, 465–468. [DOI] [PubMed] [Google Scholar]
- Shoji, M., Ratnayake, D. B., Shi, Y., Kadowaki, T., Yamamoto, K., Yoshimura, F., Akamine, A., Curtis, M. A. & Nakayama, K. (2002). Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148, 1183–1191. [DOI] [PubMed] [Google Scholar]
- Shoji, M., Naito, M., Yukitake, H., Sato, K., Sakai, E., Ohara, N. & Nakayama, K. (2004). The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol Microbiol 52, 1513–1525. [DOI] [PubMed] [Google Scholar]
- Slakeski, N., Margetts, M., Moore, C., Czajkowski, L., Barr, I. G. & Reynolds, E. C. (2002). Characterization and expression of a novel Porphyromonas gingivalis outer membrane protein, Omp28. Oral Microbiol Immunol 17, 150–156. [DOI] [PubMed] [Google Scholar]
- Stapleton, P. D. & Taylor, P. W. (2002). Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog 85, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulou, P. G., Galicia, J. C., Benakanakere, M. R., Garcia, C. A., Potempa, J. & Kinane, D. F. (2009). Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol 9, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yoshimura, F., Takahashi, K., Tani, H. & Suzuki, T. (1988). Detection of fimbriae and fimbrial antigens on the oral anaerobe Bacteroides gingivalis by negative staining and serological methods. J Gen Microbiol 134, 2713–2720. [DOI] [PubMed] [Google Scholar]
- Ton-That, H., Marraffini, L. A. & Schneewind, O. (2004). Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta 1694, 269–278. [DOI] [PubMed] [Google Scholar]
- Tsukiji, S. & Nagamune, T. (2009). Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. ChemBioChem 10, 787–798. [DOI] [PubMed] [Google Scholar]
- Vanterpool, E., Roy, F. & Fletcher, H. M. (2004). The vimE gene downstream of vimA is independently expressed and is involved in modulating proteolytic activity in Porphyromonas gingivalis W83. Infect Immun 72, 5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool, E., Roy, F. & Fletcher, H. M. (2005a). Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect Immun 73, 3971–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool, E., Roy, F., Sandberg, L. & Fletcher, H. M. (2005b). Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect Immun 73, 1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool, E., Roy, F., Zhan, W., Sheets, S. M., Sangberg, L. & Fletcher, H. M. (2006). VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology 152, 3383–3389. [DOI] [PubMed] [Google Scholar]
- Veith, P. D., Chen, Y. Y. & Reynolds, E. C. (2004). Porphyromonas gingivalis RgpA and Kgp proteinases and adhesins are C terminally processed by the carboxypeptidase CPG70. Infect Immun 72, 3655–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas, D. & Ke, N. (2001). Detection and quantitation of radiolabeled proteins and DNA in gels and blots. Curr Protoc Mol Biol Appendix 3, Appendix 3A. [DOI] [PubMed] [Google Scholar]
- Wang, M., Liang, S., Hosur, K. B., Domon, H., Yoshimura, F., Amano, A. & Hajishengallis, G. (2009). Differential virulence and innate immune interactions of type I and II fimbrial genotypes of Porphyromonas gingivalis. Oral Microbiol Immunol 24, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, T. L., Ellen, R. P., Lacroix, J. M., Sinnadurai, S. & Mittelman, M. W. (1997). Effects of metronidazole on Porphyromonas gingivalis biofilms. J Periodontal Res 32, 473–477. [DOI] [PubMed] [Google Scholar]
- Yoshimura, F., Murakami, Y., Nishikawa, K., Hasegawa, Y. & Kawaminami, S. (2009). Surface components of Porphyromonas gingivalis. J Periodontal Res 44, 1–12. [DOI] [PubMed] [Google Scholar]
- Zaas, D. W., Duncan, M., Rae, W. J. & Abraham, S. N. (2005). The role of lipid rafts in the pathogenesis of bacterial infections. Biochim Biophys Acta 1746, 305–313. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. (2009). I-TASSER: Fully automated protein structure prediction in CASP8. Proteins 77 (Suppl. 9), 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]