Abstract
Among pathogenic strains of Listeria monocytogenes, the σB transcription factor has a pivotal role in the outcome of food-borne infections. This factor is activated by diverse stresses to provide general protection against multiple challenges, including those encountered during gastrointestinal passage. It also acts with the PrfA regulator to control virulence genes needed for entry into intestinal lumen cells. Environmental and nutritional signals modulate σB activity via a network that operates by the partner switching mechanism, in which protein interactions are controlled by serine phosphorylation. This network is well characterized in the related bacterium Bacillus subtilis. A key difference in Listeria is the presence of only one input phosphatase, RsbU, instead of the two found in B. subtilis. Here, we aim to determine whether this sole phosphatase is required to convey physical, antibiotic and nutritional stress signals, or if additional pathways might exist. To that end, we constructed L. monocytogenes 10403S strains bearing single-copy, σB-dependent opuCA–lacZ reporter fusions to determine the effects of an rsbU deletion under physiological conditions. All stresses tested, including acid, antibiotic, cold, ethanol, heat, osmotic and nutritional challenge, required RsbU to activate σB. This was of particular significance for cold stress activation, which occurs via a phosphatase-independent mechanism in B. subtilis. We also assayed the effects of the D80N substitution in the upstream RsbT regulator that activates RsbU. The mutant had a phenotype consistent with low and uninducible phosphatase activity, but nonetheless responded to nutritional stress. We infer that RsbU activity but not its induction is required for nutritional signalling, which would enter the network downstream from RsbU.
INTRODUCTION
Some strains of Listeria monocytogenes cause food-borne infections with high mortality among susceptible human populations, and the σB general stress transcription factor contributes to this pathogenicity in two ways (reviewed by Sleator et al., 2009). First, its action enhances survival of L. monocytogenes during food processing, and may contribute to subsequent growth in refrigerated, minimally processed foods. Second, σB is important at every stage of food-borne infection, including gastrointestinal passage with its multiple stresses, crossing the intestinal epithelium, and later intracellular events that promote replication and spread to deeper tissues. Notably, key virulence genes are co-controlled by σB and the PrfA regulator, both in batch-grown cells and during gastrointestinal infection of mice (Hain et al., 2008; Kazmierczak et al., 2003; Milohanic et al., 2003; Toledo-Arana et al., 2009). Example genes include bsh, which contributes to bile salt resistance; argR, the product of which controls expression of the arginine deiminase acid stress genes; and inlA, needed for attachment and entry into intestinal lumen cells. Consistent with the role of σB in redirecting transcription during intestinal passage (Toledo-Arana et al., 2009), loss of σB activity decreases virulence in guinea pigs infected via the gastrointestinal route, but not intravenously (Garner et al., 2006). Because the stress responses controlled by σB form part of an integrated suite of systems that govern the interaction of Listeria with diverse environments (Chaturongakul et al., 2008; Sleator et al., 2009), it is important to understand the signals and pathways that modulate σB activity.
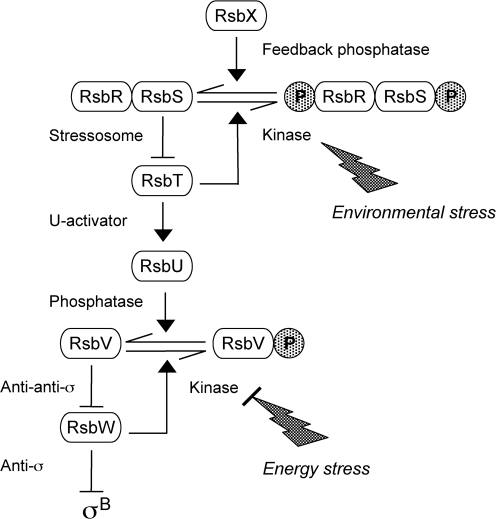
Within the order Bacillales, to which L. monocytogenes belongs, σB activity is controlled at the post-translational level by a signalling network that operates via the partner switching mechanism (for reviews see Hecker et al., 2007; Price, 2010). This mechanism relies on serine and threonine phosphorylation to manage interactions among a conserved set of regulatory proteins. Partner switching has been extensively studied in the related bacterium Bacillus subtilis, in which it controls the general stress factor σB and the sporulation factor σF (see Igoshin et al., 2007, and references therein). Given the strong similarities between the regulators of the two organisms (Hecker et al., 2007; Pané-Farré et al., 2005; Price, 2010), the model of the L. monocytogenes network shown in Fig. 1 is derived from the B. subtilis σB example.
Fig. 1.
Model of the σB signalling network in L. monocytogenes, derived from that of the B. subtilis network (see text), but with only one input phosphatase, RsbU. The large stressosome complex is simplified to show only the core components RsbR, RsbS and RsbT. We present evidence that environmental (or physical) stresses enter the network upstream from RsbU. These stresses are thought to induce RsbT kinase activity within the stressosome, leading to phosphorylation of the RsbR and RsbS antagonists; this phosphorylation frees RsbT to bind and activate RsbU by direct protein–protein interaction. By contrast, our data suggest that energy (or nutritional) stresses enter the network downstream from RsbU; these stresses would diminish RsbW kinase activity as ATP levels fall. In this scheme, the RsbX feedback phosphatase has no direct role in stress response but returns the system to its ground state following an environmental stress.
In this model, the primary regulator is the RsbW anti-σ factor, and its association with σB is governed by the phosphorylation state of the RsbV anti-anti-σ. This state reflects the balance between the serine kinase activity of RsbW and the serine phosphatase activity of RsbU. In unstressed cells, the RsbV anti-anti-σ is phosphorylated and is unable to interact with RsbW, which then sequesters σB in an inactive complex. When the RsbU phosphatase is activated by stress, dephosphorylated RsbV binds RsbW to force the release of σB. RsbW in essence switches partners, alternatively binding RsbV or σB.
Stress activation of the RsbU phosphatase relies on a second partner switch, in which the RsbT activator alternates binding its RsbU target or a multi-protein stressosome complex, depending on the phosphorylation state of the complex. RsbT is a paralogue of RsbW, and it possesses kinase activity toward its RsbR and RsbS substrates within the stressosome, which are paralogues of RsbV. In unstressed cells, the stressosome binds RsbT, preventing its interaction with the RsbU phosphatase. By an as yet unknown mechanism, stress leads to activation of the RsbT kinase, phosphorylation of the stressosome and release of RsbT, which then functions as a positive regulatory subunit of the RsbU phosphatase.
This model is consistent with genetic, biochemical and structural analysis of the B. subtilis signalling network (Hecker et al., 2007; Price, 2010), but many of its features remain to be tested in L. monocytogenes. Furthermore, the B. subtilis network has two input phosphatases that are differentially required for response to energy (RsbP) or environmental stress (RsbU), as well as a third, phosphatase-independent pathway responsible for σB induction following chill stress (Brigulla et al., 2003). It is not yet clear whether these different classes of stress all activate L. monocytogenes σB via its single RsbU phosphatase (Glaser et al., 2001) or whether additional signalling pathways remain to be discovered. The only studies to address this issue used cell viability or RT-PCR to assay the impact of an rsbT null allele on network performance (Chaturongakul & Boor, 2004, 2006). Moreover, these prior studies often relied on 5 min of centrifugation to shift unstressed cells to a stressed growth condition. This treatment places cells in an uncertain physiological state and also induces an indeterminate stress that activates σB even in control cultures (Chan et al., 2007; Chaturongakul & Boor, 2006). Thus, previous results, although providing useful information, are qualitative in nature and do not allow evaluation of key network alterations.
Here, we apply stresses directly to exponentially growing L. monocytogenes cells and indirectly assay σB activity by means of a reporter fusion stably integrated into the chromosome at an ectopic site. We use this approach to determine the phenotypes of a large, in-frame deletion mutation lacking RsbU phosphatase activity, and of a missense mutation predicted to abolish the kinase activity of RsbT while leaving partner binding unaffected. The results support a model in which response to environmental stress requires induction of the RsbU phosphatase, and that an additional input for energy stress enters the network downstream from RsbU.
METHODS
Bacterial strains and genetic methods.
L. monocytogenes 10403S and its derivatives are detailed in Table 1; the plasmids used for construction of these strains are detailed in Table 2. Standard recombinant DNA methods and electroporation were used as described by Sambrook et al. (1989), and conjugal transfer of plasmids was as described in Lauer et al. (2002). Strains were grown in brain heart infusion (BHI; Difco) at 37 °C unless specified otherwise. Chloramphenicol (10 μg ml−1), streptomycin (200 μg ml−1) or kanamycin (30 μg ml−1) was incorporated as needed.
Table 1.
L. monocytogenes strains used in this study
| Strain | Genotype | Reference or construct* |
|---|---|---|
| 10403S | Wild-type | Portnoy et al. (1988) |
| PL001 | rsbUΔ1 | pLJH2→10403S |
| PL002 | rsbT1 (RsbTD80N) | pLJH1→10403S |
| PL013 | tRNAArg : : pLJH4 | pLJH4→10403S |
| PL014 | rsbT1 tRNAArg : : pLJH4 | pLJH4→PL002 |
| PL015 | rsbUΔ1 tRNAArg : : pLJH4 | pLJH4→PL001 |
| PL016 | sigB : : km | LMA2B (Becker et al., 1998) |
| PL017 | sigB : : Km tRNAArg : : pLJH4 | pLJH4→PL016 |
*Arrow indicates transformation from donor to recipient.
Table 2.
Plasmid constructs
| Plasmid | Primer or fragment used* | Construction† |
|---|---|---|
| pLJH1 (rsbT1) | T-1 5′ TGCGGATCCATTCCAGGAAGATTTGC 3′ | pKSV7 BamHI–PstI |
| T-2 5′ GGACCTTTGTTTTTGGCAAC 3′ | ||
| T-3 5′ GTTGCCAAAAACAAAGGTCC 3′ | ||
| T-4 5′ TTCCTGCAGAGCAATTTCACTACGTA 3′ | ||
| pLJH2 (rsbUΔ1) | U-1 5′ CTCGGATCCACGTTTAGCGGAGTAC 3′ | pKSV7 BamHI–PstI |
| U-2 5′ CACTTCTTCTTATTTAGTTTTTTCCATTTTTTTCTAGCT 3′ | ||
| U-3 5′ ACTAAATAAGAAGAAGTGAAG 3′ | ||
| U-4 5′ AGGCTGCAGAGGAAGTTGTAATGTAA 3′ | ||
| pLJH3 | F 5′ CTTCCTGCAGAATGTAACACTGTCTTT 3′ | pPL2 PstI–BamHI |
| R 5′ TTTGGATCCTTTATAAGTCTTCGTTAC 3′ | ||
| pLJH4 | BglII–BamHI from pHK77, encoding spoVG–lacZ fusion | pLJH3 SacI–BamHI |
| pLJH5 (rsbRA) | F 5′ GAAAGGATCCGATGTATAAAGATTTTGCA 3′ | pET-15b BamHI–BlpI |
| R 5′ CCCGCTCAGCTCACCCCTCTTTTTCTAC 3′ | ||
| pLJH6 (rsbS) | F 5′ GGTGGATCCTGTGGGGATACCAATCTTA 3′ | pET-15b BamHI–BlpI |
| R 5′ GAATGCTCAGCTCATTCCCCCAATTCCTG 3′ | ||
| pLJH7 (rsbT) | F 5′ GGGGGATCCCATGACATTCCAATCCTGT 3′ | pET-15b BamHI–BlpI |
| R 5′ CTAGCTCAGCTTACCGAACCCATTTCGT 3′ |
*Restriction endonuclease sites used for cloning are denoted by underlined italic type.
†Vector and sites used: pKSV7 (Smith & Youngman, 1992); pPL2 (Lauer et al., 2002); pET15b (Novagen).
The missense mutations in rsbT and the in-frame deletion in rsbU were made by using a four-primer method of site-directed mutagenesis (Ho et al., 1989); these were then substituted for the wild-type alleles by a two-step replacement procedure (Camilli et al., 1993). For the rsbT1 missense allele, we altered the 80th triplet to encode asparagine instead of aspartate (D80N). For the rsbUΔ1 allele, we fused two fragments (−514 to +9 and +997 to +1530 relative to the ATG initiation triplet) to create a 987 bp deletion that removed triplets 4–332. These PCR products were cloned into the BamHI and PstI sites of pUC19, and the constructs were confirmed by sequencing. The BamHI–PstI fragments were then subcloned into the pKSV7 vector (Smith & Youngman, 1992) to make pLJH1 (rsbT1) and pLJH2 (rsbUΔ1). The constructs were electroporated into L. monocytogenes 10403S, then integrated into the chromosome, excised and cured (Camilli et al., 1993). Strains bearing the desired rsbT1 or rsbUΔ1 alleles were identified among the chloramphenicol-sensitive colonies by amplifying and sequencing the region of interest.
To provide an indirect assay for σB activity, we constructed a reporter fusion in the pPL2 vector, which integrates into the L. monocytogenes chromosome at the phage PSA attachment site (Lauer et al., 2002). The resultant pLJH4 plasmid carries an operon fusion between the σB-dependent opuCA promoter of L. monocytogenes (Chan et al., 2007; Kazmierczak et al., 2003) and a lacZ reporter. As the first step of this construction, the opuCA promoter region (−127 to +38 relative to the GTG initiation triplet) was cloned into the PstI and BamHI sites of pPL2 to make pLJH3. To ensure efficient translation in L. monocytogenes, a fused spoVG–lacZ gene was taken from pHK77 (Kim et al., 2006). pHK77 was digested with BglII, blunted with the Klenow fragment (New England Biolabs), then digested with BamHI to release spoVG–lacZ. This fragment was subcloned into the blunted SacI and overhanging BamHI sites of pLJH3 to yield pLJH4.
pLJH4 was electroporated into Escherichia coli SM10 (Simon et al., 1983) and was transferred by conjugation with recipient L. monocytogenes 10403S (wild-type), PL001 (rsbUΔ1), PL002 (rsbT1) and PL016 (sigB : : km), essentially as described by Lauer et al. (2002). In our modified procedure, 500 μl of donor and recipient cultures were mixed and the cells were collected by centrifugation rather than by filtration. The pellet was spread on a BHI plate and incubated for 2 h at 30 °C. Cells were harvested in 2.5 ml BHI and aliquots were spread directly onto BHI plates containing streptomycin (200 μg ml−1) and chloramphenicol (7.5 μg ml−1); these plates were then incubated as described by Lauer et al. (2002). pLJH4 integrates within a tRNAArg gene, reconstituting it in the process; this ectopic site is distant from the opuCA region, which remains as per the wild-type. For each strain, the tRNAArg region of integration and the eight gene sigB operon were sequenced to ensure the genetic manipulations introduced no unwanted alterations.
Purification of L. monocytogenes RsbR, RsbS and RsbT.
Plasmids encoding wild-type or mutant Rsb proteins in the pET15b vector (Novagen) were transformed into E. coli BL21 (DE3). Expression was induced by IPTG and proteins were purified under native conditions on nickel affinity columns (Qiagen), according to the manufacturer's protocol. Concentrations were determined by using the Protein Assay Reagent (Bio-Rad Laboratories).
In vitro kinase assays.
Phosphorylation assays were performed in a 100 μl reaction volume containing kinase assay buffer [50 mM Tris/HCl (pH 7.6), 50 mM KCl, 10 mM MgCl2, 1 mM DTT and 0.1 mM EDTA], 2 μM unlabelled ATP and 5 μCi [γ-32P]ATP. Purified RsbR or RsbS substrate was included at 20 μM final concentration, and wild-type or mutant RsbT kinase was included at 0.2 μM. Reactions were initiated by the addition of the protein substrate and incubated at 30 °C. For qualitative assays, 15 μl samples were removed after 30 min and the reactions were terminated by addition of 4 μl 5× sample loading buffer. Samples were heated at 100 °C for 5 min and separated on 15 % SDS-polyacrylamide gels; these were stained with Coomassie blue, dried and exposed to X-ray film. For quantitative assays, 10 μl samples were removed at the indicated times and the reactions were terminated by addition of 2.5 μl 5× sample loading buffer. Samples were heated, separated on gels, stained and dried as described for the qualitative assays, but here the amount of [32P]-labelled RsbR or RsbS was determined by using a Molecular Dynamics Storm imaging system and ImageQuaNT 4.1 software under volume quantification mode (GE Healthcare Bio-Sciences).
Stress experiments and β-galactosidase accumulation assays.
For all stresses tested, cells were grown and assayed in at least two independent experiments; representative results are shown. Strains were first grown to mid-exponential phase in shake flasks containing 1/10 volume of BHI broth at 37 °C, unless otherwise indicated. Cultures were then diluted 1 : 25 into fresh BHI medium and allowed to grow to early exponential phase before stress was imposed. Growth was monitored by using a Klett–Summerson colorimeter with red transmission filter. In all stress experiments, growth continued throughout the sampling period.
For environmental stress, NaCl (0.5 M final concentration), ethanol (4 % final, v/v) or glacial acetic acid (pH 5.3 final) was added to growing cultures. For envelope stress, vancomycin (2 μg ml−1 final), bacitracin (20 μg ml−1 final) or nisin (150 μg ml−1 final) was added (Sigma). The nisin concentration was higher than anticipated given the MIC of about 4 μg ml−1 determined by Mota-Meira et al. (2000), who used pure nisin A in their experiments, but is in line with the amounts employed by Begley et al. (2006), who used the same commercial preparation as here. This preparation contained 2.5 % nisin in skimmed milk powder with an unspecified amount of salt. Stock nisin solutions were prepared in 0.02 M HCl and clarified by centrifugation in a microfuge for 15 min at 6000 g. Control experiments showed the small amount of added HCl had no effect on pH of the medium or on fusion activity. For heat stress, cells were shifted from 25 to 42 °C, and for cold stress from 37 to 7 °C. For energy stress, carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) dissolved in ethanol was added to a final concentration of 10 μM. An equivalent amount of ethanol lacking CCCP was added to control cultures.
Samples were collected at the times indicated and assayed for β-galactosidase activity largely as described by Miller (1972), but with modification for L. monocytogenes. It was important not to cool samples on ice, a treatment that induces the general stress response. Cells were washed with Z buffer (Miller, 1972) at room temperature, collected in a microfuge after centrifugation at 6000 g for 1 min at room temperature, then resuspended in Z buffer. Cells were permeablized by vortexing for 30 s in the presence of SDS and chloroform, then incubated at 28 °C with o-nitrophenyl β-d-galactopyranoside substrate. Reactions were stopped by adding 0.5 ml of 1 M Na2CO3, and the mixes were centrifuged to remove cellular interference before reading absorbance at 420 nm (A420). Protein levels were determined by using the Bio-Rad Protein Assay reagent. Activity was defined as ΔA420×1000 min−1 (mg protein)−1.
RESULTS
A D80N substitution significantly reduces RsbT kinase activity in vitro
According to the model shown in Fig. 1, diverse environmental stresses trigger the RsbT kinase to phosphorylate the RsbR and RsbS stressosome components, freeing RsbT to bind and activate the RsbU phosphatase. Active RsbU then elicits the general stress response by removing the serine phosphate from the RsbV anti-anti-σ factor. We tested this model by assaying the effects of two mutations. One was an in-frame deletion in rsbU (rsbUΔ1) that would abrogate RsbU phosphatase activity. The other was a missense alteration in rsbT (rsbT1) predicted to affect kinase function specifically. This D80N substitution in the predicted G1 box of RsbT (Kang et al., 1998) should render RsbU phosphatase activity uninducible by environmental stress but would leave a residual phosphatase activity in mutant cells, thus providing a useful contrast with the rsbU null strain.
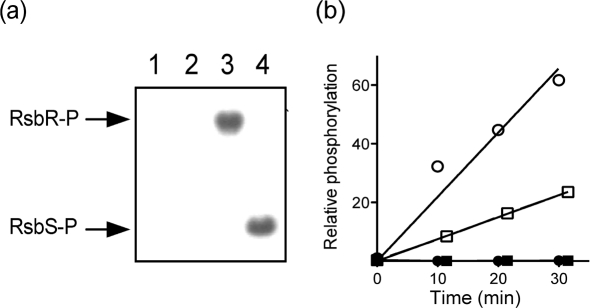
To determine the effect of the D80N substitution on kinase activity, we performed in vitro assays of wild-type and mutant RsbT enzymes by using their postulated RsbR and RsbS substrates. Purified wild-type RsbT readily transferred the γ-labelled phosphate from ATP to either RsbR or RsbS (Fig. 2a), as predicted by the model. By contrast, in the quantitative assay, the purified D80N form of RsbT failed to manifest detectable transfer (Fig. 2b), indicating that this substitution decreased activity to less than 1 % of the wild-type. Based on the properties of the analogous D78N substitution of B. subtilis (Kang et al., 1998), we presume that D80N does not significantly alter interaction of RsbT with either of its alternative binding partners, the RsbU phosphatase or the RsbR–RsbS stressosome complex. We therefore used a strain encoding this substitution at its rsbT locus to probe the role of kinase activity in vivo. In accordance with our expectation that the mutant would retain residual phosphatase activity, in subsequent assays (see below) the level of reporter expression in unstressed cells was indeed low, but was nonetheless consistently higher than that of the rsbU null mutant.
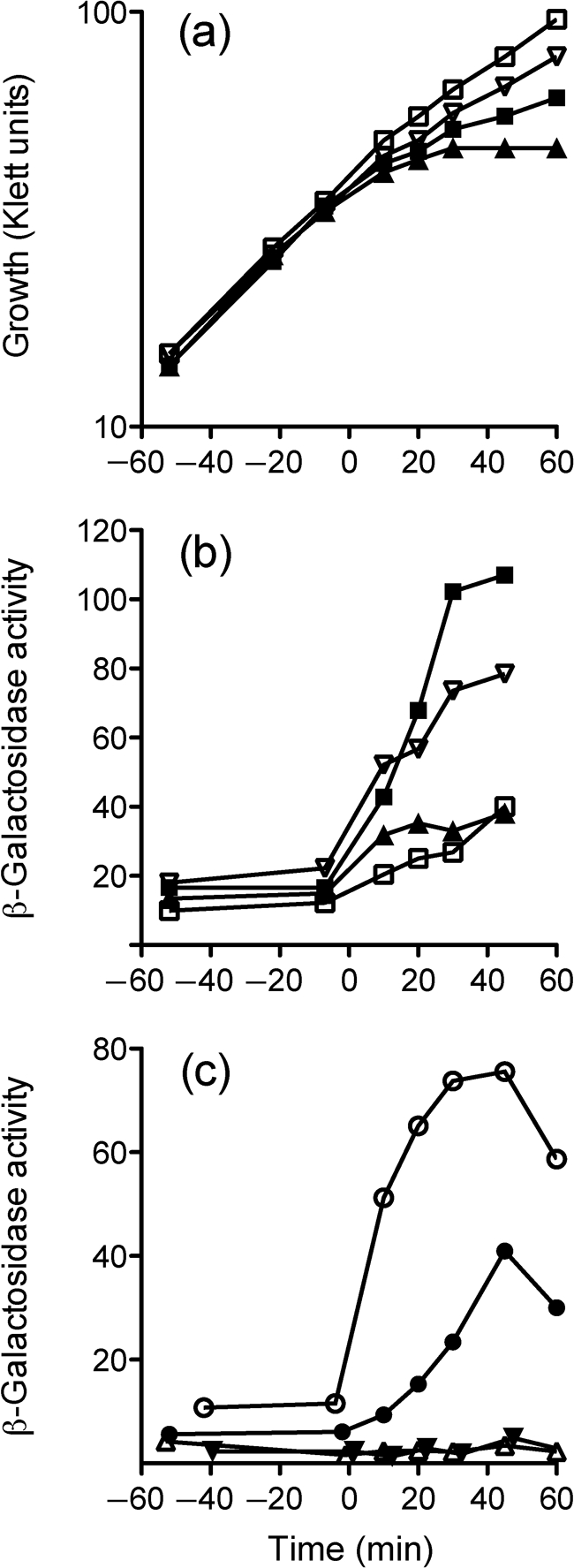
Fig. 2.
The D80N alteration significantly decreases RsbT kinase activity in vitro. (a) Purified proteins were mixed with [γ-32P]ATP and incubated as shown: lane 1, RsbT alone; lane 2, RsbR+RsbS; lane 3, RsbR+RsbT; lane 4, RsbS+RsbT. Reactions were stopped at 30 min, and proteins were then separated by PAGE and exposed to X-ray film. Arrows on the left indicate RsbR or RsbS mobility. (b) Purified RsbT (open symbols) or RsbTD80N (closed symbols) was incubated with RsbR (circles) or RsbS (squares) in the presence of [γ-32P]ATP. Samples were taken at the indicated times, separated by PAGE and analysed by phosphoimaging.
RsbT and RsbU are required for full opuCA induction in response to acid, ethanol and salt stress
We used an opuCA–lacZ reporter fusion to assay σB activity indirectly under physiological conditions. This is a well-characterized σB-dependent promoter in L. monocytogenes, and the fragment we chose renders lacZ expression completely σB-dependent under the growth conditions used. It was specifically designed to remove the upstream σB-independent promoter that is activated in response to cold shock, and that also contributes to opuCA expression at higher temperatures (Chan et al., 2007). We compared lacZ expression in the wild-type with that in sigB null, rsbUΔ1 null and rsbT1 (D80N) mutants as different stresses were applied to actively growing cells. We first tested the effect of salt, ethanol or acid addition, which are environmental stresses that are present in some foods or found during gastrointestinal passage.
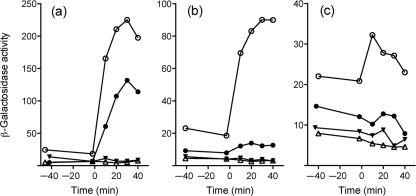
Addition of salt to exponentially growing cells caused a rapid, ten-fold increase in reporter gene activity (Fig. 3a), whereas ethanol (Fig. 3b) or acid addition (Fig. 3c) elicited more modest increases. These increases were absent in the sigB and rsbUΔ1 null mutants, indicating complete dependence on σB and the RsbU phosphatase. The rsbT1 alteration had a similar deleterious effect in most of the experiments reported here and below. By contrast, in the salt stress experiment the rsbT1 alteration decreased response by only a factor of two relative to the wild-type (Fig. 3a). This result could imply the existence of an RsbT-independent pathway of σB activation in response to salt. However, another explanation is that the mutant RsbT kinase might regain partial activity in vivo under conditions of osmotic stress. Missense mutations with this phenotype are designated osmoremedial and are common in bacteria and yeasts (Csonka, 1989). Clearly, the basis of this rsbT1 phenotype warrants further investigation. Nonetheless, from these assays we conclude that RsbU phosphatase activity is required for the environmental stress response, and that RsbT kinase activity contributes to the induction of that response.
Fig. 3.
RsbU phosphatase and RsbT kinase activities are required for full induction of opuCA–lacZ by salt, ethanol or acid stress. (a) NaCl (0.5 M final concentration), (b) ethanol (4 % final, v/v) or (c) acid (pH 5.3 final) stress was imposed on exponentially growing cells at time 0, and β-galactosidase accumulation was assayed at the indicated times. ○, Wild-type strain (PL13); •, rsbT1 (PL14); ▵, sigB : : km (PL17); ▾, rsbUΔ1 (PL15).
RsbT and RsbU are required for induction in response to wall-acting antibiotics
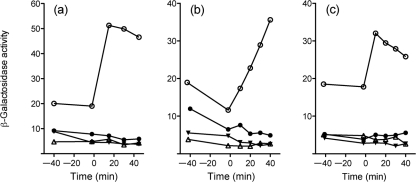
Salt, ethanol and acid stress can affect cell envelope function. Likewise, the antibiotics vancomycin and bacitracin act by inhibiting different steps of the undecaprenyl cycle in the cytoplasmic membrane, thus disrupting cell-wall synthesis (Bugg & Walsh, 1992). Vancomycin binds to Lipid II on the outer leaflet of the membrane, preventing pentapeptide cross-linking, and bacitracin inhibits dephosphorylation of undecaprenyl pyrophosphate. Because these antibiotics are found in the soil environment and are known to activate B. subtilis σB moderately in vivo (Cao et al., 2002; Mascher et al., 2003), we tested their effects on expression of the opuCA–lacZ reporter fusion. Addition of sublethal levels of vancomycin or bacitracin induced fusion expression two- to threefold, and this induction was absent in the sigB null, rsbUΔ1 null and rsbT1 mutants (Fig. 4a, b).
Fig. 4.
RsbU and RsbT are required for opuCA–lacZ induction by wall-acting antibiotics. (a) Vancomycin (2 μg ml−1 final concentration), (b) bacitracin (20 μg ml−1 final) or (c) nisin (150 μg ml−1 final) was added to exponentially growing cells at time 0. ○, Wild-type strain (PL13); •, rsbT1 (PL14); ▵, sigB : : km (PL17); ▾, rsbUΔ1 (PL15).
Nisin is an important lantibiotic used in food preservation. Like vancomycin, nisin binds to Lipid II and inhibits cell-wall synthesis, but its additional pore-forming ability has potent effects on membrane integrity as well (Wiedemann et al., 2001). L. monocytogenes σB appears to have a role in nisin resistance, although there is some disagreement regarding its contribution (Begley et al., 2006; Palmer et al., 2009). We therefore investigated whether nisin influenced σB activity. Addition of nisin led to a twofold induction of opuCA–lacZ fusion expression (Fig. 4c), and this induction was dependent on σB and its upstream regulators. In this experiment, we cannot rule out an effect of the salt present in commercial nisin preparations, and this salt cannot be separated easily from the small nisin peptide. However, in contrast with the salt stress experiment shown in Fig. 3(a), here we note that the rsbT1 mutant responds much like the sigB and rsbUΔ1 null mutants. From this, we speculate that the induction shown in Fig. 4(c) largely reflects the effect of nisin addition, and the contaminating salt has little effect. We conclude that vancomycin and bacitracin (and possibly nisin) induce σB activity, and this induction is dependent on RsbU phosphatase and RsbT kinase activity.
RsbT and RsbU are required for induction in response to heat and cold stress
Heat and cold are additional stresses that L. monocytogenes encounters in the food environment, and both stresses are known to induce σB activity (Becker et al., 1998). Heat or cold induced reporter fusion activity at least three- to fourfold in wild-type, and no induction was apparent in the sigB null, rsbUΔ1 null or rsbT1 mutants (Fig. 5). This lack of induction was not due to a growth defect, because the mutant strains grew as well as the wild-type under both heat and cold stress conditions (data not shown). The three- to fourfold induction noted in the wild-type may be an underestimate. In the heat stress experiment shown in Fig. 5(a), peak induction was defined solely by the 10 min time point. In the cold stress experiment shown in Fig. 5(b), induction was still increasing when the experiment was terminated at 40 h, before stationary phase induction could occur.
Fig. 5.
RsbU and RsbT are required for opuCA–lacZ induction by heat or cold. (a) Heat stress was imposed at time 0 by shifting exponentially growing cells from 25 to 42 °C. (b) Cold stress was imposed by a shift from 37 to 7 °C at time 0. ○, Wild-type strain (PL13); •, rsbT1 (PL14); ▵, sigB : : km (PL17); ▾, rsbUΔ1 (PL15).
These experiments confirm the work of Becker et al. (1998) and extend their study to show that both heat and cold induction strongly depend upon RsbU phosphatase and RsbT kinase activity. This dependence parallels the B. subtilis network with regard to the transmission of heat stress signals, which are known to require RsbT and RsbU (Akbar et al., 1997; Voelker et al., 1995). However, it is notably different with regard to the transmission of cold stress signals, which are phosphatase-independent in B. subtilis (Brigulla et al., 2003).
RsbT and RsbU are required for full response to energy stress
L. monocytogenes lacks the RsbP phosphatase that B. subtilis requires for energy-stress activation of σB (Vijay et al., 2000). Nonetheless, L. monocytogenes is reported to induce σB following energy stress generated by glucose starvation, or by an uncoupler of oxidative phosphorylation (Chaturongakul & Boor, 2006). However, in these previous experiments, the physiological state of the cells was uncertain and few time points were taken after stress was imposed. We therefore performed time-course experiments on wild-type and mutant cells bearing the opuCA–lacZ reporter in order to determine the magnitude of the response.
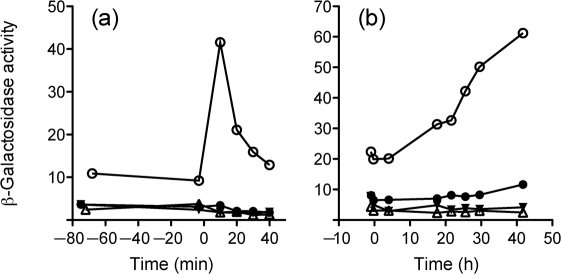
To establish a defined energy stress, we used CCCP, an uncoupler of oxidative phosphorylation that discharges the pH gradient (Harold, 1972). For the initial experiments, we added CCCP to wild-type cells growing exponentially in BHI medium, at final concentrations of 1–200 μM. The results of one such experiment are shown in Fig. 6(a, b). Here, increasing the concentration of CCCP from 5 to 25 μM had an increasing impact on growth (Fig. 6a), and the highest reporter induction was observed at 10 μM (Fig. 6b). Induction was significantly reduced at 25 μM, which we attributed to decreased biosynthetic capability in the energy-starved cells. In other experiments, CCCP concentrations above 25 μM had even more severe effects on growth (data not shown). Based on these results, we chose 10 μM CCCP for the experiment detailed in Fig. 6(c). Here, the reporter fusion was induced seven- to eightfold in the wild-type, and this induction was absent in the rsbUΔ1 and sigB null mutants. These mutants also had a very low level of basal (or pre-induction) expression in exponentially growing cells. By contrast, the rsbT1 mutant manifested a seven- to eightfold induction, similar to the wild-type; coupled with its intermediate level of basal expression, this resulted in decreased maximum expression. We conclude that CCCP is an effective inducer of σB activity, and that this induction requires RsbU phosphatase activity. Based on our results with the rsbT1 mutant, RsbT kinase activity does not significantly affect the fold induction in response to energy stress, but does contribute to setting the basal level of σB activity in unstressed cells.
Fig. 6.
RsbU but not RsbT is required for opuCA–lacZ induction in response to energy stress. (a) Growth of wild-type strain PL13 following addition of the CCCP uncoupler at time 0. □, CCCP control (0 μM), containing 0.25 % ethanol; ▿, 5 μM CCCP; ▪, 10 μM CCCP; ▴, 25 μM CCCP. (b) Induction of opuCA–lacZ fusion activity in the wild-type cultures shown in (a). (c) CCCP (10 μM) was added to exponentially growing cells at time 0. ○, Wild-type strain (PL13); •, rsbT1 (PL14); ▵, sigB : : km (PL17); ▾, rsbUΔ1 (PL15).
DISCUSSION
Single-copy lacZ reporter fusions integrated at an ectopic site possess several advantages for regulatory studies. These include the ability to isolate the regulatory region of interest from the influence of nearby promoters and other cis-acting sites, while leaving the wild-type region intact; the capacity to study promoter activity in actively growing cells; and a facile assay for this activity. Despite these advantages, it appears that such lacZ fusions have been rarely used in L. monocytogenes (Kim et al., 2006; Shin et al., 2010). This may reflect a limitation of the technology, which requires genetic manipulation to create each fusion. Thus, for studies involving the expression of multiple genes, RNA-based approaches have been the methods of choice. For our purposes here, however, use of a single opuCA–lacZ reporter at an ectopic site revealed previously unknown features of the σB signalling network.
Our results are consistent with the model for σB regulation shown in Fig. 1, and extend it in two new ways. First, we show that the single RsbU phosphatase of L. monocytogenes is necessary for response to all stresses tested, including cold stress. This phosphatase dependence stands in sharp contrast with the B. subtilis network, in which cold activation can occur in the absence of both input phosphatases and the RsbV anti-anti-σ (Brigulla et al., 2003). Thus, the cold signalling pathways must differ between the two organisms. Second, although the RsbU phosphatase of L. monocytogenes is required for full response to energy stress, the phenotype of the rsbT1 allele implies that the signal of energy depletion enters the system at a point downstream from RsbU, as explained below.
In B. subtilis the RsbP phosphatase has a sensory PAS domain and is required for energy stress response, but the question of its regulatory contribution has yet to be resolved (Brody et al., 2009; Vijay et al., 2000). RsbP is more active than RsbU in unstressed cells growing in rich medium, and thus sets the basal level for σB by countering the action of the RsbW serine kinase (Eymann & Hecker, 2001). Notably, RsbW has a Km for ATP in the physiological range, and its activity is thought to decrease as cells undergo energy depletion (Delumeau et al., 2002). It is not presently known if RsbP itself is regulated by energy stress, or if it instead provides a fixed activity that overtakes RsbW activity as ATP levels fall. In this latter case, RsbP could respond to another signal, perhaps unrelated to energy stress, that serves to adjust the basal level for σB (Brody et al., 2009). It may thus be unnecessary to have a dedicated phosphatase such as RsbP in order to efficiently respond to energy stress.
The data reported here are compatible with a model in which the single RsbU phosphatase of L. monocytogenes counters the action of the RsbW kinase and is therefore required for the energy stress response, but does not itself modulate that response. As shown in Fig. 6(c), the rsbUΔ1 mutant, in which RsbU activity is absent, cannot respond to energy stress. By contrast, the rsbT1 mutant, in which RsbU activity is predicted to be low as well as refractory to signals normally entering via RsbT, still responds. Moreover, the fold induction of the response is essentially the same in both the wild-type and the rsbT1 mutant. We infer from these results that a signal sufficient for the energy stress response enters the network downstream from RsbT, and probably downstream from RsbU. The simplest explanation is that – as is the case for B. subtilis – L. monocytogenes RsbW has a Km for ATP in the physiological range and can sense energy depletion. Partner switching kinases belong to the GHLK superfamily and are related to bacterial histidine protein kinases (Dutta & Inouye, 2000). Notably, such kinases that function in general stress networks (RsbW-like) differ from their cousins that function in sporulation networks (SpoIIAB-like) in the G1 box, which is thought to comprise part of the nucleotide binding fold. RsbW kinases manifest an imperfect box (DxGxS88 in B. subtilis and L. monocytogenes) whereas SpoIIAB kinases have a canonical box (DxGxG85 in B. subtilis). This may to some extent explain the five-log difference in Km for ATP: 0.9 mM for RsbW versus 4.4 nM for SpoIIAB (Delumeau et al., 2002; Ho et al., 2003). Thus, the ability to sense energy depletion may be a general property of RsbW kinases.
On the other hand, ATP levels do not fall during environmental stress in B. subtilis (Voelker et al., 1995), so the activity of the RsbU phosphatase must increase to activate the general stress response. We suggest that a similar situation occurs in L. monocytogenes. All environmental responses tested, including acid, antibiotic, cold, ethanol, heat and salt, were abolished in the mutant bearing the rsbU deletion, and all but one were significantly diminished in the mutant bearing the rsbT D80N alteration. The sole exception – salt stress – was reduced but not eliminated by this alteration. We do not yet know if this partial dependence represents an RsbT-independent route of activation, such as an effect of salt stress on energy metabolism, which would enter the system via RsbW, or possibly a recovery of RsbT kinase activity in the mutant under conditions of osmotic stress. Nonetheless, our results suggest that all the other environmental stress signals – and at least a substantial portion of the salt stress signal – enter the network upstream from RsbT, probably via the large, cytoplasmic stressosome.
In B. subtilis the in vitro-assembled stressosome is a 1.8 MDa complex containing 40 monomers of the RsbR co-antagonist and 20 of the RsbS antagonist, which together form a core that can bind 20 RsbT monomers (Marles-Wright et al., 2008). When purified from cell extracts, the complex contains four additional paralogues of the RsbR family, which are presumed to interchange freely with RsbR in vivo (Delumeau et al., 2006; Gaidenko et al., 2006; Kim et al., 2004). Although the external signals that activate the environmental response are known, the molecular nature of the intracellular signal (or signals) sensed by the stressosome remains unknown. Significantly, the L. monocytogenes genome encodes stressosome components similar to those found in B. subtilis, including multiple RsbR paralogues (Losi, 2004; Pané-Farré et al., 2005). It is therefore likely that the internal activating signals are similar in the two organisms. Based on the ability of the antibiotics vancomycin and bacitracin to induce σB activity in L. monocytogenes, we speculate that the stressosome directly or indirectly monitors envelope function as a means of detecting diverse physical stresses, such as heat, salt and ethanol challenge, and that cold stress signals may also be detected via a similar mechanism. Because growth at refrigerator temperatures constitutes the principal threat of food-borne Listeria infection, the mechanism by which cold signals enter the partner switching network is of considerable practical significance.
Acknowledgments
We thank Daniel Portnoy and Richard Calendar for providing L. monocytogenes 10403S, E. coli SM10, pPL2 and pKSV7; A. L. Sonenshein for pHK77; and Andrew Benson for L. monocytogenes LMA2B. This research was supported by a grant from the Western Institute of Food Safety and Security and by Public Health Service grant GM42077 from the National Institute of General Medical Sciences.
Abbreviations
BHI, brain–heart infusion
CCCP, carbonyl cyanide m-chlorophenylhydrazone
References
- Akbar, S., Kang, C. M., Gaidenko, T. A. & Price, C. W. (1997). Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol 24, 567–578. [DOI] [PubMed] [Google Scholar]
- Becker, L. A., Cetin, M. S., Hutkins, R. W. & Benson, A. K. (1998). Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol 180, 4547–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley, M., Hill, C. & Ross, R. P. (2006). Tolerance of Listeria monocytogenes to cell envelope-acting antimicrobial agents is dependent on SigB. Appl Environ Microbiol 72, 2231–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigulla, M., Hoffmann, T., Krisp, A., Völker, A., Bremer, E. & Völker, U. (2003). Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J Bacteriol 185, 4305–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody, M. S., Stewart, V. & Price, C. W. (2009). Bypass suppression analysis maps the signalling pathway within a multidomain protein: the RsbP energy stress phosphatase 2C from Bacillus subtilis. Mol Microbiol 72, 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg, T. D. & Walsh, C. T. (1992). Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep 9, 199–215. [DOI] [PubMed] [Google Scholar]
- Camilli, A., Tilney, L. G. & Portnoy, D. A. (1993). Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol 8, 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, M., Wang, T., Ye, R. & Helmann, J. D. (2002). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol Microbiol 45, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Chan, Y. C., Boor, K. J. & Wiedmann, M. (2007). σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol 73, 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul, S. & Boor, K. J. (2004). RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol 70, 5349–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul, S. & Boor, K. J. (2006). σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol 72, 5197–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul, S., Raengpradub, S., Wiedmann, M. & Boor, K. J. (2008). Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol 16, 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka, L. N. (1989). Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev 53, 121–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delumeau, O., Lewis, R. J. & Yudkin, M. D. (2002). Protein–protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J Bacteriol 184, 5583–5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delumeau, O., Chen, C. C., Murray, J. W., Yudkin, M. D. & Lewis, R. J. (2006). High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 188, 7885–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, R. & Inouye, M. (2000). GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25, 24–28. [DOI] [PubMed] [Google Scholar]
- Eymann, C. & Hecker, M. (2001). Induction of σB-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol Lett 199, 221–227. [DOI] [PubMed] [Google Scholar]
- Gaidenko, T. A., Kim, T. J., Weigel, A. L., Brody, M. S. & Price, C. W. (2006). The blue-light receptor YtvA acts in the environmental stress signaling pathway of Bacillus subtilis. J Bacteriol 188, 6387–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, M. R., Njaa, B. L., Wiedmann, M. & Boor, K. J. (2006). Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun 74, 876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser, P., Frangeul, L., Buchrieser, C., Rusniok, C., Amend, A., Baquero, F., Berche, P., Bloecker, H., Brandt, P. & other authors (2001). Comparative genomics of Listeria species. Science 294, 849–852. [DOI] [PubMed] [Google Scholar]
- Hain, T., Hossain, H., Chatterjee, S. S., Machata, S., Volk, U., Wagner, S., Brors, B., Haas, S., Kuenne, C. T. & other authors (2008). Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold, F. M. (1972). Conservation and transformation of energy by bacterial membranes. Bacteriol Rev 36, 172–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker, M., Pané-Farré, J. & Völker, U. (2007). SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol 61, 215–236. [DOI] [PubMed] [Google Scholar]
- Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Ho, M. S., Carniol, K. & Losick, R. (2003). Evidence in support of a docking model for the release of the transcription factor σF from the antisigma factor SpoIIAB in Bacillus subtilis. J Biol Chem 278, 20898–20905. [DOI] [PubMed] [Google Scholar]
- Igoshin, O. A., Brody, M. S., Price, C. W. & Savageau, M. A. (2007). Distinctive topologies of partner-switching signaling networks correlate with their physiological roles. J Mol Biol 369, 1333–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C. M., Vijay, K. & Price, C. W. (1998). Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol Microbiol 30, 189–196. [DOI] [PubMed] [Google Scholar]
- Kazmierczak, M. J., Mithoe, S. C., Boor, K. J. & Wiedmann, M. (2003). Listeria monocytogenes σB regulates stress response and virulence functions. J Bacteriol 185, 5722–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. J., Gaidenko, T. A. & Price, C. W. (2004). A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J Mol Biol 341, 135–150. [DOI] [PubMed] [Google Scholar]
- Kim, H. J., Mittal, M. & Sonenshein, A. L. (2006). CcpC-dependent regulation of citB and lmo0847 in Listeria monocytogenes. J Bacteriol 188, 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer, P., Chow, M. Y., Loessner, M. J., Portnoy, D. A. & Calendar, R. (2002). Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184, 4177–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi, A. (2004). The bacterial counterparts of plant phototropins. Photochem Photobiol Sci 3, 566–574. [DOI] [PubMed] [Google Scholar]
- Marles-Wright, J., Grant, T., Delumeau, O., van Duinen, G., Firbank, S. J., Lewis, P. J., Murray, J. W., Newman, J. A., Quin, M. B. & other authors (2008). Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322, 92–96. [DOI] [PubMed] [Google Scholar]
- Mascher, T., Margulis, N. G., Wang, T., Ye, R. W. & Helmann, J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol Microbiol 50, 1591–1604. [DOI] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Milohanic, E., Glaser, P., Coppée, J. Y., Frangeul, L., Vega, Y., Vázquez-Boland, J. A., Kunst, F., Cossart, P. & Buchrieser, C. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47, 1613–1625. [DOI] [PubMed] [Google Scholar]
- Mota-Meira, M., LaPointe, G., Lacroix, C. & Lavoie, M. C. (2000). MICs of mutacin B-Ny266, Nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob Agents Chemother 44, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, M. E., Wiedmann, M. & Boor, K. J. (2009). σB and σL contribute to Listeria monocytogenes 10403S response to the antimicrobial peptides SdpC and nisin. Foodborne Pathog Dis 6, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pané-Farré, J., Lewis, R. J. & Stülke, J. (2005). The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J Mol Microbiol Biotechnol 9, 65–76. [DOI] [PubMed] [Google Scholar]
- Portnoy, D. A., Jacks, P. S. & Hinrichs, D. J. (1988). Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167, 1459–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, C. W. (2010). General stress response in Bacillus subtilis and related Gram positive bacteria. In Bacterial Stress Responses, 2nd edn, in press. Edited by Storz, G. & Hengge, R.. Washington, DC. : American Society for Microbiology.
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Shin, J.-H., Kim, J., Kim, S.-M., Kim, S., Lee, J.-C., Ahn, J.-M. & Cho, J.-Y. (2010). σB-dependent protein induction in Listeria monocytogenes during vancomycin stress. FEMS Microbiol Lett 308, 94–100. [DOI] [PubMed] [Google Scholar]
- Simon, R., Priefer, U. & Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1, 784–791. [Google Scholar]
- Sleator, R. D., Watson, D., Hill, C. & Gahan, C. G. (2009). The interaction between Listeria monocytogenes and the host gastrointestinal tract. Microbiology 155, 2463–2475. [DOI] [PubMed] [Google Scholar]
- Smith, K. & Youngman, P. (1992). Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74, 705–711. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., Loh, E., Gripenland, J., Tiensuu, T. & other authors (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. [DOI] [PubMed] [Google Scholar]
- Vijay, K., Brody, M. S., Fredlund, E. & Price, C. W. (2000). A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol Microbiol 35, 180–188. [DOI] [PubMed] [Google Scholar]
- Voelker, U., Voelker, A., Maul, B., Hecker, M., Dufour, A. & Haldenwang, W. G. (1995). Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol 177, 3771–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, I., Breukink, E., van Kraaij, C., Kuipers, O. P., Bierbaum, G., de Kruijff, B. & Sahl, H. G. (2001). Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276, 1772–1779. [DOI] [PubMed] [Google Scholar]