Abstract
During cycling between the tick vector and a mammal, the Lyme disease spirochaete Borrelia burgdorferi must coordinate expression of outer-surface proteins (Osps) A and B to quickly respond to environmental changes. The pathogen abundantly produces OspA/B in the tick, but represses their expression during mammalian infection. This paper reports a regulatory structure, consisting of two sequences flanking the ospAB promoter, that is required for enhancing ospA expression in B. burgdorferi grown in vitro, but repressing its expression during murine infection. Deletion or replacement of either the upstream or downstream sequence of the ospAB promoter caused a significant decrease in ospA expression in vitro, but a dramatic increase during murine infection. Fusion of either sequence with the flaB reporter promoter led to increased expression of an ospA reporter gene in vitro, but a decrease in the murine host. Furthermore, simultaneous fusion of both sequences with the reporter promoter showed a synergistic effect in enhancing expression of the ospA reporter in vitro, but repressing its expression during murine infection. Taken together, the results demonstrate that the regulatory structure functions oppositely in the two different environments and potentially provides B. burgdorferi with a molecular mechanism to quickly adapt to the distinct environments during its enzootic life cycle.
INTRODUCTION
Outer-surface proteins (Osps) A and B, encoded by a two-gene operon (Howe et al., 1986), are the surface antigens most abundantly produced by the Lyme disease spirochaete Borrelia burgdorferi in engorged and unfed Ixodes ticks (de Silva et al., 1996; Ohnishi et al., 2001; Schwan et al., 1995; Schwan & Piesman, 2000). In response to a fresh blood meal, B. burgdorferi downregulates OspA/B and upregulates OspC and other Osps, a process that prepares the spirochaete for infection of a mammal (Fingerle et al., 2007; Grimm et al., 2004; Pal et al., 2004b; Stewart et al., 2006). Repressing ospAB expression during mammalian infection is critical for B. burgdorferi to evade the immune system, cause persistent infection and maintain the enzootic cycle, as OspA and OspB, even expressed at a low level, may ultimately induce a strong specific humoral response owing to their high immunogenicity. The specific response can impose tremendous pressure on the pathogen or clear the infection (Strother et al., 2007; Xu et al., 2008a). Even though the anti-OspA/B response may not effectively target spirochaetes with low OspA/B production in mammalian tissues, once acquired by the tick vector, the pathogen dramatically upregulates OspA/B and becomes extremely vulnerable to anti-OspA/B antibodies in a blood meal (de Silva et al., 1997; Tsao et al., 2001, 2004), potentially leading to the eradication of the organism and consequently to discontinuation of the enzootic cycle.
Expression of the ospAB operon is driven by a σ70-dependent promoter (Sohaskey et al., 1999); thus, B. burgdorferi cannot shut off this major sigma factor in order to achieve the downregulation of OspA/B. Sohaskey et al. (1999) showed that deletion of the T-rich region immediately upstream of the ospAB promoter resulted in greatly reduced expression of a chloramphenicol acetyltransferase reporter gene in B. burgdorferi grown in vitro. Caimano et al. (2005) showed that the downregulation of OspA requires RpoS production and proposed a model for RpoS-dependent in vivo repression of ospA, which involves an unknown repressor protein or an unknown cofactor that can bind RpoS and an unidentified sequence within the ospAB promoter (Caimano et al., 2005). Later, the group further suggested that the T-rich region immediately upstream of the ospAB promoter may serve in this capacity (Caimano et al., 2007).
In spite of this progress, it remains largely unknown how B. burgdorferi can so effectively down- and upregulate ospAB during its enzootic cycle. The present study focused on sequences both upstream and downstream of the ospAB promoter, and successfully identified two cis elements, which are involved in ospAB regulation by B. burgdorferi grown in vitro as well as during murine infection.
METHODS
Previously generated strains and constructs used in the current study.
Clones 13A and 13A/E22/C and the ospA mutant, ΔospA, were generated previously (Xu et al., 2007a, b, 2008b). The constructs pBBE22-CpospA and pBBE22-ospA′ were constructed in an earlier study (Xu et al., 2008b). The shuttle vector pBBE22 and the B. burgdorferi B31 clone 5A11 were kindly provided by S. Norris (Purser & Norris, 2000; Purser et al., 2003). The features of these clones and constructs are summarized in Supplementary Table S1, available with the online version of this paper.
Creation of the constructs pBBE22-ospA1 to pBBE22-ospA7.
The seven constructs were generated as illustrated in Fig. 1. Briefly, three amplicons, ospA1 (1085 bp), ospA2 (958 bp) and ospA3 (925 bp), were generated by PCR using forward primers P1F, P2F and P3F (Supplementary Table S2), respectively, and a common reverse primer, P1R, with borrelial DNA as template. The resulting products were digested with the restriction enzymes BamHI and XbaI, purified, and cloned into the shuttle vector pBBE22, digested with the same enzymes. The inserts and their flanking regions were sequenced to ensure they were arranged as designed. The three constructs were designated pBBE22-ospA1, pBBE22-ospA2 and pBBE22-ospA3.
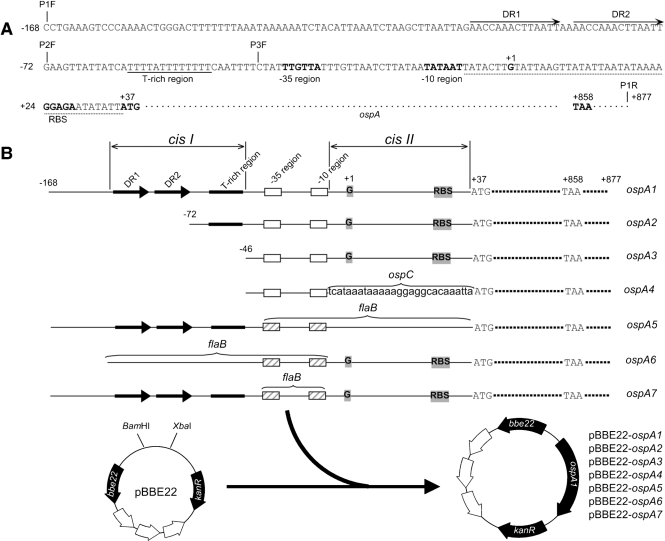
Fig. 1.
Generation of constructs. (A) Potential regulatory sequences upstream of the coding region of the ospAB operon, including a pair of direct repeats, DR1 and DR2 (labelled with long arrows), a T-rich region (underlined with a solid line), and a sequence between the −10 region and the start codon ATG (underlined with a dotted line). The −35 and −10 regions, putative ribosome-binding site (RBS), start codon ATG, and stop codon TAA (all in bold type) are indicated. The +1 marks the previously identified transcriptional initiation site (Jonsson et al., 1992). The amplification start sites of primers P1F, P2F, P3F and P1R, designed for amplifying the fragments ospA1, ospA2 and ospA3, are marked with vertical lines. (B) Generation of constructs pBBE22-ospA1 to pBBE22-ospA7. Three amplicons, ospA1, ospA2 and ospA3, were generated by PCR using forward primers P1F, P2F and P3F, respectively, and a common reverse primer P1R, with borrelial DNA as a template, and cloned into the shuttle vector pBBE22. The fragments ospA4, ospA5, ospA6 and ospA7 were created as illustrated in Fig. 2 and were cloned into pBBE22. The DRs and the T-rich region are collectively designated cisI. cisII represents the sequence between the −10 region and the start codon.
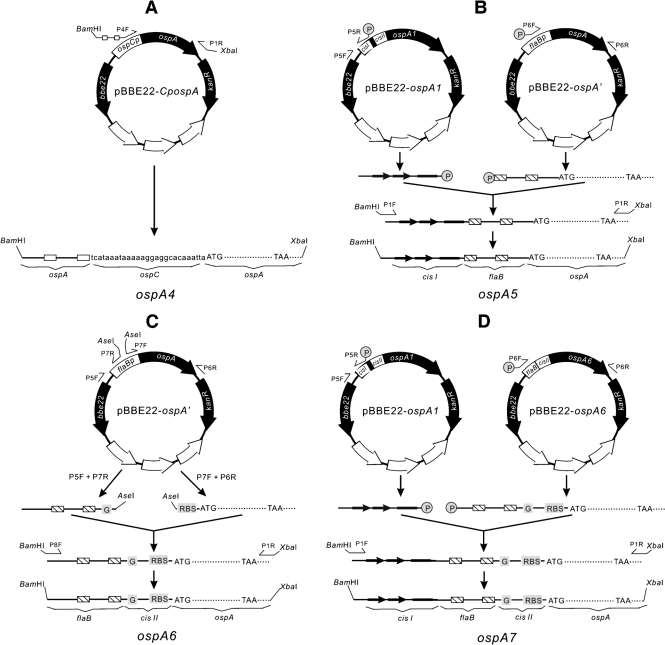
As illustrated in Fig. 2(A), the fragment ospA4 was directly amplified from the construct pBBE22-CpospA with use of primers P4F and P1R (Supplementary Table S2). The long primer, P4F, incorporated the −35 and −10 regions of the ospAB promoter, and the sequence between them (Supplementary Table S2).
Fig. 2.
Creation of fragments ospA4 (A), ospA5 (B), ospA6 (C) and ospA7 (D). Details of the constructions are described in Methods.
To generate the fragment ospA5 (Fig. 2B), two amplicons were generated from the construct pBBE22-ospA1 with use of primers P5F and P5R, and from the construct pBBE22-ospA′ with primers P6F and P6R (Supplementary Table S2), respectively. The resulting amplicons were fused via blunt-end ligation and further amplified with use of primers P1F and P1R to introduce BamHI and XbaI restriction enzyme sites at the ends.
To construct ospA6 (Fig. 2C), two amplicons were generated from the construct pBBE22-ospA′ with the use of primer pairs P5F and P7R, and P7F and P6R (Supplementary Table S2), respectively. The resulting amplicons were digested with AseI, ligated and amplified using primers P8F and P1R, to introduce BamHI and XbaI restriction enzyme sites at the ends.
To create the fragment ospA7 (Fig. 2D), two amplicons were generated from the construct pBBE22-ospA1 with the use of primers P5F and P5R, and from the construct pBBE22-ospA6 using primers P6F and P6R, respectively. The resulting amplicons were fused via blunt-end ligation and further amplified with the use of primers P1F and P1R to introduce BamHI and XbaI restriction enzyme sites at the ends.
Fragments ospA4, ospA5, ospA6 and ospA7 were cloned into pBBE22, to complete the creation of the constructs pBBE22-ospA4, pBBE22-ospA5, pBBE22-ospA6 and pBBE22-ospA7 (Fig. 1B). The inserts and flanking regions were sequenced to ensure that the constructs were created as designed.
Transformation of B. burgdorferi and selection of transformants.
Constructs were electroporated into the ΔospA mutant; the resulting transformants were screened and analysed for plasmid content as described previously (Xu et al., 2005). Restoration of OspA production was verified using immunoblots probed with a mixture of FlaB and OspA mAbs, as described in an earlier study (Xu et al., 2008b).
Constructs were also electroporated into clone 13A spirochaetes; the resulting transformants were screened and selected as described above, but immunoblotting was not performed because overwhelming OspA production from the native ospA copy potentially masked the contribution from introduced constructs.
Mouse infection.
Severe combined immunodeficiency (SCID) mice (BALB/c background; age 4–8 weeks; provided by the LSU Division of Laboratory Animal Medicine) were given a single intradermal/subcutaneous injection of 104 spirochaetes. Animals were examined for the development of arthritis at 2 day intervals, starting at day 7, and sacrificed 1 month post-inoculation. If no significant joint swelling was noted, heart, joint and skin samples were subjected to spirochaetal isolation as described previously (Xu et al., 2005). If joint swelling was apparent, skin, joint and heart specimens were collected for RNA preparation. RNA was converted to cDNA by reverse transcription (RT) and quantified for the mRNA copy numbers of flaB and ospA by quantitative PCR (qPCR) as described previously (Liang et al., 2004a). The animal procedures described here were approved by the Institutional Animal Care and Use Committee at Louisiana State University.
Statistical analysis.
Data were first analysed by using a one-way analysis of variance when an experiment involved more than two groups. A two-tailed Student t test was used to compare two treatments and calculate P-values. Calculated P-values of ≤0.05 were considered significant.
RESULTS
Generation of constructs
A total of seven constructs, pBBE22-ospA1 to pBBE22-ospA7, were generated from the shuttle vector pBBE22. pBBE22 was modified from pBSV2 by inserting a copy of bbe22, a gene that codes for a nicotinamidase essential for the basic survival of B. burgdorferi in mammalian hosts (Purser et al., 2003; Stewart et al., 2001). Because the B. burgdorferi B31 clone 13A, which lacks lp25 and lp56 (Xu et al., 2007a), was used as the parental strain in the current study, constructs harbouring bbe22 were required for restoration of infectivity of its derivatives.
Deletion of the direct repeats and the T-rich region leads to decreased ospA expression in vitro but increased expression in mice
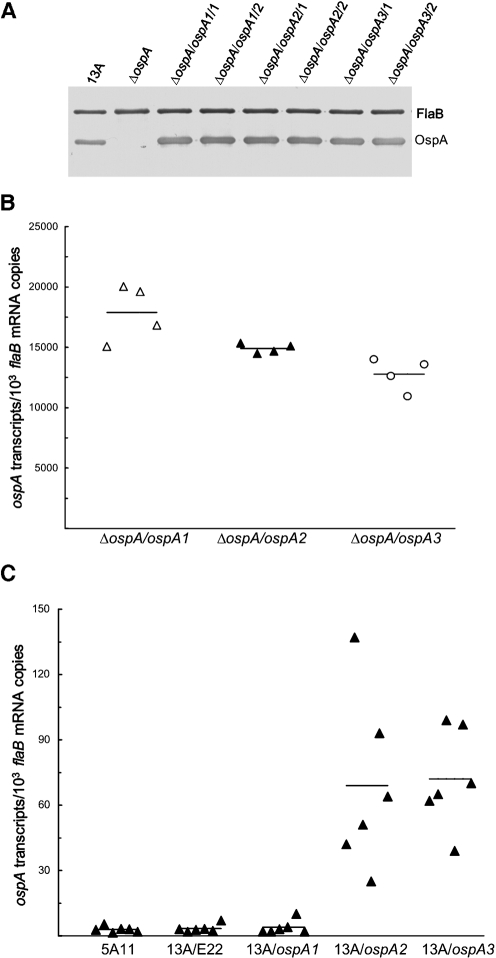
Three constructs, pBBE22-ospA1 to pBBE22-ospA3, were first electroporated into ΔospA; between 11 and 20 transformants were obtained from transformation with each construct. Plasmid content analyses led to the selection of two clones containing each construct. The six clones, ΔospA/ospA1/1, ΔospA/ospA1/2, ΔospA/ospA2/1, ΔospA/ospA2/2, ΔospA/ospA3/1 and ΔospA/ospA3/2, shared the same plasmid content as ΔospA, which had lost cp9, lp5, lp21, lp28-1, lp25 and lp56 (Xu et al., 2008b). An immunoblot analysis showed that all transformants abundantly produced OspA (Fig. 3A).
Fig. 3.
The presence of DRs and the T-rich region enhances ospA expression in vitro but reduces expression in mice. (A) Restoration of OspA synthesis resulting from introduction of a construct. 13A, ΔospA, ΔospA/ospA1/1, ΔospA/ospA1/2, ΔospA/ospA2/1, ΔospA/ospA2/2, ΔospA/ospA3/1 and ΔospA/ospA3/2 spirochaetes were harvested at late-exponential phase and subjected to immunoblot analysis, probed with a mixture of FlaB and OspA mAbs. (B) Deletion of cisI and the T-rich region causes reduced ospA expression in vitro. ΔospA/ospA1/1, ΔospA/ospA1/2, ΔospA/ospA2/1, ΔospA/ospA2/2, ΔospA/ospA3/1 and ΔospA/ospA3/2 spirochaetes were harvested at late-exponential phase in duplicate. RNA was prepared and analysed for ospA and flaB expression by RT-qPCR. The expression activity is presented as ospA mRNA copy numbers per 103 flaB transcripts. For comparison, the subgroups were combined into three groups: ΔospA/ospA1/1 and ΔospA/ospA1/2, ΔospA/ospA2/1 and ΔospA/ospA2/2, and ΔospA/ospA3/1 and ΔospA/ospA3/2. The mean copy numbers (horizontal lines) for each group are presented. (C) The sequence of DRs is involved in repressing ospA expression in vivo. Subgroups of three SCID mice were inoculated with clone 13A/ospA1/1, 13A/ospA1/2, 13A/ospA2/1, 13A/ospA2/2, 13A/ospA3/1 or 13A/ospA3/2, and an additional 12 mice were infected with clones 5A11 and 13A/E22/C. All animals were euthanized 1 month later. RNA was extracted from skin specimens and quantified for flaB and ospA mRNAs by RT-qPCR. Data are presented as ospA transcripts per 103 flaB mRNA copies in five groups (genotypes including 5A11, 13A/E22, 13A/ospA1, 13A/ospA2 and 13A/ospA3). The means (horizontal lines) for each group are also presented.
To precisely compare the ospA expression activity driven by each construct, the ΔospA/ospA1/1, ΔospA/ospA1/2, ΔospA/ospA2/1, ΔospA/ospA2/2, ΔospA/ospA3/1 and ΔospA/ospA3/2 spirochaetes were harvested at late-exponential phase (108 cells ml−1). RNA was prepared and analysed for ospA and flaB mRNA copies by RT-qPCR. As shown in Fig. 3(B), the pBBE22-ospA1 spirochaetes accumulated 17 % and 29 % more ospA transcripts than the genotypes pBBE22-ospA2 (P<0.05) and pBBE22-ospA3 (P=9.2×10−3), respectively. Moreover, the genotype pBBE22-ospA2 produced 14 % more ospA mRNA than the genotype pBBE22-ospA3 (P=0.02). These data indicate that the presence of the T-rich region increases ospA expression and that the inclusion of the sequence of direct repeats (DRs) further enhances expression in B. burgdorferi grown in vitro. The current study was unable to rule out whether the T-rich region and DRs functioned as a single regulatory element or two independent units, so they were collectively designated cisI.
Next, groups of three SCID mice were inoculated with clone ΔospA/ospA1/1, ΔospA/ospA1/2, ΔospA/ospA2/1, ΔospA/ospA2/2, ΔospA/ospA3/1 or ΔospA/ospA3/2. Only 2 of the 18 inoculated animals were infected when mice were euthanized 1 month post-inoculation (data not shown). The inability to fully restore infectivity of ΔospA prevented us from using its derivatives in animal studies.
Fortunately, the native ospAB operon is not expressed by B. burgdorferi during mammalian infection; therefore its presence should not affect the behaviour of an introduced construct that may drive ospA expression. The constructs pBBE22-ospA1 to pBBE22-ospA3 were electroporated into 13A spirochaetes. Six clones, 13A/ospA1/1, 13A/ospA1/2, 13A/ospA2/1, 13A/ospA2/2, 13A/ospA3/1 and 13A/ospA3/2, were selected for the study as they had the same plasmid content, including having lost cp9, lp21 and lp5, in addition to lp25 and lp56. Subgroups of three SCID mice were inoculated with each of these clones. An additional 12 mice were challenged with clone 5A11 or 13A/E22/C as controls. Clone 5A11 contains 20 plasmids, lacks lp5, and is considered to have a wild-type phenotype (Purser & Norris, 2000); clone 13A/E22/C has the same plasmid content as the six test clones and was generated via introduction of pBBE22 into clone 13A in our previous study (Xu et al., 2007b). The two controls were used to assess whether the lack of cp9, lp21, lp5, lp25 and lp56 affected ospA expression in mice. Evidence of joint swelling in all 30 mice occurred within 12 days after inoculation and quickly developed into severe arthritis (data not shown), indicating that all the clones were infectious in SCID mice.
All mice were euthanized 1 month post-inoculation; RNA was prepared from skin specimens and assessed for the relative copy numbers of ospA and flaB mRNAs by RT-qPCR. As shown in Fig. 3(C), both 5A11 and 13A/E22/C spirochaetes expressed ospA at a baseline level (P=0.67), indicating that the absence of cp9, lp21, lp5, lp25 and lp56 does not affect ospA regulation in vivo. There was also no significant difference in ospA expression detected in the genotypes 13A/E22 and 13A/ospA1 (P=0.74). When a gene is expressed at an extremely low level, RT-qPCR data may only reflect ‘noise’. This could be the reason why the 13A/ospA1 spirochaetes did not show increased ospA expression although they contained a construct-source ospA copy in addition to the native gene. Nevertheless, these data indicate that the presence of the native ospA copy does not interfere with data interpretation in our system.
When the three genotypes harbouring an ospA construct were compared for ospA expression in skin, the genotype 13A/ospA1 accumulated ospA transcripts at a 17.3- and 18.0-fold lower level than the genotypes 13A/ospA2 (P=2.9×10−3) and 13A/ospA3 (P=2.7×10−5), respectively (Fig. 3C); but there was no significant difference in ospA expression between the latter two genotypes (P=0.86). Similar ospA expression patterns were detected for these genotypes when RNA prepared from the heart and joint specimens was analysed (data not shown). These data indicated that the sequence upstream of the ospAB promoter, which includes at least DRs, functions as a cis element critical for repressing ospA expression in mice. Although our data are not indicative of the T-rich region being a part of the regulatory element, our study cannot rule out its involvement, as the deletion of the DRs alone may completely destroy the integrity of the unit. This is a reason why both the T-rich region and the DRs are collectively termed cisI.
Replacement of cisII with the corresponding sequence of the ospC gene leads to decreased ospA mRNA accumulation in vitro but increased accumulation in mice
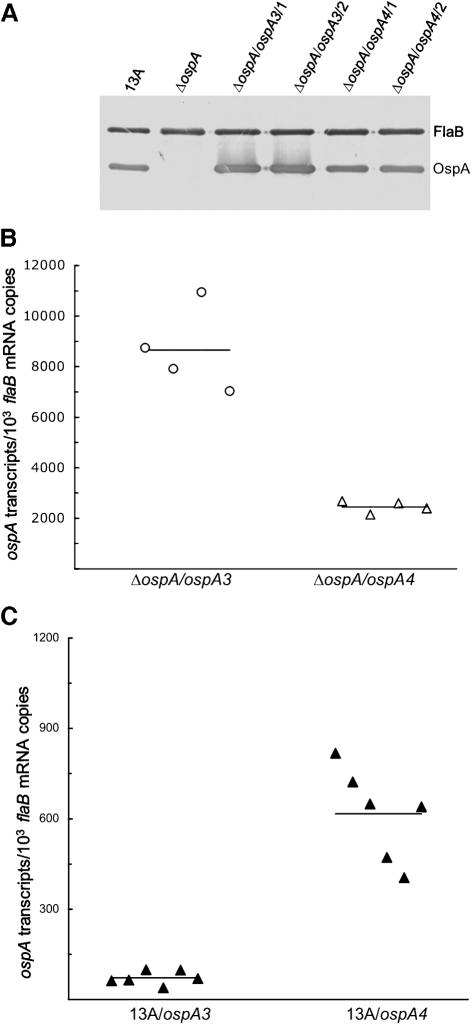
Surprisingly, the construct pBBE22-ospA3 carried the minimum ospAB promoter but drove significant ospA expression in murine tissues, albeit initiating much lower expression than the flaB promoter that was fused with the ospA reporter gene (Xu et al., 2008a), leading us to hypothesize that other sequences adjacent to the ospAB promoter may also be involved in repression of ospA expression. To examine this hypothesis, we focused on the sequence between the minimal ospAB promoter and the start codon ATG, which was collectively designated cisII. To this end, construct pBBE22-ospA4 was generated as illustrated in Figs 1 and 2. Within this construct, cisII was replaced with the corresponding region of ospC (the sequence between the −10 region of the ospC promoter and the start codon of the ospC gene), which was selected based on a previous study showing its non-involvement in gene regulation (Xu et al., 2007a). The construct was first electroporated into ΔospA to examine whether this modification affected the activity of the ospA promoter in vitro. Two clones, ΔospA/ospA4/1 and ΔospA/ospA4/2, which shared the same plasmid content as ΔospA, were selected for the study. An immunoblot analysis showed that the construct conferred OspA production at a level slightly lower than that of pBBE22-ospA3 (Fig. 4A).
Fig. 4.
The sequence cisII contributes to increased ospA expression in vitro but decreased expression in mice. (A) Restoration of OspA synthesis resulting from introduction of pBBE22-ospA4. 13A, ΔospA, ΔospA/ospA3/1, ΔospA/ospA3/2, ΔospA/ospA4/1 and ΔospA/ospA4/2 spirochaetes were harvested at late-exponential phase and subjected to immunoblot analysis, probed with a mixture of FlaB and OspA mAbs. (B) The sequence cisII significantly contributes to ospA mRNA accumulation in vitro. ΔospA/ospA3/1, ΔospA/ospA3/2, ΔospA/ospA4/1 and ΔospA/ospA4/2 spirochaetes were harvested at late-exponential phase in duplicate; RNA was extracted and analysed for ospA and flaB expression by RT-qPCR. The expression activity is presented as ospA mRNA copy numbers per 103 flaB transcripts in two groups (combining the subgroups), ΔospA/ospA3/1 and ΔospA/ospA3/2, and ΔospA/ospA4/1 and ΔospA/ospA4/2. The mean copy numbers (horizontal lines) for each group are presented. (C) The sequence cisII contributes to the downregulation of ospA expression in vivo. Subgroups of three mice were inoculated with clone 13A/ospA3/1, 13A/ospA3/2, 13A/ospA4/1 or 13A/ospA4/2. Mice were euthanized 1 month later; RNA was extracted from skin specimens and quantified for flaB and ospA expression by RT-qPCR. Data are presented as ospA transcripts per 103 flaB mRNA copy numbers in two groups (genotypes including 13A/ospA3 and 13A/ospA4). The means (horizontal lines) for each group are also presented.
Next, RNA was prepared and analysed for ospA and flaB expression by RT-qPCR. Clone ΔospA/ospA3 produced 72 % more ospA transcripts than ΔospA/ospA4 (P=3.3×10−4) (Fig. 4B). Clearly, the replacement of cisII with the corresponding sequence of the ospC gene led to a significant reduction in ospA expression in vitro. However, this piece of data alone does not necessarily suggest that the modification reduces the activity of the ospAB promoter. Alternatively, cisII may play an important role in enhancing ospA expression in vitro.
To examine the influence of cisII on ospA expression in vivo, pBBE22-ospA4 was electroporated into 13A spirochaetes. Two clones, 13A/ospA4/1 and 13A/ospA4/2, shared the same plasmid content as the genotypes 13A/ospA1, 13A/ospA2 and 13A/ospA3, and were inoculated into six SCID mice. An additional six mice were inoculated with clone 13A/ospA3/1 or 13A/ospA3/2 as a control. Mice were euthanized 1 month post-inoculation, and RNA was prepared from skin specimens and analysed. As shown in Fig. 4(C), the genotype 13A/ospA4 produced 8.6-fold more ospA transcripts than the genotype 13A/ospA3 (P=6.5×10−6). Similar ospA expression patterns were noted for these genotypes when heart and joint RNA preparations were analysed (data not shown). The data indicated that the ospA promoter can drive active gene expression in the murine host once cisII is substituted, thus suggesting that cisII contributes to the downregulation of ospA expression in the murine host.
Confirmation of the role of both cisI and cisII in enhancing ospA expression in vitro but reducing its expression in mice by using the flaB reporter promoter
In a previous study, we created a construct, pBBE22-ospA′, by fusing the flaB promoter with a promoterless ospA, demonstrating that it drove active ospA expression both in vitro and in vivo (Xu et al., 2008a). In the current study, we used the same system to confirm the role of cisI and cisII in ospA regulation through generation of three constructs, pBBE22-ospA5 to pBBE22-ospA7. In pBBE22-ospA5, cisI was directly fused with the flaB promoter, aimed at confirmation of the influence of cisI on the activity of a downstream promoter. In pBBE22-ospA6, cisII was inserted between the flaB promoter and the ospA reporter gene, a construct that would allow us to further examine whether cisII can influence downstream gene expression. In pBBE22-ospA7, cisI and cisII were, respectively, inserted immediately upstream and downstream of the flaB reporter promoter, in order to investigate whether the two cis elements have a synergistic effect on gene regulation.
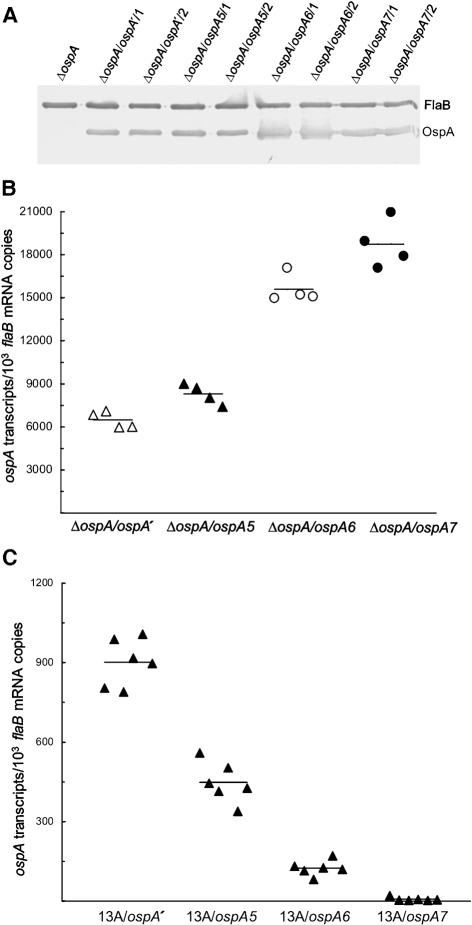
These three constructs were first electroporated into ΔospA to investigate whether the two cis elements contribute to ospA regulation in vitro. As a control, pBBE22-ospA′ was also electroporated into ΔospA. Eight clones, namely ΔospA/ospA5/1, ΔospA/ospA5/2, ΔospA/ospA6/1, ΔospA/ospA6/2, ΔospA/ospA7/1, ΔospA/ospA7/2, ΔospA/ospA′/1 and ΔospA/ospA′/2, were selected for the study as they had the same plasmid content as ΔospA. An immunoblot analysis indicated that all three flaB promoter derivatives drove abundant OspA production (Fig. 5A).
Fig. 5.
Confirmation of the role of cisI and cisII in enhancing ospA expression in vitro but reducing expression in vivo. (A) Restoration of OspA synthesis resulting from introduction of pBBE22-ospA5, pBBE22-ospA6 or pBBE22-ospA7. ΔospA, ΔospA/ospA′/1, ΔospA/ospA′/2, ΔospA/ospA5/1, ΔospA/ospA5/2, ΔospA/ospA6/1, ΔospA/ospA6/2, ΔospA/ospA7/1 and ΔospA/ospA7/2 spirochaetes were harvested at late-exponential phase and subjected to immunoblot analysis, probed with a mixture of FlaB and OspA mAbs. (B) Confirmation of the role of cisI and cisII in enhancing ospA expression in vitro. ΔospA/ospA′/1, ΔospA/ospA′/2, ΔospA/ospA5/1, ΔospA/ospA5/2, ΔospA/ospA6/1, ΔospA/ospA6/2, ΔospA/ospA7/1 and ΔospA/ospA7/2 spirochaetes were harvested at late-exponential phase in duplicate. RNA was extracted and analysed for ospA and flaB mRNA accumulation by RT-qPCR. The expression activity is presented as ospA mRNA copy numbers per 103 flaB transcripts and compared in four groups by combining the subgroups ΔospA/ospA′/1 and ΔospA/ospA′/2, ΔospA/ospA5/1 and ΔospA/ospA5/2, ΔospA/ospA6/1 and ΔospA/ospA6/2, and ΔospA/ospA7/1 and ΔospA/ospA7/2. The mean copy numbers (horizontal lines) for each group are also presented. (C) Confirmation of the contributions of cisI and cisII to the downregulation of ospA expression in vivo. Subgroups of three mice were inoculated with clone 13A/ospA′/1, 13A/ospA′/2, 13A/ospA5/1, 13A/ospA5/2, 13A/ospA6/1, 13A/ospA6/2, 13A/ospA7/1 or 13A/ospA7/2 and euthanized 1 month later. RNA was extracted from skin specimens and quantified for flaB and ospA expression by RT-qPCR. Data are presented as ospA transcripts per 103 flaB mRNA copies in four groups (genotypes including 13A/ospA′, 13A/ospA5, 13A/ospA6 and 13A/ospA7). The means (horizontal lines) for each group are also presented.
RNA was prepared from spirochaetes harvested at late-exponential phase and analysed by RT-qPCR. As shown in Fig. 5(B), the genotype ΔospA/ospA′ accumulated 28 % and 141 % less ospA transcripts than the genotypes ΔospA/ospA5 (P=0.008) and ΔospA/ospA6 (P=4.1×10−6), respectively, confirming the role of both cis elements in enhancing ospA expression in vitro. Moreover, the genotype ΔospA/ospA6 produced 88 % more ospA mRNA than the genotype ΔospA/ospA5 (P=2.1×10−5), indicating that cisII increases ospA accumulation more effectively than cisI. Finally, the genotype ΔospA/ospA7 accumulated 189 %, 126 % and 20 % more ospA mRNA than the genotypes ΔospA/ospA′ (P=9.3×10−6), ΔospA/ospA5 (P=2.7×10−5) and ΔospA/ospA6 (P<0.02), respectively, indicating that cisI and cisII function synergistically in enhancing ospA expression in cultivated B. burgdorferi.
To confirm the role of both cisI and cisII in ospA regulation in vivo, the four constructs were electroporated into 13A spirochaetes. Eight clones, 13A/ospA5/1, 13A/ospA5/2, 13A/ospA6/1, 13A/ospA6/2, 13A/ospA7/1, 13A/ospA7/2, 13A/ospA′/1 and 13A /ospA′/2, were selected to challenge subgroups of three SCID mice. These clones contained the same plasmids as the genotypes 13A/ospA1, 13A/ospA2, 13A/ospA3 and 13A/ospA4. Mice were euthanized 1 month post-inoculation; skin RNA was prepared and analysed. As shown in Fig. 5(C), the genotype 13A/ospA5 accumulated twofold less ospA transcripts than the control 13A/ospA′ (P=2.9×10−9), thus confirming the role of cisI in repressing ospA expression in vivo. The genotype 13A/ospA6 accumulated 7.3 times less ospA transcripts than the control 13A/ospA′ (P=2.2×10−9), confirming the contribution of cisII to the downregulation of ospA expression. When the genotypes 13A/ospA5 and 13A/ospA6 were compared, the former expressed 4.1-fold higher ospA transcripts than the latter (P=9.7×10−8), indicating that cisII more effectively contributes to ospA repression than cisI.
When both cisI and cisII were simultaneously fused with the flaB promoter, upstream and downstream, respectively, their contribution to ospA repression was further confirmed (Fig. 5C): the genotype 13A/ospA7 produced 64 times less ospA transcripts than the genotype 13A/ospA5 (P=6.1×10−8). It expressed 18-fold less ospA mRNA than the genotype 13A/ospA6 (P=2.0×10−6). Overall, the collective effect of the two cis elements reduced ospA expression by 129-fold (P=3.5×10−10), when 13A/ospA′ and 13A/ospA7 were compared.
DISCUSSION
Although controversies surround the exact functions of OspA and OspB in the tick, abundant production of these surface lipoproteins is crucial for B. burgdorferi to colonize the vector during its natural life cycle (Battisti et al., 2008; Neelakanta et al., 2007; Pal et al., 2004a; Yang et al., 2004). Moreover, the downregulation of the ospAB operon is critical for the pathogen to effectively evade the immune system, cause persistent infection, and maintain the enzootic cycle (Strother et al., 2007; Tsao et al., 2001, 2004; Xu et al., 2008a). The molecular mechanisms by which B. burgdorferi dramatically changes OspA/B expression during cycling between the two distinct hosts remained largely unknown, until this study carefully analysed the elements surrounding the ospAB promoter, showing involvement of sequences both upstream and downstream of the promoter in the regulation of ospA expression.
During the past 3 years, we have generated three ospAB mutants, and all lack the expected infectivity following complementation. These mutants were thinner and less motile than the wild-type, growing very poorly while aggregating at the bottom of culture tubes during in vitro cultivation, essentially consistent with a report by Sadziene et al. (1995) describing severe growth defects of OspAB-deficient B. burgdorferi. In contrast, we successfully inactivated ospC, dbpAB, and rpoS without difficulty using the same system, readily restoring these mutants to full infectivity (Shi et al., 2008; Xu et al., 2007a). Because both OspA and OspB are such highly produced outer-surface lipoproteins in cultivated B. burgdorferi, inactivation of the ospAB operon may cause a severe compromise in the integrity of the outer-surface lipoprotein layer, consequently severely affecting spirochaete survival. The ospAB mutants selected most likely underwent dramatic changes in gene expression to compensate for the loss of OspA/B. To date, three groups have generated infectious ospAB or ospB mutants (Battisti et al., 2008; Neelakanta et al., 2007; Yang et al., 2004), but concluded differing roles for OspA/B during the life cycle of B. burgdorferi in the tick vector. While different tick-feeding techniques used in their studies could be an explanation for these disparities, as discussed by Battisti et al. (2008), the fact that each group used different mutants should also be considered, as the current study shows derivatives of the ospAB mutant producing inconsistent results following inoculation into mice. A recent study by He et al. (2008) reported two phenotypes of the ospA mutant: one with constitutive ospC expression and the other having lost the ability to induce ospC expression in response to environmental changes. Our ospAB mutant exhibits a third phenotype, the ospC expression pattern of wild-type B. burgdorferi (Xu et al., 2008b).
Deletion of the DRs resulted in decreased ospA expression in vitro, and further removal of the T-rich region led to an additional reduction, suggesting that both sequences are involved in positive regulation of ospA expression in vitro. However, our study could not determine whether DRs and the T-rich region function independently or act collectively as a single regulatory element; hence they were designated cisI. The identification of the cis element predicts the existence of regulatory protein(s) called activator(s). Further identification of these unknown regulator(s) should help solve the puzzle of whether DRs and the T-rich region constitute a single regulatory unit. To confirm the regulatory function of cisI, it was used to replace the upstream sequence of the flaB promoter and was shown to significantly increase expression of an ospA reporter gene in vitro.
Exactly opposite to what was observed in vitro, deletion of the DRs caused a significant increase in ospA expression in the murine host, indicating that the sequence is involved in repressing ospA expression in vivo. Although further removal of the T-rich region did not affect ospA expression in murine tissues, our study could not rule out whether the region and DRs collectively form a regulatory element because deletion of DRs alone may destroy its function. The regulatory function of cisI, which includes both the DRs and the T-rich region, in mammalian hosts was further confirmed by using the flaB reporter promoter. cisI is likely to function as an operator, whose identification predicts the existence of a regulator named repressor. Further identification of the unknown regulator should help determine whether the T-rich region is a part of the operator of B. burgdorferi during mammalian infection.
Sohaskey et al. (1999) showed that deletion of the T-rich region results in a great reduction in expression of a chloramphenicol acetyltransferase reporter gene in vitro, while Caimano et al. (2007) suggested that this sequence may be involved in the downregulation of ospA/B in the mammalian host. The current study clearly confirmed the findings of Sohaskey et al. (1999) when B. burgdorferi was grown in vitro. However, we could not confirm the T-rich region as a part of the regulatory element of this spirochaete during mammalian infection.
Because the downstream sequence, collectively called cisII, of the ospAB promoter contains the transcriptional initiation site and ribosome-binding site, it cannot simply be deleted in order to investigate its regulatory function. The replacement of cisII caused a significant decrease in ospA expression in vitro, indicating a role of the sequence in enhancing expression. This role was further confirmed via fusion of cisII with the flaB reporter promoter. An opposite function of cisII was revealed in B. burgdorferi during murine infection. Like cisI, cisII may function as an operator interacting with an as-yet-unidentified repressor and contribute to ospAB downregulation in the mammalian host. Given that activator sites are usually located upstream of the −10 region (Collado-Vides et al., 1991) and that cisII is sited between the −10 region and the start codon, with most of its sequence being transcribed into mRNA, it may be involved in post-transcriptional rather than transcriptional regulation (Lucchetti-Miganeh et al., 2008), when B. burgdorferi is grown in vitro. The RNA sequence transcribed from cisII may interact with an as-yet-unidentified RNA-binding protein, and consequently lead to protection of ospAB transcripts when B. burgdorferi is grown in vitro. RNA-binding protein may also have an opposite function, which is to facilitate RNA turnover and subsequently cause gene downregulation (Lucchetti-Miganeh et al., 2008). From this point of view, cisII does not necessarily function as an operator in B. burgdorferi during murine infection, but instead its RNA sequence may interact with a specific RNA-binding protein and, as a consequence, facilitate mRNA degradation. Interestingly, our previous study noted that the 3′ sequence (ospB portion) of ospAB mRNA is more stable than the 5′ (ospA) portion in murine tissues but less stable when spirochaetes are grown in vitro (Liang et al., 2004a). It remains to be addressed whether this differential degradation of ospA and ospB portions of the bicistronic mRNA is due to a function of cisII.
The identification of the regulatory structure provided insights into the complexity of ospAB regulation. During in vitro cultivation, this currently unidentified activator is induced, which may enhance ospAB expression through an interaction with cisI. To further increase expression, the in vitro environment may also induce an unknown RNA-binding protein, which binds to the specific RNA sequence transcribed from cisII and protects ospAB mRNA. In contrast, the mammalian environment may induce two unidentified repressors, which bind to cisI and cisII, respectively, and shut off the ospAB operon. The mammalian milieu may also induce an unknown RNA-binding protein, which binds to the RNA sequence of cisII and facilitates mRNA degradation to further minimize OspA/B production. One could speculate that the unknown activator and repressor may be the same regulators, whose function is environment-dependent, and that the same RNA-binding protein may protect ospAB mRNA in vitro but facilitate decay during mammalian infection. Future studies should help to resolve these interesting issues.
A previous study reported that the downregulation of ospA is induced by IgG molecules (Hodzic et al., 2005), in contrast to our earlier study showing that B. burgdorferi effectively represses ospA expression during infection of both SCID and immunocompetent mice (Liang et al., 2004a). The current study clarified the issue of whether IgG is required for the induction of the downregulation. When cisI was removed and cisII was replaced, the ospA promoter efficiently initiated ospA expression as the flaB promoter drove the ospA reporter gene in SCID mice. Moreover, when one of the elements was fused with the core flaB promoter, expression of the ospA reporter gene was dramatically reduced, and when both sequences were fused with the promoter, the reporter was essentially shut off. Given that IgG molecules were absent under each of these situations, the current study allowed us to conclude that the mammalian environment alone sufficiently induces downregulation of the ospAB operon.
Both OspA and OspB are highly immunogenic, so even a low level of expression may stimulate a significant humoral response. Although the responses may not effectively target spirochaetes with low OspA and OspB production in mammalian tissues, once the organisms are acquired by a tick vector, the surface antigens are greatly upregulated and consequently become effective targets in a blood meal. Thus, concealing OspA and OspB from the adaptive immune system is crucial for maintaining the enzootic cycle. The current study demonstrated that this ability depends on the newly identified cis elements. Even more importantly, the dual functions of these regulatory sequences can potentially provide B. burgdorferi with a greater advantage for quick adaptation to diverse environments during its enzootic life cycle travelling between the two distinct hosts. During persistent infection of a mammal, many outer-surface lipoproteins, including both RpoS- and σ70-dependent gene products, are actively produced (Liang et al., 2002, 2004b; Miller & Stevenson, 2006). Abundant surface lipoprotein expression is required for stabilizing the outer membrane of B. burgdorferi against innate defences (Xu et al., 2008b). Once acquired by the vector, however, B. burgdorferi quickly downregulates rpoS (Caimano et al., 2007) and consequently RpoS-dependent genes, such as dbpA, dbpB, ospF and bbk32 (Eggers et al., 2004; He et al., 2007; Hubner et al., 2001), as well as some σ70-dependent genes such as vlsE and some erp genes (Bykowski et al., 2006). A simple shift of cisI and cisII from the repressing mode to the activating function would allow B. burgdorferi to boost OspA/B production, effectively maintain the integrity of its outer-surface lipoprotein layer, and ultimately adapt to the new environment.
Acknowledgments
The authors would like to thank S. Norris for providing pBBE22 and B31 5A11. This work was supported in part by AI077733, AR053338, and RR020159 from the National Institutes of Health. F. T. L. is the recipient of an Arthritis Foundation Investigators Award.
Abbreviations
DR, direct repeat
Osp, outer-surface protein
SCID, severe combined immunodeficiency
RT, reverse transcription
qPCR, quantitative PCR
Footnotes
Two supplementary tables with details of the constructs, clones and plasmids used in this study are available with the online version of this paper.
References
- Battisti, J. M., Bono, J. L., Rosa, P. A., Schrumpf, M. E., Schwan, T. G. & Policastro, P. F. (2008). Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect Immun 76, 5228–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykowski, T., Babb, K., von Lackum, K., Riley, S. P., Norris, S. J. & Stevenson, B. (2006). Transcriptional regulation of the Borrelia burgdorferi antigenically variable VlsE surface protein. J Bacteriol 188, 4879–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano, M. J., Eggers, C. H., Gonzalez, C. A. & Radolf, J. D. (2005). Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J Bacteriol 187, 7845–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano, M. J., Iyer, R., Eggers, C. H., Gonzalez, C., Morton, E. A., Gilbert, M. A., Schwartz, I. & Radolf, J. D. (2007). Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol 65, 1193–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides, J., Magasanik, B. & Gralla, J. D. (1991). Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev 55, 371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva, A. M., Telford, S. R., III, Brunet, L. R., Barthold, S. W. & Fikrig, E. (1996). Borrelia burgdorferi OspA is an arthropod-specific transmission-blocking Lyme disease vaccine. J Exp Med 183, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva, A. M., Fish, D., Burkot, T. R., Zhang, Y. & Fikrig, E. (1997). OspA antibodies inhibit the acquisition of Borrelia burgdorferi by Ixodes ticks. Infect Immun 65, 3146–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers, C. H., Caimano, M. J. & Radolf, J. D. (2004). Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J Bacteriol 186, 7390–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle, V., Goettner, G., Gern, L., Wilske, B. & Schulte-Spechtel, U. (2007). Complementation of a Borrelia afzelii ospC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int J Med Microbiol 297, 97–107. [DOI] [PubMed] [Google Scholar]
- Grimm, D., Tilly, K., Byram, R., Stewart, P. E., Krum, J. G., Bueschel, D. M., Schwan, T. G., Policastro, P. F., Elias, A. F. & Rosa, P. A. (2004). Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc Natl Acad Sci U S A 101, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M., Boardman, B. K., Yan, D. & Yang, X. F. (2007). Regulation of expression of the fibronectin-binding protein BBK32 in Borrelia burgdorferi. J Bacteriol 189, 8377–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M., Oman, T., Xu, H., Blevins, J., Norgard, M. V. & Yang, X. F. (2008). Abrogation of ospAB constitutively activates the Rrp2-RpoN-RpoS pathway (σN-σS cascade) in Borrelia burgdorferi. Mol Microbiol 70, 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodzic, E., Tunev, S., Feng, S., Freet, K. J. & Barthold, S. W. (2005). Immunoglobulin-regulated expression of Borrelia burgdorferi outer surface protein A in vivo. Infect Immun 73, 3313–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, T. R., LaQuier, F. W. & Barbour, A. G. (1986). Organization of genes encoding two outer membrane proteins of the Lyme disease agent Borrelia burgdorferi within a single transcriptional unit. Infect Immun 54, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner, A., Yang, X., Nolen, D. M., Popova, T. G., Cabello, F. C. & Norgard, M. V. (2001). Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A 98, 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, M., Noppa, L., Barbour, A. G. & Bergstrom, S. (1992). Heterogeneity of outer membrane proteins in Borrelia burgdorferi: comparison of osp operons of three isolates of different geographic origins. Infect Immun 60, 1845–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F. T., Nelson, F. K. & Fikrig, E. (2002). Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med 196, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F. T., Caimano, M. J., Radolf, J. D. & Fikrig, E. (2004a). Borrelia burgdorferi outer surface protein (osp) B expression independent of ospA. Microb Pathog 37, 35–40. [DOI] [PubMed] [Google Scholar]
- Liang, F. T., Yan, J., Mbow, M. L., Sviat, S. L., Gilmore, R. D., Mamula, M. & Fikrig, E. (2004b). Borrelia burgdorferi changes its surface antigenic expression in response to host immune responses. Infect Immun 72, 5759–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetti-Miganeh, C., Burrowes, E., Baysse, C. & Ermel, G. (2008). The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154, 16–29. [DOI] [PubMed] [Google Scholar]
- Miller, J. C. & Stevenson, B. (2006). Borrelia burgdorferi erp genes are expressed at different levels within tissues of chronically infected mammalian hosts. Int J Med Microbiol 296 (Suppl. 40), 185–194. [DOI] [PubMed] [Google Scholar]
- Neelakanta, G., Li, X., Pal, U., Liu, X., Beck, D. S., DePonte, K., Fish, D., Kantor, F. S. & Fikrig, E. (2007). Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog 3, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi, J., Piesman, J. & de Silva, A. M. (2001). Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc Natl Acad Sci U S A 98, 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, U., Li, X., Wang, T., Montgomery, R. R., Ramamoorthi, N., Desilva, A. M., Bao, F., Yang, X., Pypaert, M. & other authors (2004a). TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119, 457–468. [DOI] [PubMed] [Google Scholar]
- Pal, U., Yang, X., Chen, M., Bockenstedt, L. K., Anderson, J. F., Flavell, R. A., Norgard, M. V. & Fikrig, E. (2004b). OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest 113, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser, J. E. & Norris, S. J. (2000). Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97, 13865–13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purser, J. E., Lawrenz, M. B., Caimano, M. J., Howell, J. K., Radolf, J. D. & Norris, S. J. (2003). A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol Microbiol 48, 753–764. [DOI] [PubMed] [Google Scholar]
- Sadziene, A., Thomas, D. D. & Barbour, A. G. (1995). Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect Immun 63, 1573–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan, T. G. & Piesman, J. (2000). Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol 38, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan, T. G., Piesman, J., Golde, W. T., Dolan, M. C. & Rosa, P. A. (1995). Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A 92, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Xu, Q., McShan, K. & Liang, F. T. (2008). Both decorin-binding proteins A and B are critical for the overall virulence of Borrelia burgdorferi. Infect Immun 76, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohaskey, C. D., Zuckert, W. R. & Barbour, A. G. (1999). The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol Microbiol 33, 41–51. [DOI] [PubMed] [Google Scholar]
- Stewart, P. E., Thalken, R., Bono, J. L. & Rosa, P. (2001). Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol 39, 714–721. [DOI] [PubMed] [Google Scholar]
- Stewart, P. E., Wang, X., Bueschel, D. M., Clifton, D. R., Grimm, D., Tilly, K., Carroll, J. A., Weis, J. J. & Rosa, P. A. (2006). Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect Immun 74, 3547–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strother, K. O., Hodzic, E., Barthold, S. W. & de Silva, A. M. (2007). Infection of mice with Lyme disease spirochetes constitutively producing outer surface proteins A and B. Infect Immun 75, 2786–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao, J., Barbour, A. G., Luke, C. J., Fikrig, E. & Fish, D. (2001). OspA immunization decreases transmission of Borrelia burgdorferi spirochetes from infected Peromyscus leucopus mice to larval Ixodes scapularis ticks. Vector Borne Zoonotic Dis 1, 65–74. [DOI] [PubMed] [Google Scholar]
- Tsao, J. I., Wootton, J. T., Bunikis, J., Luna, M. G., Fish, D. & Barbour, A. G. (2004). An ecological approach to preventing human infection: vaccinating wild mouse reservoirs intervenes in the Lyme disease cycle. Proc Natl Acad Sci U S A 101, 18159–18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Seemanapalli, S. V., Lomax, L., McShan, K., Li, X., Fikrig, E. & Liang, F. T. (2005). Association of linear plasmid 28-1 with an arthritic phenotype of Borrelia burgdorferi. Infect Immun 73, 7208–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., McShan, K. & Liang, F. T. (2007a). Identification of an ospC operator critical for immune evasion of Borrelia burgdorferi. Mol Microbiol 64, 220–231. [DOI] [PubMed] [Google Scholar]
- Xu, Q., Seemanaplli, S. V., McShan, K. & Liang, F. T. (2007b). Increasing the interaction of Borrelia burgdorferi with decorin significantly reduces the 50 percent infectious dose and severely impairs dissemination. Infect Immun 75, 4272–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., McShan, K. & Liang, F. T. (2008a). Modification of Borrelia burgdorferi to overproduce OspA or VlsE alters its infectious behaviour. Microbiology 154, 3420–3429. [DOI] [PubMed] [Google Scholar]
- Xu, Q., McShan, K. & Liang, F. T. (2008b). Essential protective role attributed to the surface lipoproteins of Borrelia burgdorferi against innate defences. Mol Microbiol 69, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. F., Pal, U., Alani, S. M., Fikrig, E. & Norgard, M. V. (2004). Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med 199, 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]