Abstract
Early-phase transmission (EPT) is a recently described model of plague transmission that explains the rapid spread of disease from flea to mammal host during an epizootic. Unlike the traditional blockage-dependent model of plague transmission, EPT can occur when a flea takes its first blood meal after initially becoming infected by feeding on a bacteraemic host. Blockage of the flea gut results from biofilm formation in the proventriculus, mediated by the gene products found in the haemin storage (hms) locus of the Yersinia pestis chromosome. Although biofilms are required for blockage-dependent transmission, the role of biofilms in EPT has yet to be determined. An artificial feeding system was used to feed Xenopsylla cheopis and Oropsylla montana rat blood spiked with the parental Y. pestis strain KIM5(pCD1)+, two different biofilm-deficient mutants (ΔhmsT, ΔhmsR), or a biofilm-overproducer mutant (ΔhmsP). Infected fleas were then allowed to feed on naïve Swiss Webster mice for 1–4 days after infection, and the mice were monitored for signs of infection. We also determined the bacterial loads of each flea that fed upon naïve mice. Biofilm-defective mutants transmitted from X. cheopis and O. montana as efficiently as the parent strain, whereas the EPT efficiency of fleas fed the biofilm-overproducing strain was significantly less than that of fleas fed either the parent or a biofilm-deficient strain. Fleas infected with a biofilm-deficient strain harboured lower bacterial loads 4 days post-infection than fleas infected with the parent strain. Thus, defects in biofilm formation did not prevent flea-borne transmission of Y. pestis in our EPT model, although biofilm overproduction inhibited efficient EPT. Our results also indicate, however, that biofilms may play a role in infection persistence in the flea.
INTRODUCTION
Yersinia pestis infection is characterized as a rapidly spreading disease that cycles between rodents and fleas (Gage & Kosoy, 2005; Pollitzer, 1954). Humans are also seriously affected, as indicated by the occurrence of three historical pandemics that killed millions (Perry & Fetherston, 1997). The transmission of Y. pestis from flea to host occurs primarily via flea bite; however, the mechanisms of transmission remain under investigation. Fleas could also play a significant role as reservoirs of infection, providing that they are capable of sustaining Y. pestis infections over extended periods of time. The most studied form of transmission of Y. pestis by fleas is the Bacot and Martin model, in which transmission depends on development of a biofilm-dependent proventricular blockage that begins to form when a flea ingests a blood meal from an infected mammalian host (Bacot & Martin, 1914; Bacot, 1915; Eskey & Haas, 1940). In this process, the bacteria multiply and produce a biofilm inside the flea that eventually forms a blockage in the proventriculus of certain fleas such as Xenopsylla cheopis, which is a species that is the major vector of plague in many foci around the world (Gage & Kosoy, 2005; Hinnebusch et al., 1996; Jarrett et al., 2004). The blockage prevents the flea from taking a full blood meal, resulting in eventual starvation and repeated attempts by starving fleas to take blood meals from multiple hosts. During these feeding attempts, it is presumed that pieces of the blockage matrix containing the bacteria are regurgitated into the host, thereby infecting them (Burroughs, 1947; Eskey & Haas, 1940; Pollitzer, 1954). The possible role of biofilms in the maintenance of Y. pestis in fleas has received little attention, although it has been suggested that they may be important for colonization of fleas by the bacteria, and of soil organisms such as nematodes and protists, which have been suggested by some to be potential reservoirs of Y. pestis in soils (Darby et al., 2002; Udovikov et al., 2009).
Although this blockage-dependent mechanism is well described and undoubtedly important, it cannot support the rapid rate of spread that characterizes many plague epidemics or epizootics, particularly those that involve transmission by fleas that require a long extrinsic incubation period of perhaps two or more weeks to become blocked, and then experience a short window of infectivity before flea death (Drancourt et al., 2006; Eisen et al., 2006; Wood et al., 2003). In addition, blockage formation is rare in many species of fleas, including those that, based on epidemiological data, are thought to be important vectors for plague (Eisen et al., 2009). For example, despite its poor blocking capability, Oropsylla montana is considered a primary vector of Y. pestis to humans in the USA, and is the major flea found on rock squirrels and ground squirrels that routinely succumb to plague epizootics. Even in blockage-forming fleas, such as X. cheopis, the rate of blockage formation is only 25–45 % (Burroughs, 1947; Eskey & Haas, 1940; Gage & Kosoy, 2005; Hinnebusch et al., 1996). Recently, Eisen et al. (2006) described an alternative form of transmission termed ‘early-phase transmission’ (EPT). In this model, fleas are fed a blood meal, and were then allowed to feed on naïve mice for 3–96 h after infection. Transmission occurs each day for at least 4 days after fleas become infected, and is efficient enough to provide an explanation of the temporal dynamics in fast-moving plague epizootics (Eisen et al., 2006, 2007b). One day, or even 4 days, provides insufficient time to form a block in X. cheopis, and this suggests that an alternative mechanism of transmission is required to explain these results.
The hms genes that create and regulate biofilms are located in three identified operons: hmsHFRS, hmsT and hmsP (Hinnebusch et al., 1996; Jones et al., 1999; Kirillina et al., 2004; Perry et al., 1990). The hmsHFRS operon is a region within the pgm locus, whose gene products are responsible for the adsorption of vast quantities of exogenous haemin and structurally similar Congo Red (CR) dye (Jackson & Burrows, 1956; Surgalla & Beesley, 1969). Together, these gene products are predicted to synthesize the β-1,6-N-acetylglucosamine-like polymers that form biofilms (Bobrov et al., 2008; Hinnebusch et al., 1996; Itoh et al., 2005; Kirillina et al., 2004; Perry et al., 1990, 2004). Synthesis of biofilm exopolysaccharide is initiated by the putative formation of a cyclic-dimeric GMP (c-di-GMP)-dependent HmsR–HmsS complex (Bobrov et al., 2008). HmsR is a putative glycosyltransferase with a conserved glycosyltransferase D,D,D35QXXRW motif, and is critical for biofilm formation and CR binding in Y. pestis (Forman et al., 2006). The amount of local c-di-GMP is proposed to be tightly regulated by the gene products of hmsT and hmsP (Bobrov et al., 2008; Kirillina et al., 2004). HmsT is a diguanylate cyclase with a signature GGDEF domain, and promotes the formation of higher levels of c-di-GMP, thus increasing biofilm production, while HmsP contains an EAL phosphodiesterase motif, and is required for degrading c-di-GMP (Bobrov et al., 2005; Jones et al., 1999; Kirillina et al., 2004; Perry et al., 2004; Simm et al., 2005). Deletion of hmsT leads to a large decrease in c-di-GMP-dependent production of the poly-β-1,6-N-acetylglucosamine-like polysaccharide, and deletion of hmsP results in overproduction of biofilm due to the loss of the encoded c-di-GMP phosphodiesterase (Bobrov et al., 2008; Kirillina et al., 2004).
Our study sought to determine whether biofilm formation was necessary for EPT. We used two different species of fleas (X. cheopis and O. montana), to investigate whether biofilm formation differed in a vector (X. cheopis) which routinely becomes blocked compared with another epidemiogically important vector (O. montana) which does not become blocked. Our results showed that biofilm was not necessary for EPT, and that the overproduction of biofilm was actually inhibitory to EPT. Moreover, studies that quantified bacterial loads in fleas indicated a potential role for biofilms in the maintenance of the bacteria in the flea.
METHODS
Bacterial growth conditions and construction of mutants.
Bacterial strains used in this study are listed in Table 1. The biofilm-defective mutants [ΔhmsR (KIM6-2118) and ΔhmsT (KIM6-2051.6+)] form white colonies on CR plates incubated at 26 °C, in contrast to the red ‘pigmented’ colonies formed by their parental Hms+ strain (KIM6+). The biofilm-overproducing strain KIM6-2090.7+ (ΔhmsP) is characterized by the formation of pigmented colonies on CR plates at both 26 and 37 °C (Bobrov et al., 2008; Jones et al., 1999; Kirillina et al., 2004; Perry et al., 2004). Although the hms locus is part of the pgm locus, none of the hms mutations interferes with the yersiniabactin-mediated iron-acquisition system of the pgm locus or affects virulence in the mammalian host (Abu Khweek et al., 2010; Bearden et al., 1997; Burrows & Jackson, 1956a, b; Fetherston et al., 1992; Lillard et al., 1999; Perry et al., 1990).
Table 1.
Bacterial strains and plasmids
Y. pestis strains marked with ‘+’ carry the wild-type 102 kb pgm locus, while those without ‘+’ have one or more mutations, or a deletion, in the pgm locus. Hms− indicates the presence of an intact pgm locus, but with deletions in the hmsHFRS, hmsT or hmsP operons; these render the strain incapable of producing wild-type levels of biofilm. HmsC indicates an intact pgm locus coupled with the loss of hmsP; this allows for temperature-independent expression of biofilm.
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Strains | ||
| KIM6+ | Pgm+ Lcr− Pla+; pPCP1, pMT1 | Fetherston et al. (1992) |
| KIM5(pCD1Ap)+ | Apr Pgm+, pPCP1+, pMT+, pCD1Ap (′yadA : : bla); derived from KIM6+ | Gong et al. (2001) |
| KIM6-2051.6+ | Hms− (ΔhmsT); Lcr− Pla+; pPCP1, pMT1 | This study |
| KIM5-2051.6(pCD1Ap)+ | Apr Hms− (ΔhmsT2090.2), pPCP1, pMT1, pCD1Ap (′yadA : : bla); derived from KIM6-2051.6 | This study |
| KIM6-2090.1+ | Cmr Hmsc (ΔhmsP : : cam2090.1) Lcr− Pla+; pPCP1, pMT1 | Kirillina et al. (2004) |
| KIM6-2090.2+ | Hmsc (ΔhmsP : : cam2090.1) Lcr− Pla+; pPCP1, pMT1; derived from KIM6-2090.1+ | This study |
| KIM6-2090.7+ | Hmsc, ΔhmsP, pPST+, pMT+, pCD−, Kmr; derived from KIM6-2090.2+ | This study |
| KIM5-2090.7(pCD1Ap)+ | Kmr Apr Hmsc (ΔhmsP2090.2) Δy2360 : : kan2093.1, pPCP1, pMT1, pCD1Ap (′yadA : : bla); derived from KIM6-2090.7 | This study |
| KIM6-2118 | Hms− (in-frame ΔhmsR2118) Lcr− Pla+; pPCP1, pMT1 | Forman et al. (2006) |
| KIM5-2118(pCD1Ap) | Apr Hms− (in-frame ΔhmsR2118) Lcr+ Pla+; pPCP1, pMT1, pCD1Ap (′yadA : : bla); derived from KIM6-2118 | This study |
| Plasmids | ||
| pKD4 | 3.3 kb, Kmr, template plasmid | Datsenko & Wanner (2000) |
| pKD46 | 6.3 kb, Apr, Red recombinase expression plasmid | Datsenko & Wanner (2000) |
| pCP20 | Suicide plasmid with temperature-sensitive replication and thermally induced expression of FLP recombinase; Apr | Datsenko & Wanner (2000) |
| pCD1Ap | 70.5 kb pCD1 with bla cassette inserted into ′yadA; 71.7 kb Lcr+ Apr | Gong et al. (2001) |
The hmsT deletion mutant (KIM6-2051.6+) was constructed using lambda red recombinase (Datsenko & Wanner, 2000). The kanamycin-resistance (Kmr) cassette was amplified from pKD4 using primer pair deltaT-5′ (5′-ATGCAGAGTAAATTGAATATGAATAGCCACTCCTACGATCGTGTAGGCTGGAGCTGCTTC-3′) and deltaT-3′ (5′-AGGGGAAGACTGTACATTTGATAATTCATCTTTAGCAAATCATATGAATATCCTCCTTAG-3′). The PCR product was electroporated into KIM6(pKD46)+, and the culture was plated on CR plates containing kanamycin. Cells that formed white colonies were checked using primers 10-2inv (5′-GTGAAATTAAACGTGCAG-3′) and 11Binv (5′-TATTTGTCGTGATGTCGG-3′) for the deletion of hmsT and the insertion of the Kmr cassette, which was eliminated by introducing pCP20 [encoding the Flp recombinase, and ampicillin (Apr) and chloramphenicol resistance (Cmr)] (Cherepanov & Wackernagel, 1995; Datsenko & Wanner, 2000). To remove pCP20, isolated colonies were streaked onto tryptose blood agar (TBA) base plates and grown at 40 °C. Individual colonies from these plates were grown in a 96-well dish at 37 °C, and then replica-plated onto TBA plates containing ampicillin and TBA containing chloramphenicol. Sensitivities to kanamycin and the presence of the pgm locus were checked as described elsewhere (Perry & Bearden, 2008). One colony, which was kanamycin-sensitive and retained the pgm locus, was selected for further analysis.
The hmsP mutant used in this study was derived from KIM6-2090.1+ (ΔhmsP : : cam2090.1). The Cmr cassette was first excised by introducing the pCP20 plasmid, as described above, creating KIM6-2090.2+ (ΔhmsP2090.1). To mark the pgm region with a Kmr cassette, red-mediated recombination (Datsenko & Wanner, 2000) was again employed to insert the Kmr cassette into y2360, which is located in the pgm locus, and encodes a putative endopeptidase. This created KIM6-2090.7+ [ΔhmsP2090.2 (Δy2360 : : kan2093.1)]. The Δy2360 : : kan2093.1 mutation was confirmed by PCR using primers previously described for confirming a similar mutation, Δy2360 : : cam2093 (Bobrov et al., 2008; Jones et al., 1999; Kirillina et al., 2004; Perry et al., 2004).
The KIM6-2051.6+ (ΔhmsT), KIM6-2090.7+ (ΔhmsP) and KIM6-2118 (in-frame ΔhmsR2118) strains were electroporated with pCD1Ap to restore the type III secretion and effector protein virulence factors, creating strains KIM5-2051.6(pCD1Ap)+, KIM5-2090.7(pCD1Ap)+ and KIM5-2118(pCD1Ap), respectively (Table 1). Transformants were then tested by using PCR and plasmid profile analysis to confirm the presence of the plasmid (Chu, 2000). To confirm virulence, 1×104 c.f.u. of each strain was inoculated subcutaneously into two 6-week-old naïve Swiss Webster mice. Upon appearance of signs of plague illness (described below), each mouse was euthanized. Each strain was then reisolated from the liver of each mouse, and glycerol stocks [0.5 ml aliquots containing ∼1×106 c.f.u. in heart infusion broth (HIB) supplemented with 10 % (v/v) glycerol] were made to use with each subsequent artificial feeding trial. This procedure was also used to confirm the virulence of each strain for each artificial feed trial performed. When required, carbenicillin was used at a concentration of 50 μg ml−1 and kanamycin at 25 μg ml−1.
To determine the Pgm+ phenotype retention rate in the fleas, 10 fleas from each artificial feeding that were infected with either the parent KIM5(pCD1Ap)+ strain or the biofilm-overproducing strain (ΔhmsP) were selected at random. A 0.1 ml aliquot of a 10−3 dilution of the flea homogenate was plated on CR plates in duplicate, and the ratio of red to white colonies was counted. In all, about 1000 colonies were counted for each group.
Infection of fleas.
Fleas were fed using an artificial feeding system described previously (Eisen et al., 2006). Briefly, on day 0, batches (150 fleas per feeder) of colony-reared O. montana or X. cheopis were allowed to feed on artificial feeders containing blood infected with approximately 1×109 c.f.u. ml−1 of KIM5(pCD1Ap)+, KIM5-2090.7(pCD1Ap)+, KIM5-2118(pCD1Ap) or KIM5-2051.6(pCD1Ap)+. To prepare infected blood, a 0.5 ml aliquot of each bacterial stock (prepared as described above) was inoculated into 60 ml HIB. The culture was grown for 16 h at 28 °C and 175 r.p.m. to ∼1×109 c.f.u. ml−1. Bacteria were collected by centrifugation, and resuspended in 60 ml defibrinated Sprague Dawley strain rat blood (Bioreclamation). The fleas were allowed to feed for 1 h, and were then examined by light microscopy. Only fed fleas with red blood in the proventriculus or mid-gut were kept, while unfed fleas were discarded. Potentially infected fleas were held at 23 °C and 85 % relative humidity for 1–4 days post-infection (p.i.). An artificial feeding of either species of flea normally results in 50–95 % of fleas taking an infectious blood meal, and of those fed fleas, 90 % are infected (Eisen et al., 2006).
Transmission of Y. pestis from fleas to mice.
On each of the 4 days p.i., pools of 10 potentially infectious fleas were added to a feeding capsule affixed to seven 6-week-old naïve Swiss Webster mice, and the fleas were allowed to feed for 1 h (Eisen et al., 2008b). The fleas were then removed by a mechanical aspirator, and examined by light microscopy to determine whether they had consumed a blood meal. Normally, 5–10 of 10 fleas take a blood meal upon the mouse (Eisen et al., 2006, 2007a, 2008a). The fed fleas were stored in individual centrifuge tubes at −80 °C until flea bacterial loads could be determined by serial dilution (Eisen et al., 2006). The mice were housed in individual cages, and monitored daily for signs of Y. pestis infection (e.g. shivering, lethargy, ruffled fur). Mice showing signs of infection were euthanized and necropsied, and slide preparations of their livers and spleens were examined for the presence of bacteria through the use of an immunofluorescent antibody to the Y. pestis F1 antigen (Chu, 2000). Mice that did not show signs of illness were euthanized 21 days after exposure to the infected fleas and tested serologically for exposure to Y. pestis. All animal procedures were approved by the Division of Vector-Borne Infectious Diseases (Centers for Disease Control and Prevention) Institutional Animal Care and Use Committee.
Statistical analysis.
The estimation of EPT efficiency was based on the number of infected fleas that fed on an individual mouse, and whether or not transmission was observed for that recipient mouse, by using the Microsoft Excel Add-In PooledInfRate version 3.0 (Biggerstaff, 2006; Eisen et al., 2006, 2007b). The resulting number is a prediction of the expected number of transmission events in a sample pool of 100 mice. The bacterial load of each flea was categorized as high (>1×104 c.f.u. per flea), low (1×101–1×104 c.f.u. per flea) or uninfected (0 c.f.u. per flea). The percentage of fleas in the high category was compared with those in the other two categories combined by using two-tailed Fisher's exact tests. Results were included from at least three artificial feeding experiments for each bacterial strain and each flea species tested. All statistical comparisons were performed using JMP statistical software (SAS Institute), and were considered statistically significant at P<0.05.
RESULTS
Biofilm is not required for EPT
When assessed in X. cheopis fleas, which routinely become blocked, EPT of Y. pestis was observed at three of the four time points in both the ΔhmsT mutant and the ΔhmsR mutant (Table 2). Transmission was demonstrated for the diguanylate cyclase mutant KIM6-2051.6(pCD1Ap)+ (ΔhmsT) on days 1–3 p.i. in 86 % (six of seven) to 57 % (four of seven) of mice exposed to infected fleas. When fleas were infected with the ΔhmsR mutant KIM6-2118(pCD1Ap), transmission events were observed in 43 % (three of seven) to 57 % (four of seven) of mice on days 1–3 p.i. of the fleas. We did not observe EPT of either the ΔhmsT or the ΔhmsR strain on day 4 p.i. Transmission efficiencies of these two mutants were comparable with those of the parent strain KIM5(pCD1Ap)+ (Table 2). Fleas infected with KIM5(pCD1Ap)+ were able to transmit infection on days 1, 2 and 4, with transmission events occurring in 14 % (one of seven) to 42 % (three of seven) of mice exposed.
Table 2.
Transmission efficiencies of X. cheopis for KIM6+ and hms mutant strains
| Strain | Day p.i. | No. of transmission events* | Estimated transmission efficiency per time point† | Estimated transmission efficiency per strain for all time points† |
|---|---|---|---|---|
| Parent KIM5(pCD1Ap)+ | 1 | 3 (7) | 7.60 (2.46–18.86) | 3.48 (1.48–7.02) |
| 2 | 2 (7) | 4.38 (0.84–13.97) | ||
| 3 | 0 (7) | 0.00 (0.00–6.24) | ||
| 4 | 1 (7) | 2.10 (0.12–10.19) | ||
| Biofilm-negative ΔhmsR KIM6-2118(pCD1Ap) | 1 | 3 (7) | 5.75 (1.6–15.74) | 5.14 (2.66–9.15) |
| 2 | 4 (7) | 9.37 (3.13–24.03) | ||
| 3 | 3 (7) | 6.09 (1.67–16.09) | ||
| 4 | 0 (7) | 0.00 (0.00–5.21) | ||
| Biofilm-reduced ΔhmsT KIM6-2051.6 | 1 | 6 (7) | 20.28 (10.22–40.28) | 9.83 (5.66–16.11) |
| 2 | 3 (7) | 8.78 (2.53–23.02) | ||
| 3 | 4 (7) | 13.04 (4.31–33.04) | ||
| (pCD1Ap)+ | 4 | 0 (7) | 0.00 (0.00–6.95) | |
| Biofilm-overproducer ΔhmsP KIM6-2090.7(pCD1Ap)+ | 1 | 0 (7) | 0.00 (0.00–5.55) | 0.00 (0.00–1.99) |
| 2 | 0 (7) | 0.00 (0.00–6.85) | ||
| 3 | 0 (7) | 0.00 (0.00–6.90) | ||
| 4 | 0 (7) | 0.00 (0.00–7.10) |
*Total number of mice tested in parentheses.
†95 % confidence interval in parentheses.
When tested with O. montana, which is a flea that rarely becomes blocked, biofilms were again not required for EPT (Table 3). Both ΔhmsT and ΔhmsR mutants were transmitted from infected fleas to naïve mice at all time points, ranging in efficiency from 14 % (one of seven mice) to 71 % (five of seven mice), and 28 % (two of seven mice) to 86 % (six of seven mice), for all instances in which infected fleas were fed on plague-susceptible mice, respectively. The parent KIM5(pCD1Ap)+ strain was transmitted on days 1, 2 and 4, with between 14 and 42 % of exposed mice becoming infected. As seen in X. cheopis, O. montana could transmit the biofilm-deficient strains at rates comparable with those of the parent strain. Although one flea-mediated EPT event, as indicated by seroconversion, was demonstrated on day 2 p.i. for the biofilm-overproducing strain, the level of transmission was significantly lower than that observed for the parent strain (Table 3).
Table 3.
Transmission efficiencies of O. montana for KIM6+ and hms mutant strains
| Strain | Day p.i. | No. of transmission events* | Estimated transmission efficiency per time point† | Estimated transmission efficiency per strain for all time points† |
|---|---|---|---|---|
| Parent KIM5(pCD1Ap)+ | 1 | 1 (7) | 1.72 (0.10–8.42) | 3.21 (1.33–6.65) |
| 2 | 3 (7) | 6.46 (1.76–18.48) | ||
| 3 | 0 (7) | 0.00 (0.00–6.14) | ||
| 4 | 2 (7) | 4.76 (0.88–15.70) | ||
| Biofilm-negative ΔhmsR KIM6-2118(pCD1Ap) | 1 | 2 (7) | 3.36 (0.63–11.06) | 7.88 (4.34–13.41) |
| 2 | 6 (7) | 18.75 (8.53–38.76) | ||
| 3 | 4 (7) | 9.69 (3.46–23.18) | ||
| 4 | 5 (7) | 14.75 (5.91–34.01) | ||
| Biofilm-reduced ΔhmsT KIM6-2051.61(pCD1Ap)+ | 1 | 5 (7) | 10.93 (4.33–26.27) | 10.85 (6.77–16.82) |
| 2 | 5 (7) | 17.33 (6.63–37.33) | ||
| 3 | 1 (7) | 2.15 (0.13–10.54) | ||
| 4 | 1 (7) | 2.55 (0.15–12.18) | ||
| Biofilm-overproducer ΔhmsP KIM6-2097.7(pCD1Ap)+ | 1 | 0 (7) | 0.00 (0.00–5.36) | 0.46 (0.03–2.23) |
| 2 | 1 (7) | 1.76 (0.11–8.49) | ||
| 3 | 0 (7) | 0.00 (0.00–5.43) | ||
| 4 | 0 (7) | 0.00 (0.00–5.81) |
*Total number of mice tested in parentheses.
†95 % confidence interval in parentheses.
Biofilm effects in the flea
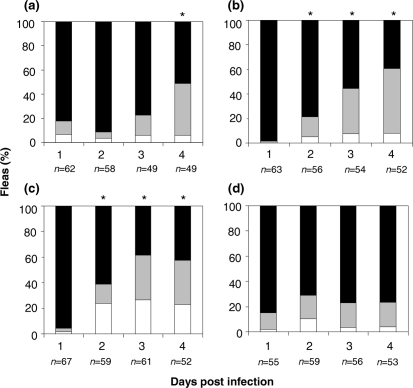
Bacterial loads were quantified for all potentially infected fleas that fed on naïve mice. Preliminary observations of the data suggested that bacterial loads in the fleas fell into two reasonably distinct groups: one group harbouring >104 c.f.u. per flea, and the other with <104 c.f.u. per flea. Accordingly, the bacterial load for each flea was then categorized as high bacterial load (>104 c.f.u. per flea), low bacterial load (1–104 c.f.u. per flea) or uninfected (Figs 1 and 2). X. cheopis infected with the parent strain KIM5(pCD1Ap)+ retained a high bacterial load over 4 days p.i. (>85 %) (Fig. 1a). In contrast, X. cheopis infected with either the biofilm-reduced strain [ΔhmsT, KIM5-2051.6(pCD1Ap)+] or the biofilm-negative strain [ΔhmsR, KIM5-2118(pCD1Ap)] experienced a decrease in bacterial load over 4 days p.i. The percentage of fleas infected with high bacterial loads of the biofilm-reduced strain was significantly decreased (P=0.003) compared with non-infected fleas and fleas harbouring low bacterial loads, on days 1–4 p.i. (93.2–68.1 %, respectively) (Fig. 1c), or found to be uninfected. The percentage of fleas infected with high bacterial loads of the biofilm-negative strain on days 3 and 4 p.i. was significantly lower than for fleas tested on day 1 p.i. (P=0.003). Bacterial loads in fleas tested on day 2 p.i. did not differ significantly from those observed in fleas tested on day 1 p.i.
Fig. 1.
Percentages of X. cheopis that were uninfected, infected with high bacterial load (>104) or infected with low bacterial load (<104) at 1–4 days p.i. Fleas received an infectious blood meal with Y. pestis: (a) parent strain, (b) biofilm-negative strain, (c) biofilm-reduced strain and (d) biofilm-overproducing strain. The infected fleas were then allowed to feed on naïve mice for 1–4 days p.i. Fleas taking blood meals on naïve mice were harvested after the blood feed, and it was determined whether their bacterial load was high (>1×104 c.f.u. per flea; black bar), low (>0, but <1×104 c.f.u. per flea; grey bar) or zero (0 c.f.u. per flea; white bar). An asterisk indicates that the percentage of fleas harbouring >1×104 c.f.u. Y. pestis was significantly different (P<0.05) from that of fleas carrying <1×104 c.f.u. of the same strain on day 1 p.i., as determined by two-tailed Fisher's exact test.
Fig. 2.
Percentages of O. montana that were uninfected, infected with high bacterial loads (>104) or infected with low bacterial loads (<104) at 1–4 days p.i. Fleas received an infectious blood meal with Y. pestis: (a) parent strain, (b) biofilm-negative strain, (c) biofilm-reduced strain and (d) biofilm-overproducing strain. The infected fleas were then allowed to feed on naïve mice for 1–4 days p.i. Fleas taking blood meals on naïve mice were harvested after the blood feed, and it was determined whether their bacterial load was high (>1×104 c.f.u. per flea; black bar), low (>0, but <1×104 c.f.u. per flea; grey bar) or zero (0 c.f.u. per flea; white bar). An asterisk indicates that the percentage of fleas harbouring >1×104 c.f.u. Y. pestis was significantly different (P<0.05) from that of fleas carrying <1×104 c.f.u. of the same strain on day 1 p.i., as determined by two-tailed Fisher's exact test.
Results with the poorly blocking species O. montana were similar to those obtained with X. cheopis (Fig. 2). O. montana infected with the parent strain experienced a gradual decrease from day 1 to day 4 p.i. in the proportion of fleas infected with high bacterial loads, with significantly fewer (P=0.001) fleas with high bacterial loads (51.0 %) on day 4 p.i. compared with those with a similar load on day 1 p.i. (82.9 %) (Fig. 2a). Fleas infected with either the biofilm-reduced or the biofilm-negative strain showed a much more dramatic decrease in the proportion of fleas with high bacterial loads, with each group experiencing a significant drop (P<0.001 and P=0.001, respectively) in fleas with a high bacterial load by day 2 p.i. (Fig. 2b, c).
Results for the biofilm-overproducing strain KIM5-2090.7(pCD1Ap)+ (ΔhmsP) were different from those for the parent and biofilm-deficient strains. In X. cheopis, bacterial loads were lower in fleas infected with this strain compared with those infected with the parent strain (Fig. 1d). To show that this effect on c.f.u. was indeed due to fewer bacteria in the flea, and not to clumping of the bacilli because of increased biofilm production, which would result in artificially low c.f.u. values, we performed quantitative PCR on flea lysates (Gabitzsch et al., 2008). Quantitative PCR assay results were highly correlated with the bacterial counts (data not shown), confirming that the bacterial loads observed in X. cheopis infected with the biofilm-overproducing strain were indeed lower than the bacterial loads in X. cheopis infected with the other strains. Moreover, this strain did not show a growth defect in O. montana, as indicated by the c.f.u. counts of bacteria in infected fleas (Fig. 2). However, unlike the other strains, the percentage of fleas with a high or low bacterial load in both X. cheopis and O. montana infected with the biofilm-overproducing strain did not significantly change over 1–4 days p.i. The proportions of fleas with high bacterial loads on days 1–4 p.i. were similar for both X. cheopis and O. montana (Figs 1d and 2d).
The ability of each strain to retain its Pgm+ status (a virulence characteristic conferred by the yersiniabactin biosynthetic and transport loci encoded in this region) was then tested. As expected, fleas harbouring the parent strain KIM5(pCD1Ap)+ contained nearly 100 % Hms+ colonies (Table 4). However, in both flea species, the biofilm-overproducing strain KIM5-2090.7(pCD1Ap)+, isolated from X. cheopis, lost the Hms+ phenotype at a higher frequency than the parent strain. Interestingly, this phenomenon was not observed in the ΔhmsP mutant cells isolated from O. montana (Table 4). Only 5–13 % of KIM6-2090.7(pCD1Ap)+ colonies isolated from X. cheopis were CR-negative at days 3 and 4 p.i., presumably indicating either a partial or an incomplete excision of the pgm locus, and suggesting that most colonies retained the hms genes required for the pigmentation phenotype; this is unlikely to explain the lack of EPT of the biofilm-overproducing strain in this flea species (Table 4). While maintaining a high proportion of Pgm+ cells (>98 %), the EPT rate for this biofilm overproducer was also low in O. montana (Table 3).
Table 4.
Percentage of Pgm+ colonies on days 1–4 p.i. of the parent or biofilm-overproducing strain from either X. cheopis or O. montana
| Flea species | Bacterial strain | Day p.i. | Mean percentage of Pgm+ colonies* |
|---|---|---|---|
| X. cheopis | Parent KIM5(pCD1Ap)+ | 1 | 99.9 (99.1–100.0) |
| 2 | 99.8 (99.5–100.0) | ||
| 3 | 99.9 (99.3–100.0) | ||
| 4 | 99.9 (99.8–100.0) | ||
| Biofilm overproducer ΔhmsP KIM6-2097.7(pCD1Ap)+ | 1 | 99.4 (98.1–100.0) | |
| 2 | 96.3 (95.0–100.0) | ||
| 3 | 87.4 (68.3–100.0) | ||
| 4 | 95.8 (78.4–100.0) | ||
| O. montana | Parent KIM5(pCD1Ap)+ | 1 | 98.6 (87.5–100.0) |
| 2 | 100.0 (100.0–100.0) | ||
| 3 | 100.0 (100.0–100.0) | ||
| 4 | 100.00 (100.0–100.0) | ||
| Biofilm overproducer ΔhmsP KIM6-2097.7(pCD1Ap)+ | 1 | 100.0 (100.0–100.0) | |
| 2 | 99.4 (95.3–100.0) | ||
| 3 | 99.5 (97.2–100.0) | ||
| 4 | 99.8 (78.4–100.0) |
*The range is given in parentheses.
DISCUSSION
EPT is the period of time during which fleas can transmit Y. pestis bacilli before a blockage can form. EPT of plague bacteria by X. cheopis and O. montana occurs for at least 4 days following the ingestion of an infectious blood meal, and it has been proposed as a mechanism to explain the temporal dynamics of rapidly moving plague epizootics in rodent populations (Eisen et al., 2006, 2007a). Our study used two biofilm-defective mutants to determine whether biofilms are necessary for EPT. Both ΔhmsT and ΔhmsR mutants are biofilm-negative in vitro. However, while the putative glycosyltransferase HmsR is essential for biofilm formation, HmsT is only one of several potential diguanylate cyclases in the Y. pestis KIM genome. Thus, it is possible that one of these other diguanylate cyclases produces c-di-GMP in the flea, allowing in vivo biofilm development. Consequently, we tested both mutants. Unlike the biofilm-dependent classic model of transmission, our results show that hms-mediated biofilm formation is not required for EPT, which likely involves an alternative mechanism. One potential alternative mechanism involves mechanical transmission, in which bacteria survive without multiplication on the external surface of the mouthparts or in the gut of the flea prior to being transmitted to a susceptible host (Burroughs, 1944; Eskey & Haas, 1940). However, this mechanism is unlikely to explain EPT because Y. pestis can only survive on mouthparts for up to 3 h, which is much shorter than the infectious time period presented here (Bibikova, 1965). Furthermore, Rose et al. (2003) have also demonstrated the lack of survivability of Y. pestis on certain environmental surfaces, indicating that one or more additional mechanisms besides mechanical transmission is likely to be involved in EPT (Rose et al., 2003). An alternative mechanism may involve aggregates of Y. pestis bacilli that develop in the anterior portion of the digestive tract of the flea (Eisen & Gage, 2009). Presumably, the formation of such aggregates might depend on biofilm formation, although our results indicate that strains that overproduce biofilm are less likely to be transmitted than parent strains. These results were somewhat surprising, and it is possible that a biofilm overproducer would form bacterial aggregates too large to pass through the mouthparts and into the bite site of a naïve mammalian host. Regardless, aggregate formation, even at the lower levels exhibited by the parent strain in our experiments, clearly is not required for EPT, as demonstrated by our experiments with biofilm-negative Y. pestis strains.
Results shown in our study provide strong evidence to suggest that biofilms are critical for maintenance of Y. pestis infection in fleas. Fleas fed biofilm-defective strains were more likely to recover from infections, and show reduced bacterial loads, compared with fleas infected with the parent strain. In this study, we found that both ΔhmsR and ΔhmsT mutants, which are biofilm-negative in vitro, did not maintain a high-bacterial-load infection in fleas to the same extent as the parent or the biofilm-overproducing strains. Kutyrev and co-workers found that an hms-negative strain is eliminated within 4 days from Nosopsyllus laeviceps fleas, the established vector for transmission of Y. pestis to voles in the natural plague foci of the former USSR (Kutyrev et al., 1992). Hinnebusch et al. (1996) noted that an hms-negative mutant was more likely to be cleared from X. cheopis than its parent strain. Moreover, our study found that the biofilm overproducer maintained its infection in the flea better than the parent strain. Over the 4 days p.i., fleas infected with the biofilm-overproducing strain maintained a constant, albeit somewhat lower, bacterial load in the flea, while fleas infected with the parent strain showed a significant drop in the percentage of fleas with a high bacterial load over 4 days p.i. (Figs 1 and 2). Taken together, these results suggest that biofilms are indeed important for Y. pestis persistence in the flea gut. Further studies will be conducted to determine whether fleas infected with a biofilm-deficient strain of Y. pestis will, over a longer period of time, experience a complete clearing of infection instead of simply a decrease in bacterial load.
Generally, compared with X. cheopis, O. montana experienced a more dramatic reduction over 4 days p.i. in the proportion of fleas assigned to the high-bacterial-load class. Since biofilm-dependent blockage is rare in O. montana compared with X. cheopis, this suggests a role for biofilms in non-blocking flea species. Ours is not the first study to document different bacterial behaviours in different flea species. Engelthaler et al. (2000) showed that Y. pestis infection primarily develops in the mid-gut of O. montana, while in X. cheopis, infection develops simultaneously in the proventriculus and the mid-gut (Engelthaler et al., 2000). Although Y. pestis bacteria seem to colonize the two flea species differently, the effect on EPT efficiency appears to be negligible, since both flea species transmit at similar rates. This is not the case with all flea species examined so far. Aetheca wagneri, Oropsylla tuberculata cynomuris, Oropsylla hirsuta and Ctenocephalides felis are not as efficient as X. cheopis and O. montana in EPT (Eisen et al., 2006, 2007b, 2008b; Wilder et al., 2008a, b). Previous studies have suggested that differences in proventriculus structure explain why some flea species are better at blocking, and therefore transmission (Konnov et al., 1979; Korzun & Nikitin, 1997). Since EPT occurs before blockage of the proventriculus, more studies are needed to determine the role of the proventriculus in EPT, and whether other anatomical differences between flea species contribute to varying transmission efficiencies.
In X. cheopis, overproduction of biofilms also limited the growth of bacteria. However, we do not believe the lack of EPT of the overproducing strain was a result of reduced numbers of bacteria in the flea. Other studies have shown that a decrease in bacterial load in individual fleas does not lead to decreases in transmission rates, and transmission of a wild-type strain has been seen with a median bacterial load of 2.65×102, which is a quantity lower than the lowest median value seen in fleas in this study (Eisen et al., 2007b, 2008a, b; Wilder et al., 2008a, b). Another possible explanation for seeing lower transmission rates could be an accumulation of non-pigmented (Δpgm) mutants that spontaneously appear in the flea gut over the time that it remains infected. Wild-type strains of Y. pestis spontaneously delete the pgm locus at a rate of 10−5 (Brubaker, 1969). In the laboratory, it has been noted that the biofilm-overproducing ΔhmsP strain has a higher accumulation of pgm excision mutants. Since the pgm locus contains the yersiniabactin genes required for siderophore-mediated iron acquisition in Y. pestis, deletion of this locus renders the bacteria attenuated with respect to plague virulence (Bearden et al., 1997; Burrows & Bacon, 1958; Fetherston et al., 1995; Une & Brubaker, 1984). While it is possible that deregulation of intracellular c-di-GMP in the ΔhmsP mutant (Bobrov et al., 2008) could contribute to a higher rate of spontaneous deletion of the pgm locus than that observed in wild-type strains, these spontaneous mutants may simply be able to outgrow biofilm-overproducing cells, and this effect may be amplified in the flea-gut environment. However, 88 % of X. cheopis infected with the biofilm-overproducing strain retained an intact pgm locus on day 4 p.i. (Fig. 1b). Thus, even if loss of CR binding was due to a deletion of a portion of or the entire pgm locus, this loss was still probably not sufficient to affect the transmission efficiency.
The synthesis of biofilms is tightly regulated by HmsP and HmsT, and levels of c-di-GMP. High levels of c-di-GMP close to an HmsR–HmsS–HmsT–HmsP complex activate biofilm formation. HmsT, characterized as a diguanylate cyclase, increases the local levels of c-di-GMP near this complex, while HmsP, a phosphodiesterase, degrades c-di-GMP (Bobrov et al., 2008). The coordinating levels of HmsT and HmsP control the amount of exopolysaccharide synthesized. Regulation of HmsT (as well as HmsR) is post-translational and dependent on temperature, resulting in lower protein levels above 30 °C, suggesting that biofilms must be important in the environment of the flea gut, which is normally at a lower temperature (Kirillina et al., 2004). Moreover, it has been demonstrated that biofilms are not critical for the pathogenesis of bubonic or pneumonic plague in mice, thus supporting the hypothesis that a function of Y. pestis biofilm production in fleas could be long-term survival of Y. pestis in its flea vectors (Abu Khweek et al., 2010; Lillard et al., 1999). The results shown here suggest that too much biofilm reduces EPT, while too little biofilm may not allow bacteria to be maintained over extended periods, a factor that could help sustain enzootic plague cycles. It is possible that this regulation is crucial for the balance between transmission and maintenance. Although not required for EPT, biofilms undoubtedly remain critical in the classic model of transmission, which occurs later in the period when fleas are infectious, and is likely to play an important role in long-term maintenance of Y. pestis infection in flea populations.
Acknowledgments
We thank Jacqueline Fetherston, PhD, for construction of mutant strains, and Kristen Van Wyk for excellent technical assistance with serology. This research was partially funded through an American Society for Microbiology Coordinating Center for Infectious Diseases (CCID) post-doctoral fellowship, and the National Institutes of Health (NIH) R01 AI025098-20.
Abbreviations
c-di-GMP, cyclic-dimeric GMP
CR, Congo Red
EPT, early-phase transmission
p.i., post-infection
References
- Abu Khweek, A., Fetherston, J. D. & Perry, R. D. (2010). Analysis of HmsH and its role in plague biofilm formation. Microbiology 156, 1424–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacot, A. W. (1915). Further notes on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 14 (Plague suppl. 4.), 774–776. [PMC free article] [PubMed] [Google Scholar]
- Bacot, A. W. & Martin, C. J. (1914). Observations on the mechanism of the transmission of plague by fleas. J Hyg (Lond) 13 (Plague suppl. 3), 423–439. [PMC free article] [PubMed] [Google Scholar]
- Bearden, S. W., Fetherston, J. D. & Perry, R. D. (1997). Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun 65, 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, V. A. (1965). Conditions for the existence of the plague microbe in fleas. Cesk Parazitol 12, 41–46. [Google Scholar]
- Biggerstaff, B. J. (2006). PooledInfRate, Version 3.0: a Microsoft Excel Add-In to Compute Prevalence Estimates from Pooled Samples. Fort Collins, CO: Centers for Disease Control and Prevention.
- Bobrov, A. G., Kirillina, O. & Perry, R. D. (2005). The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett 247, 123–130. [DOI] [PubMed] [Google Scholar]
- Bobrov, A. G., Kirillina, O., Forman, S., Mack, D. & Perry, R. D. (2008). Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol 10, 1419–1432. [DOI] [PubMed] [Google Scholar]
- Brubaker, R. R. (1969). Mutation rate to nonpigmentation in Pasteurella pestis. J Bacteriol 98, 1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs, A. L. (1944). The flea Malareus telchinum a vector of P. pestis. Proc Soc Exp Biol Med 55, 10–11. [Google Scholar]
- Burroughs, A. L. (1947). Sylvatic plague studies: the vector efficiency of nine species of fleas compared with Xenopsylla cheopis. J Hyg (Lond) 45, 371–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows, T. W. & Bacon, G. A. (1958). The effects of loss of different virulence determinants on the virulence and immunogenicity of strains of Pasteurella pestis. Br J Exp Pathol 39, 278–291. [PMC free article] [PubMed] [Google Scholar]
- Burrows, T. W. & Jackson, S. (1956a). The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br J Exp Pathol 37, 570–576. [PMC free article] [PubMed] [Google Scholar]
- Burrows, T. W. & Jackson, S. (1956b). The virulence-enhancing effect of iron on nonpigmented mutants of virulent strains of Pasteurella pestis. Br J Exp Pathol 37, 577–583. [PMC free article] [PubMed] [Google Scholar]
- Cherepanov, P. P. & Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. [DOI] [PubMed] [Google Scholar]
- Chu, M. C. (2000). Laboratory Manual of Plague Diagnostics. Geneva: Centers for Disease Control and Prevention, and World Health Organization.
- Darby, C., Hsu, J. W., Ghori, N. & Falkow, S. (2002). Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A. & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drancourt, M., Houhamdi, L. & Raoult, D. (2006). Yersinia pestis as a telluric, human ectoparasite-borne organism. Lancet Infect Dis 6, 234–241. [DOI] [PubMed] [Google Scholar]
- Eisen, R. J. & Gage, K. L. (2009). Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res 40, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, R. J., Bearden, S. W., Wilder, A. P., Montenieri, J. A., Antolin, M. F. & Gage, K. L. (2006). Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci U S A 103, 15380–15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, R. J., Lowell, J. L., Montenieri, J., Bearden, S. W. & Gage, K. L. (2007a). Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae). J Med Entomol 44, 672–677. [DOI] [PubMed] [Google Scholar]
- Eisen, R. J., Wilder, A. P., Bearden, S. W., Montenieri, J. & Gage, K. L. (2007b). Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol 44, 678–682. [DOI] [PubMed] [Google Scholar]
- Eisen, R. J., Borchert, J. N., Holmes, J. L., Amatre, G., Van Wyk, K., Enscore, R. E., Babi, N., Atiku, L. A., Wilder, A. P. & other authors (2008a). Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am J Trop Med Hyg 78, 949–956. [PubMed] [Google Scholar]
- Eisen, R. J., Holmes, J. L., Schotthoefer, A. M., Vetter, S. M., Montenieri, J. A. & Gage, K. L. (2008b). Demonstration of early-phase transmission of Yersinia pestis by the mouse flea, Aetheca wagneri (Siphonaptera: Ceratophylidae), and implications for the role of deer mice as enzootic reservoirs. J Med Entomol 45, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Eisen, R. J., Eisen, L. & Gage, K. L. (2009). Studies of vector competency and efficiency of North American fleas for Yersinia pestis: state of the field and future research needs. J Med Entomol 46, 737–744. [DOI] [PubMed] [Google Scholar]
- Engelthaler, D. M., Hinnebusch, B. J., Rittner, C. M. & Gage, K. L. (2000). Quantitative competitive PCR as a technique for exploring flea–Yersina pestis dynamics. Am J Trop Med Hyg 62, 552–560. [DOI] [PubMed] [Google Scholar]
- Eskey, C. R. & Haas, V. H. (1940). Plague in the western part of the United States. Public Health Bulletin 254, 1–83. [Google Scholar]
- Fetherston, J. D., Schuetze, P. & Perry, R. D. (1992). Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol 6, 2693–2704. [DOI] [PubMed] [Google Scholar]
- Fetherston, J. D., Lillard, J. W., Jr & Perry, R. D. (1995). Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol 177, 1824–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman, S., Bobrov, A. G., Kirillina, O., Craig, S. K., Abney, J., Fetherston, J. D. & Perry, R. D. (2006). Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152, 3399–3410. [DOI] [PubMed] [Google Scholar]
- Gabitzsch, E. S., Vera-Tudela, R., Eisen, R. J., Bearden, S. W., Gage, K. L. & Zeidner, N. S. (2008). Development of a real-time quantitative PCR assay to enumerate Yersinia pestis in fleas. Am J Trop Med Hyg 79, 99–101. [PubMed] [Google Scholar]
- Gage, K. L. & Kosoy, M. Y. (2005). Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol 50, 505–528. [DOI] [PubMed] [Google Scholar]
- Gong, S., Bearden, S. W., Geoffroy, V. A., Fetherston, J. D. & Perry, R. D. (2001). Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect Immun 69, 2829–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, B. J., Perry, R. D. & Schwan, T. G. (1996). Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273, 367–370. [DOI] [PubMed] [Google Scholar]
- Itoh, Y., Wang, X., Hinnebusch, B. J., Preston, J. F., III & Romeo, T. (2005). Depolymerization of β-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol 187, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S. & Burrows, T. W. (1956). The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br J Exp Pathol 37, 570–576. [PMC free article] [PubMed] [Google Scholar]
- Jarrett, C. O., Deak, E., Isherwood, K. E., Oyston, P. C., Fischer, E. R., Whitney, A. R., Kobayashi, S. D., DeLeo, F. R. & Hinnebusch, B. J. (2004). Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis 190, 783–792. [DOI] [PubMed] [Google Scholar]
- Jones, H. A., Lillard, J. W., Jr & Perry, R. D. (1999). HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145, 2117–2128. [DOI] [PubMed] [Google Scholar]
- Kirillina, O., Fetherston, J. D., Bobrov, A. G., Abney, J. & Perry, R. D. (2004). HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54, 75–88. [DOI] [PubMed] [Google Scholar]
- Konnov, N. P., Anisimov, P. I., Kondrashkina, K. I., Sinichkina, A. A., Luk'ianova, A. D. & Demchenko, T. A. (1979). The proventriculus of the Xenopsylla cheopis flea studied by scanning electron microscopy. Parazitologiia 13, 26–28. [PubMed] [Google Scholar]
- Korzun, V. M. & Nikitin, A. (1997). Asymmetry of the chaetae in fleas (Citellophilus tesquorum) as a possible marker of their capacity for blocking. Med Parazitol (Mosk) 1, 34–36 (in Russian). [PubMed] [Google Scholar]
- Kutyrev, V. V., Filippov, A. A., Oparina, O. S. & Protsenko, O. A. (1992). Analysis of Yersinia pestis chromosomal determinants Pgm+ and Psts associated with virulence. Microb Pathog 12, 177–186. [DOI] [PubMed] [Google Scholar]
- Lillard, J. W., Jr, Bearden, S. W., Fetherston, J. D. & Perry, R. D. (1999). The haemin storage (Hms+) phenotype of Yersinia pestis is not essential for the pathogenesis of bubonic plague in mammals. Microbiology 145, 197–209. [DOI] [PubMed] [Google Scholar]
- Perry, R. D. & Bearden, S. W. (2008). Isolation and confirmation of Yersinia pestis mutants exempt from select agent regulations. Curr Protoc Microbiol 11, 5B.2.1–5B.2.12. [DOI] [PubMed] [Google Scholar]
- Perry, R. D. & Fetherston, J. D. (1997). Yersinia pestis – etiologic agent of plague. Clin Microbiol Rev 10, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. D., Pendrak, M. L. & Schuetze, P. (1990). Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol 172, 5929–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. D., Bobrov, A. G., Kirillina, O., Jones, H. A., Pedersen, L., Abney, J. & Fetherston, J. D. (2004). Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol 186, 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitzer, R. (1954). Plague. In World Health Organization Monograph Series, no. 22. Geneva: World Health Organization.
- Rose, L. J., Donlan, R., Banerjee, S. N. & Arduino, M. J. (2003). Survival of Yersinia pestis on environmental surfaces. Appl Environ Microbiol 69, 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm, R., Fetherston, J. D., Kader, A., Romling, U. & Perry, R. D. (2005). Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol 187, 6816–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surgalla, M. J. & Beesley, E. D. (1969). Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol 18, 834–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udovikov, A. I., Grigor'eva, G. V., Tolokonnikova, S. I., Iakovlev, S. A., Tarasov, M. A. & Sludskii, A. A. (2009). Role of burrow microbiocenosis in plague enzootia. Med Parazitol (Mosk) 2, 44–46. [PubMed] [Google Scholar]
- Une, T. & Brubaker, R. R. (1984). Roles of V antigen in promoting virulence and immunity in yersiniae. J Immunol 133, 2226–2230. [PubMed] [Google Scholar]
- Wilder, A. P., Eisen, R. J., Bearden, S. W., Montenieri, J. A., Gage, K. L. & Antolin, M. F. (2008a). Oropsylla hirsuta (Siphonaptera: Ceratophyllidae) can support plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus) by early-phase transmission of Yersinia pestis. Vector Borne Zoonotic Dis 8, 359–367. [DOI] [PubMed] [Google Scholar]
- Wilder, A. P., Eisen, R. J., Bearden, S. W., Montenieri, J. A., Tripp, D. W., Brinkerhoff, R. J., Gage, K. L. & Antolin, M. F. (2008b). Transmission efficiency of two flea species (Oropsylla tuberculata cynomuris and Oropsylla hirsuta) involved in plague epizootics among prairie dogs. EcoHealth 5, 205–212. [DOI] [PubMed] [Google Scholar]
- Wood, J. W., Ferrell, R. J. & Dewitte-Avina, S. N. (2003). The temporal dynamics of the fourteenth-century Black Death: new evidence from English ecclesiastical records. Hum Biol 75, 427–448. [DOI] [PubMed] [Google Scholar]