Abstract
Mycobacterium tuberculosis serine/threonine protein kinases (STPKs) are key regulators of growth and metabolism; however, evidence for their roles in virulence is limited. In a preliminary screen based on comparative expression between strains H37Rv and H37Ra, six STPK genes, pknD, pknG, pknH, pknJ, pknK and pknL, showed higher expression in H37Rv. In the second screen, STPK expression was analysed in H37Rv-infected human macrophages. Interestingly, significant expression of pknK was detected only at 18 h post-infection, suggesting its involvement in early infection events. We have investigated the roles of PknK in vitro and in vivo. PknK levels were induced under stationary phase and deletion of pknK resulted in increased resistance of the mutant to acidic pH, hypoxia, oxidative and stationary-phase stresses in vitro. These results, together with the increased survival of the ΔpknK strain during persistent infection in mice, reveal a role for PknK in adaptive mechanisms that slow the growth of mycobacteria. A novel finding of this study was the inhibition of growth of ΔpknK strain during acute infection in mice that correlated with the significant upregulation of tumour necrosis factor as well as the simultaneous downregulation of interleukin-12p40, interferon-γ and induced nitric oxide synthase transcripts. Finally, we provide evidence for the localization of PknK during infection and discuss its implications in pathogenesis.
INTRODUCTION
Historically, tuberculosis has been a serious infectious disease, with significant adverse effects on the human population, and it remains so even today. A hallmark of the disease is the ability of Mycobacterium tuberculosis to persist in the host for years and to reactivate under conditions of immune suppression. The situation is worsened by the increasing incidence of multi-drug-resistant strains. It is therefore imperative to identify novel M. tuberculosis antigens/targets for the development of new effective anti-tubercular drugs and vaccines.
The success of M. tuberculosis in establishing a niche inside the host resides primarily in its adaptability to the macrophage environment and evasion of the host defence system. Recent studies on phospho-serine/threonine/tyrosine protein signalling in bacteria have established a role for these proteins as key components of distinct, but well-coordinated, signal transduction pathways regulating host–pathogen interactions and intracellular survival (Cozzone, 2005; Zhang et al., 1992). These kinases are capable of transducing subtle changes in the environment, both intracellular and extracellular, by the rapid phosphorylation of several cellular proteins, specifically on serine/threonine or tyrosine residues. M. tuberculosis possesses 11 ‘eukaryotic-like’ STPKs (PknA–L) and one tyrosine kinase (Av-Gay & Everett, 2000; Bach et al., 2009; Cole et al., 1998). Of these, nine are transmembrane proteins and are classified in three groups (PknA/B/L, PknF/I/J, PknD/E/H), while the other two, PknG and PknK, are soluble proteins that closely resemble the eukaryotic STPKs (Av-Gay & Everett, 2000; Narayan et al., 2007).
All mycobacterial STPKs have been biochemically characterized, mostly by in vitro studies, in terms of their kinase activity and the identification of cognate target proteins (Chao et al., 2009; Greenstein et al., 2005; Jang et al., 2010). Several studies have established that mycobacterial STPKs regulate a variety of metabolic processes, including cell shape and cell division (Chaba et al., 2002; Dasgupta et al., 2006, Kang et al., 2005, 2008; Thakur & Chakraborti, 2006, 2008), sugar uptake (Deol et al., 2005), metabolic enzymes (Molle et al., 2006; Veyron-Churlet et al., 2009), nitrogen homeostasis (Cowley et al., 2004; O'Hare et al., 2008), transcription (Cohen-Gonsaud et al., 2009; Greenstein et al., 2007; Kumar et al., 2009; Park et al., 2008; Sharma et al., 2006) and biofilm formation (Gopalaswamy et al., 2008). Despite such extensive research, very little is known about STPK expression and function during M. tuberculosis growth and survival in human macrophages. Besides PknG (Cowley et al., 2004), only PknH and PknI (Gopalaswamy et al., 2009; Papavinasasundaram et al., 2005) have been implicated in M. tuberculosis survival, adaptation and growth in vivo. PknG is of particular interest since the ectopic expression of Mtb- or BCG-PknG in Mycobacterium bovis BCG, or non-pathogenic mycobacteria, promotes survival by inhibiting phagosome–lysosome fusion (Scherr et al., 2009; Tiwari et al., 2009; Walburger et al., 2004); however, this is yet to be demonstrated in M. tuberculosis. The possibility of mycobacterial STPKs interacting with host signalling proteins is intriguing and will have a profound impact on our understanding of the mechanisms of mycobacterial virulence, persistence, and reactivation.
This study aimed to identify candidate STPK genes that may be involved in the survival of mycobacteria in human macrophages, and thus may contribute to the pathogenesis of the tubercle bacillus. Using a transcriptomic approach in two separate preliminary screens, we analysed the differential expression of the 11 STPK genes in M. tuberculosis H37Rv (virulent) versus H37Ra (avirulent) strains and during H37Rv infection in human macrophages. The screen identified six STPK genes, two of which encode less well-characterized STPKs (PknK and L) that showed higher levels of expression in H37Rv compared to H37Ra and were also expressed during infection. Because of its close resemblance to eukaryotic kinases and its high expression during exponential growth of H37Rv in culture and at 18 h post-infection in human macrophages, we were specifically interested in investigating the role of PknK in mycobacterial pathogenesis. Our results implicate PknK in the regulation of M. tuberculosis growth under in vitro stress conditions. Further, we elucidate a role for PknK during infection in mice and present evidence that PknK not only plays a key role during early events of infection by modulating host immunity, but also modulates the growth of M. tuberculosis during the persistent phase of infection. These findings identify PknK as a component of mycobacterial signalling pathways regulating early and late events in tuberculosis infection and have implications in host–pathogen interactions.
METHODS
Bacterial strains, plasmids and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains JM109 and HB101 were used as host strains for all molecular manipulations. M. tuberculosis H37Rv and H37Ra strains were analysed for differences in STPK gene expression. Mycobacterium smegmatis mc2155 was used for the isolation and propagation of recombinant mycobacteriophage for mutagenesis of pknK.
Table 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Description | Source/reference |
|---|---|---|
| Strains | ||
| M. tuberculosis H37Rv | Virulent laboratory strain | ATCC* |
| M. tuberculosis H37Ra | Avirulent laboratory strain | CWRU† |
| M. smegmatis mc2155 | ept-1 | AECOM‡ |
| E. coli HB101 | F–, Δ(gpt-proA)62 leuB6 glnV44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20(Strr) xyl-5 mtl-1 recA13 thi-1 | UCSF§ |
| E. coli JM109 | e14− (McrA−) recA1 endA1 gyrA96 thi-1hsdR17 supE44 relA1 Δ(lac-proAB) | Stratagene |
| E. coli BL21(DE3) | F–, ompT hsdSB ( |
Invitrogen |
| LIX11 | H37Rv ΔpknK | This study |
| LIX16 | LIX11 : : pYA1580 | This study |
| Plasmids | ||
| pGEM-T Easy | AmpR, TA cloning vector for PCR products | Promega |
| pET SUMO | KanR, 6xHis-SUMO expression vector | Invitrogen |
| pYUB854 | HygR, cosmid vector used in construction of the phasmid carrying the allelic-exchange substrate | Bardarov et al. (2002) |
| pMV306 | KanR, mycobacterial integrating vector | Stover et al. (1991) |
| pSH276 | Rv0195 gene in pQE40ΔDHFR | Haydel & Clark-Curtiss (2004) |
| pSH337 | clpC gene in pGEM-T | Haydel & Clark-Curtiss (2004) |
| pSH317 | devR gene in pQE40ΔDHFR | Haydel & Clark-Curtiss (2004) |
| pSH281 | Rv2027c gene in pQE40ΔDHFR | Haydel & Clark-Curtiss (2004) |
| pSH334 | regX3 gene in pZERO/Km | Haydel & Clark-Curtiss (2004) |
| pYA1620 | pknA gene (C-term) in pGEM-T Easy | This study |
| pYA1537 | pknB gene in pGEM-T Easy | This study |
| pYA1538 | pknD gene in pGEM-T Easy | This study |
| pYA1539 | pknE gene in pGEM-T Easy | This study |
| pYA1540 | pknF gene in pGEM-T Easy | This study |
| pYA1541 | pknG gene in pGEM-T Easy | This study |
| pYA1621 | pknH gene (N-term) in pGEM-T Easy | This study |
| pYA1542 | pknI gene in pGEM-T Easy | This study |
| pYA1543 | pknJ gene in pGEM-T Easy | This study |
| pYA1544 | pknK gene (N-term) in pGEM-T Easy | This study |
| pYA1545 | pknK gene (C-term) in pGEM-T Easy | This study |
| pYA1546 | pknL gene in pGEM-T Easy | This study |
| pYA1556 | Full-length pknK gene in pET SUMO | This study |
| pYA1619 | 3′ and 5′ flanking regions of pknK gene cloned in the multiple cloning sites of pYUB854 | This study |
| pYA1580 | pknK gene+300 bp upstream region cloned in XbaI/HindIII sites in pMV306 | This study |
*ATCC no. 25618, American Type Culture Collection.
†ATCC no. 25177, obtained from Dr Richard F. Silver, Case Western Reserve University, OH, USA.
‡Dr William R. Jacobs, Jr, Albert Einstein College of Medicine, NY, USA.
§Dr Herbert Boyer, University of California, San Francisco, CA, USA.
E. coli cultures were grown in Luria–Bertani (LB) broth or on LB agar plates at 37 °C. M. tuberculosis cultures were cultivated in Middlebrook 7H9 medium (Difco) supplemented with 0.05 % Tween 80 (Tw) and ADS [0.5 % albumin/0.2 % dextrose (glucose)/0.085 % saline] (henceforth referred to as 7H9-Tw-ADS), or on Middlebrook 7H10 agar plates at 37 °C. M. smegmatis cultures were grown at 37 °C in Middlebrook 7H9 or LB medium supplemented with 0.05 % Tween 80 or on LB agar plates. For stress studies, M. tuberculosis cultures (OD600 ∼0.2–0.4) were harvested, washed with 7H9 basal medium, resuspended in 7H9-Tw-ADS medium whose pH had been adjusted to 5.5 (acidic stress) or spiked with 5 mM hydrogen peroxide (oxidative stress), and grown aerobically. For hypoxia studies, M. tuberculosis cultures were grown in Dubos Tween-Albumin medium (Difco) to OD600 ∼0.6, diluted to starting OD600 ∼0.05 and subjected to hypoxia in sealed tubes as described previously (Saini et al., 2004). Antibiotics were used at the following concentrations (μg ml−1): ampicillin (Amp), 100; kanamycin (Kan), 25; hygromycin (Hyg), 150 or 100 for E. coli and mycobacteria, respectively. Primer sequences used in this study are listed in Supplementary Table S1, available with the online version of this paper.
RNA isolation and quantitative RT-PCR (qRT-PCR).
RNA isolation and qRT-PCR were performed as described previously (Malhotra et al., 2009). RNA (100 ng) was reverse-transcribed into cDNA to serve as the template in a real-time PCR using gene-specific primers (Supplementary Table S1) and SYBR green dye (Bio-Rad). Expression analyses were performed with RNA from three independent experiments. For each experiment, the calculated cycle threshold (Ct) of the test gene was normalized to the Ct of the internal control 16S rRNA gene (amplified from the same samples), before calculating the fold change of the test samples using the iQ5 expression software (Bio-Rad).
SCOTS analysis.
For analysis of STPK gene expression during H37Rv infection of human macrophages, selective capture of transcribed sequences (SCOTS) (Graham & Clark-Curtiss, 1999) was performed. Previously developed SCOTS-generated cDNA mixtures (Graham & Clark-Curtiss, 1999) were used as probes for hybridization with STPK genes. Briefly, all STPK genes were amplified and cloned into pGEM-T Easy vector (Promega) to generate plasmids pYA1537 through pYA1546, pYA1620 and pYA1621 (Table 1). For pknA, pknH and pknK genes, fragments encoding C- and N-terminal regions were amplified separately. Equal amounts of experimental and control plasmid DNA (pSH276, pSH337, pSH317, pSH281 and pSH334) (Haydel & Clark-Curtiss, 2004) were digested with appropriate enzymes, resolved on an agarose gel and transferred onto nylon membranes for hybridization with DIG-labelled cDNA probe mixtures. Detection was done using anti-DIG antibody conjugated with alkaline phosphatase and the CDP-Star chemiluminescent substrate (Roche). Based on intensity values, the expression of genes was designated low (+), moderate (++/+++) or high (++++).
Purification of PknK, generation of polyclonal anti-PknK antibodies and immunoblot analysis.
The pknK ORF (Rv3080c, 3333 bp) was amplified and cloned into pET SUMO expression vector (Invitrogen) to generate plasmid pYA1556. Subsequently, pYA1556 was transformed into E. coli BL21(DE3) to induce expression. Recombinant PknK with an N-terminal 6xHis-SUMO tag was purified from IPTG-induced cultures by affinity chromatography using the ProBond Ni-Chelating resin (Invitrogen) according to the manufacturer's instructions.
For antibody generation, 100 μg of purified PknK protein was emulsified with Freund's incomplete adjuvant (Sigma) and injected subcutaneously into two New Zealand White rabbits. Rabbits were given two booster injections at 3-week intervals and sera were collected 10 days after the second boost. Equal amounts of M. tuberculosis cell lysates, prepared as described previously (Malhotra et al., 2009), were resolved on a 6 or 10 % SDS-PAGE gel, transferred onto a nitrocellulose membrane and immunoblotted with anti-PknK antibodies (1 : 2500). Detection was done using anti-rabbit IgG-HRP (1 : 10 000, Sigma) and LumiGLO chemiluminescent substrate (KPL).
Construction of ΔpknK mutant and complemented mutant strains of M. tuberculosis.
The pknK (Rv3080c) gene was deleted in M. tuberculosis H37Rv and replaced with a Hyg resistance cassette by allelic exchange using the specialized transducing mycobacteriophage system as previously described (Bardarov et al., 2002). The 5′ and 3′ flanking regions of the pknK gene were amplified and cloned into pYUB854 (Bardarov et al., 2002) to create pYA1619. pYA1619 and phAE87 phage DNA were digested with PacI, ligated and packaged using a λGigaPack III packaging extract (Stratagene) followed by transduction into E. coli HB101. The phasmid DNA from the transductants was electroporated into M. smegmatis mc2155 and plated for mycobacteriophage plaques at 30 °C. A high-titre phage lysate, prepared from one temperature-sensitive phage plaque, was used to infect H37Rv as described previously (Bardarov et al., 2002). Hygromycin-resistant colonies were selected and confirmed by Southern hybridization. The resulting H37Rv ΔpknK mutant strain was designated LIX11. For complementation of the mutant, the pknK gene plus the 300 bp upstream sequence, containing the putative pknK promoter region, was cloned into the integrating vector pMV306 (Stover et al., 1991) to generate pYA1580 and electroporated into LIX11 electrocompetent cells. The complemented strain was designated LIX16.
Electron microscopy.
For scanning electron microscopy (SEM), colonies of M. tuberculosis H37Rv and LIX11 from Middlebrook 7H10 agar plates were fixed (2 % glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.2), washed and treated with 1 % osmium tetroxide. The cells were dehydrated in acetone and coated with gold before scanning by a 360FE scanning electron microscope (Leica-Cambridge) at 10 kV accelerating voltage. The cells were measured using the software EDS 2006 (IXRF Systems). At least 75 measurements in different samples were made to determine the size of the respective bacilli; means±se were calculated. For transmission electron microscopy (TEM), cells from exponential-phase cultures of wild-type H37Rv and LIX11, from two independent experiments, were fixed with glutaraldehyde, embedded in Spurr's epoxy resin according to standard microscopy methods and stained with 1 % aqueous uranyl acetate and lead citrate before observation under a Philips CM12 transmission electron microscope.
In vitro growth determinations.
For comparison of the in vitro growth rates, wild-type H37Rv, and strains LIX11 and LIX16, were cultured aerobically at 37 °C and growth was monitored in 7H9-Tw-ADS medium for 30 days, in acid/oxidative stress medium for 5 days or in sealed tubes for 20 days (in the case of hypoxia). Serial dilutions of the cultures at various time points were plated on Middlebrook 7H10 agar plates and incubated at 37 °C for 3 weeks before quantification of c.f.u.
Low-dose-aerosol mouse infection studies.
All animal experiments and procedures were conducted according to protocols approved by the Arizona State University Institutional Animal Care and Use Committee. Mycobacterial stocks for infection were cultured in 7H9-Tw-ADS medium and quantified by assessment of c.f.u. prior to infection. For preparation of the inoculum, each stock was passed through a 30G needle at least 10 times to obtain a single-cell suspension. Bacterial suspensions of strains H37Rv, LIX11 and LIX16 thus prepared were diluted in PBS and used to infect 6- to 8-week old female C57BL/6 mice (Charles River laboratories) with ∼100–200 c.f.u./lung. Aerosol infections were performed essentially as described previously (Kelly et al., 1996), using a Glas-Col Airborne infection apparatus. In two separate, but identically conducted, experiments, growth of M. tuberculosis strains was monitored from day 1 to 4 weeks post-infection (acute phase, experiment 1) and between 4 and 12 weeks (chronic phase, experiment 2). At day 1 post-infection, lung tissues were collected from two mice in each group to verify the exact infectious dose. At specific time points, five animals per strain were euthanized by carbon dioxide asphyxiation to assess growth and survival of M. tuberculosis strains during infection in lungs and spleens. The cardiac lobe of the lungs was dissected, kept in RNAlater (Ambion) at 4 °C for subsequent RNA extraction and the remaining lung was homogenized and assessed for c.f.u.
Cytokine measurement in lung tissues.
Lung tissues from four mice infected with the wild-type or the LIX11 mutant were pooled and processed for RNA isolation as described by Malhotra et al. (2009). Total RNA (1 μg) was converted to cDNA and real-time PCR with SYBR green was used to measure the expression of interleukin (IL)-12p40, tumour necrosis factor (TNF)-α, interferon (IFN)-γ and inducible nitric oxide synthase (iNOS) using primer sequences and parameters as described previously (Gonzales & Orlando, 2008; Overbergh et al., 1999). β-Actin mRNA was used as an internal normalization control and fold change of the transcripts in LIX11-infected mice was determined relative to the expression in H37Rv-infected mice.
In vitro and in vivo localization of PknK in M. tuberculosis.
PknK localization was determined in vitro by Western blot analysis of the whole-cell lysate and the cell wall, cell membrane, culture filtrate, cytosolic and soluble cell wall protein fractions of M. tuberculosis. In vivo localization of PknK was done using immunoelectron microscopy (IEM). Peripheral blood was obtained from healthy human subjects specifically for research. This research was conducted according to a protocol approved by the Institutional Review Board of Arizona State University and permission was granted by all subjects. Human macrophage infections were performed as described elsewhere (Graham & Clark-Curtiss, 1999; Hou et al., 2002), except that macrophages were grown attached to Thermanox coverslips (Nunc) and association with bacteria was allowed for 4 h. On day 1 post-infection, cells on the coverslips were fixed with 2 % paraformaldehyde and 0.5 % glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) and prepared for electron microscopy as described previously (Dasgupta et al., 2000). For immunolabelling, the sections were blocked in buffer [0.1 M sodium phosphate (pH 7.0), 0.15 M sodium chloride and 1 % BSA], followed by incubation with anti-PknK antibody (1 : 20) overnight at 4 °C. After washing, the grids were incubated with anti-rabbit IgG conjugated to 20 nm gold particles (1 : 100; MP Biomedical) and stained for TEM.
Statistical analysis.
Statistical analyses were done using two-way ANOVA with Bonferroni post-tests whereever applicable. Significance of differences between the mutant and wild-type bacilli in animal studies was assessed by Student's t-test. All tests were done using GraphPad Prism 5.0 software. Differences with a P-value <0.05 were considered significant.
RESULTS
Expression profiling of M. tuberculosis STPK genes in culture and during infection in human macrophages
Using qRT-PCR, we analysed the differential expression of STPK genes in exponential-phase cultures of H37Rv and H37Ra. The basal level of expression of STPK genes in H37Rv and H37Ra is shown in Supplementary Fig. S1. For each gene, the ratio of its expression in H37Rv versus H37Ra was calculated and is listed in Table 2. Six STPK genes (pknD, pknG, pknH, pknJ, pknK and pknL) were expressed at higher levels (ratio >1) in H37Rv compared to H37Ra, with the difference being significant (P<0.01) for pknD, pknK and pknL (Table 2, bold type).
Table 2.
M. tuberculosis STPK gene expression in virulent and avirulent strains during exponential growth and in H37Rv-infected human macrophages, and published data on expression in macrophages
The six STPK genes (pknD, pknG, pknH, pknJ, pknK and pknL) that were expressed at higher levels (ratio >1) in H37Rv compared with H37Ra are shown in bold type.
| Rv no. | Gene | Expression in broth cultures by qRT-PCR H37Rv/H37Ra* | Expression in H37Rv-infected human PBMC-derived macrophages, determined by SCOTS† at: | Expression during growth in macrophages (published data)‡ | ||
|---|---|---|---|---|---|---|
| 18 h | 48 h | 110 h | ||||
| Rv0015c | pknA | 0.82 | + | − | − | Upregulation in THP1 cells (Singh et al., 2006) |
| Rv0014c | pknB | 0.76 | ++++ | ++++ | ++++ | Constitutive expression in murine alveolar and THP1 macrophages (Av-Gay et al., 1999; Singh et al., 2006) |
| Rv0931c | pknD | 2.4 (P<0.001) | +++ | ++++ | +++ | nd |
| Rv1743 | pknE | 0.9 | +++ | − | − | nd |
| Rv1746 | pknF | 0.71 | ++++ | − | ++++ | nd |
| Rv0410c | pknG | 1.27 | − | ++++ | ++++ | nd |
| Rv1266c | pknH | 1.2 | − | + | − | nd |
| Rv2914c | pknI | 0.72 | − | − | − | Decreased expression in THP1 cells (Singh et al., 2006) |
| Rv2088 | pknJ | 1.1 | + | − | − | nd |
| Rv3080c | pknK | 1.5 (P<0.001) | ++++ | − | − | nd |
| Rv2176 | pknL | 1.6 (P<0.01) | ++++ | ++++ | +++ | nd |
*Expressed as a ratio of H37Rv/H37Ra. P-values are given where difference is significant.
†Expression of genes was designated low (+), moderate (++/+++) or high (++++), based on intensity values. −, No expression.
‡ND, No data.
Given the limitations in obtaining considerable amounts of both human macrophages and bacterial RNA from infected human macrophages, expression profiling of STPK genes in human macrophages was done using SCOTS-purified cDNA probes, previously made in our laboratory from H37Rv-infected macrophages at 18 h, 48 h and 110 h post-infection (Graham & Clark-Curtiss, 1999). As seen in Table 2, transcripts of all STPK genes except pknI were detected during infection. Transcripts specific to pknB, pknD and pknL were constitutively expressed at all time points studied, while others exhibited differential expression. The expression of clpC, Rv2027c, regX3, devR and Rv0195 served as controls as described previously (Haydel & Clark-Curtiss, 2004) (data not shown). In Table 2, we have compiled the SCOTS results for STPK genes with current data on their expression in macrophages, and the lack of information on STPK gene expression in macrophages is strikingly evident. Based on the observations that pknK is expressed at significantly higher levels in H37Rv compared to H37Ra (Table 2, Supplementary Fig. S1, P<0.001) and only at 18 h of infection in human macrophages (Table 2), we hypothesized that PknK played a role in the early establishment of infection in macrophages. Further characterization of PknK was carried out to test this hypothesis.
Construction of ΔpknK and complemented mutant strains of M. tuberculosis
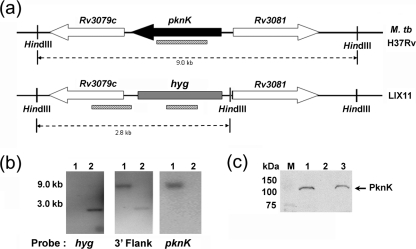
We constructed an M. tuberculosis H37Rv ΔpknK strain, designated here LIX11, through allelic exchange with a hygromycin-resistance gene, using the specialized transducing mycobacteriophage system (Bardarov et al., 2002). The genome organization of pknK (Rv3080c), its adjacent genes in the H37Rv chromosome and the changes in the mutant strain are illustrated in Fig. 1(a). The hygromycin-resistance cassette flanked by res sites replaced the pknK gene in a double-crossover event, resulting in complete deletion of the pknK gene. The mutant was verified by Southern hybridization using probes specific for the hygromycin gene, the 3′ flanking region of pknK and pknK itself (Fig. 1b). Allelic exchange in LIX11 was confirmed by the absence or presence of a hybridization signal with pknK- and hyg-specific probes, respectively (Fig. 1b). Deletion of pknK in LIX11 was complemented by transforming the mutant with pYA1580, which provides a single functional copy of pknK transcribed from a putative pknK promoter to generate the complemented strain LIX16. Western blot analysis confirmed the absence of a band corresponding to PknK in the LIX11 mutant (Fig. 1c, lane 2) and its presence in the wild-type and complemented mutant strains (Fig. 1c; lanes 1 and 3, respectively). The blot was stained with Ponceau S in order to confirm that an equal amount of protein was loaded and that the protein had not degraded (data not shown).
Fig. 1.
Construction of an M. tuberculosis ΔpknK strain by allelic replacement. (a) Schematic representation of the chromosomal locus of pknK and its adjacent genes in the wild-type H37Rv and the LIX11 mutant strain, in which the pknK gene is deleted and replaced with a hygromycin-resistance cassette. The striped bars indicate the position of the DNA probes used for Southern blot analysis and the dashed lines indicate the expected size of the hybridization with the specific probes. (b) Southern blot confirmation of the LIX11 mutant strain. Chromosomal DNA was isolated from strains H37Rv (lane 1) and LIX11 (lane 2), digested with HindIII and hybridized with probes specific for the hyg, pknK and 3′ flank regions. (c) Western blot analysis of PknK (∼119 kDa, arrow) in the whole-cell lysates of M. tuberculosis H37Rv (lane 1), LIX11 mutant (lane 2) and LIX16 complemented mutant (lane 3) strains. M, molecular size markers.
Deletion of pknK affects cell size and cell wall composition
The morphology of broth-grown wild-type and mutant bacilli was compared by SEM and TEM. Although no differences were noted in colony morphology on solid media, SEM analyses revealed that the mutant bacilli were significantly smaller in length (1.7±0.034 μm, n=75; P<0.001) compared to the wild-type (2.3±0.064 μm, n=75; n refers to the number of independent measurements). TEM cross-sections of exponentially grown wild-type and mutant cells were similar, except for the dense staining and contrast that was observed associated with the cell wall of wild-type cells but was absent in a majority of the mutant cells (arrows, Fig. 2). These results suggest that PknK function may affect cell size and cell wall composition in M. tuberculosis.
Fig. 2.
TEM analysis of M. tuberculosis H37Rv (a) and the LIX11 mutant strain (b), showing differences in the staining pattern of the cell wall. Wild-type H37Rv and the LIX11 mutant were cultivated in Middlebrook 7H9-Tw-ADS medium to exponential phase (OD600 0.3–0.4) and processed for electron microscopy in two separate experiments. One representative picture is shown. Scale bars, 0.2 μm. Arrows indicate dense granular staining present in the cell wall of the wild-type, but absent in LIX11.
PknK slows the growth of M. tuberculosis during stationary phase
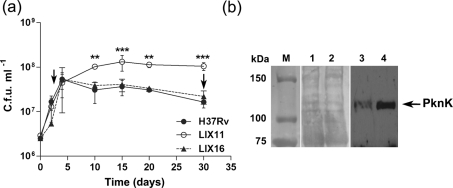
The growth characteristics of strains H37Rv, LIX11 and LIX16 were analysed by measuring viable cell counts during growth in broth cultures over a period of 30 days. All three strains grew similarly during the exponential phase; however, a significant increase in c.f.u. of the mutant bacilli was observed from day 10 onwards. At day 15, a 3.6-fold increase (P<0.001) in viable counts was determined for LIX11 compared to H37Rv (Fig. 3a). It appears that while the wild-type entered into stationary phase between days 5 and 10 and maintained a steady level of c.f.u. for the next 10 days, strain LIX11 did not show a typical entry into stationary phase, but continued to grow and maintained higher c.f.u. consistently through days 10 to 30 (Fig. 3a). The growth of LIX16 was similar to that of H37Rv, indicating that the phenotype of LIX11 was indeed due to the deletion of pknK. To correlate PknK production as a function of growth of H37Rv, we determined PknK levels by Western blot analysis of H37Rv cell lysates prepared from samples at days 3 and 30, representing exponential and late stationary phases, respectively. Equal amounts of total protein from day 3 and day 30 lysates, as seen in the Ponceau S stained blot (Fig. 3b, lanes 1 and 2, respectively), were analysed with anti-PknK antibody. PknK was significantly induced in lysates from late stationary phase (Fig. 3b, lane 4) compared to exponential phase (Fig. 3b, lane 3). It is apparent from these observations that induced levels of PknK in stationary phase either directly or indirectly activate cellular targets or pathways that slow the growth of mycobacteria.
Fig. 3.
Effect of pknK deletion on growth of M. tuberculosis in culture. (a) Aerobic growth of H37Rv wild-type, LIX11 mutant and LIX16 complemented strain in 7H9-Tw-ADS medium was monitored for 30 days by measuring viable cell counts. The results presented are the mean±sd from three independent growth experiments. ** and *** represent P<0.01 and P<0.001, respectively, for the differences between the mutant and wild-type strain at a particular time point. Arrows indicate the time points, representing exponential phase (day 3) and late stationary phase (day 30), respectively, during growth at which samples were taken and analysed for PknK protein levels. (b) Western blot analysis of PknK protein in H37Rv cell lysates at days 3 and 30. The Ponceau S stained blot shows equal loading of total protein from day 3 and 30 samples (lanes 1 and 2, respectively). PknK production is induced in stationary phase (lane 4) compared to exponential phase (lane 3). M, molecular size markers.
Based on these results, we also investigated growth and survival of M. tuberculosis strains H37Rv, LIX11 and LIX16 under in vitro stress conditions: acidic (pH 5.5), oxidative (5 mM hydrogen peroxide) and hypoxia (Supplementary Fig. S2). By day 5, LIX11 was significantly resistant, compared to wild-type H37Rv, in terms of c.f.u., to acidic pH (3.58-fold; P<0.05), 5 mM hydrogen peroxide (6.48-fold; P<0.001), and hypoxia (3.0-fold; P<0.001) (Supplementary Fig. S2). Though the fold changes seem modest, the increased resistance of the mutant to stress conditions examined in this study, including stationary-phase stress (Fig. 3a), was statistically significant and was obtained consistently over three independent experiments. The reversion of the phenotype to wild-type levels upon complementation of the mutant further supports our observations. Together, these results suggest a role for PknK in the regulation of cellular events that promote bacterial adaptation to environmental changes, a function that may be critical to survival in vivo.
Roles of PknK during acute and persistent M. tuberculosis infection in mice
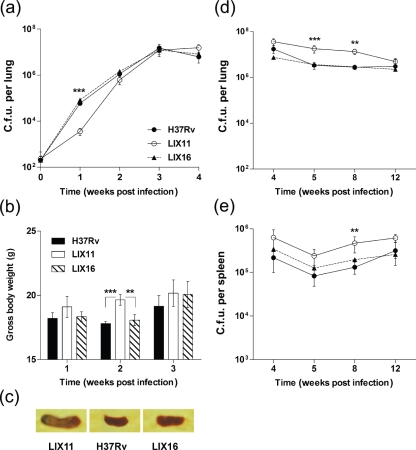
The relevance of PknK-mediated growth adaptation was evaluated in individual low-dose aerosol infections with strains H37Rv, LIX11 and LIX16 in C57BL/6 mice. Growth was measured between day 1 (deposition) and week 4 (peak of acute phase), and between weeks 4 and 12 (chronic phase) in two separate experiments. During the acute phase of infection, the intracellular growth of LIX11 was significantly inhibited at 1 week post-infection in mouse lungs by ∼16.5-fold (P<0.001) compared to H37Rv or LIX16 (Fig. 4a). However, by week 4, all three strains reached similar c.f.u. levels, indicating a function for PknK during early adaptive events in the host. To assess whether the lag in the growth of LIX11 at week 1 post-infection was due to inherent reduced infectivity of the mutant, we analysed uptake and intracellular growth of all three strains in an in vitro model of infection using human macrophages. No differences were observed between the three strains in their abilities to infect and survive in the macrophages (data not shown), suggesting that the phenotype of LIX11 in mice was specifically due to an effect of the in vivo environment. We also monitored the total body weight of infected animals as an index of inflammation and pathology. Mice infected with LIX11 exhibited a significant increase in gross body weights compared to the wild-type at both week 1 (5 %) and week 2 post-infection (10 %, P<0.001), but by week 3, all animals had similar weights (Fig. 4b). The ability of all the strains to disseminate to spleens was similar and occurred by week 2. Although spleens infected with LIX11 exhibited higher inflammation at week 3 post-infection (Fig. 4c), no differences in the viable counts of the three strains were detected in the spleens (data not shown). Thus, a defect during early growth in lungs specifically at week 1 and increased inflammation were the hallmarks of acute infection with the M. tuberculosis ΔpknK strain.
Fig. 4.
Growth and survival of M. tuberculosis strains H37Rv, LIX11 and LIX16 in immunocompetent mice. Virulence of the wild-type, LIX11 and LIX16 strains was assessed in aerosol-infected C57BL/6 mice during the acute phase of infection (experiment 1; day 1 through 4 weeks). (a) Viable counts in lungs. For the sake of clarity, day 1 c.f.u. levels are plotted as time=0. (b) Increase in gross body weight over time. Statistical differences between LIX11 versus wild-type or LIX16 strains at week 2 post-infection are shown. ** and *** denote P<0.01 and P<0.001 respectively. (c) Spleen inflammation observed at 3 weeks post-infection with all three strains. One representative picture is shown. (d, e) Growth during the chronic phase of infection (experiment 2; 4 to 12 weeks) was assessed by viable counts in lungs (d) and in spleens (e). Note that the initial day 1 c.f.u. levels were similar in both experiments. Data presented are the mean c.f.u. (±sd) obtained from five mice per group. ** and *** represent P<0.01 and P<0.001, respectively, for the differences in the growth of LIX11 compared to H37Rv in the lungs and spleens of infected animals.
In the second experiment, H37Rv grew exponentially in the lungs for 4 weeks as in experiment 1 (Fig. 4a), after which the c.f.u. levels decreased and stabilized, indicating a chronic or persistent infection (Fig. 4d). Approximately 5.3- and 4.8-fold higher c.f.u. (P<0.01) were recovered from LIX11-infected mice at 5 and 8 weeks post-infection, respectively, than from mice infected with H37Rv (Fig. 4d). In spleens, a similar increase of 3.56-fold (P<0.01) in viable counts of the mutant strain was observed at 8 weeks post-infection (Fig. 4e). In both experiments, the LIX16 complemented strain was able to restore the parental phenotype. Reproducible results from a third experiment, wherein c.f.u. levels were measured at weeks 1, 2, 3, 4 and 8 weeks post-infection, further confirmed our observations (data not shown). Collectively, these results indicate that PknK is required during early stages of infection in lungs and modulates the intracellular survival of mycobacteria during the chronic phase of infection in both lungs and spleens.
PknK modulates host immune effectors during early infection
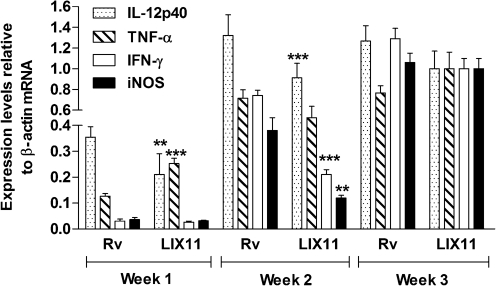
To decipher the role of PknK in early infection events, we compared the mRNA levels of key pro-inflammatory cytokines, IL-12p40, TNF-α, IFN-γ and iNOS, by qRT-PCR over 3 weeks in lung tissues pooled from groups of four mice infected with the wild-type or mutant strain. Transcript levels of TNF-α were twofold higher (P<0.001) at week 1 post-infection in LIX11-infected mice compared to the wild-type-infected mice (Fig. 5). In contrast, expression of IL-12p40, a key effector of Th1-mediated immunity, was 1.66-fold lower (P<0.01) in LIX11-infected mice (Fig. 5). At week 2, LIX11-infected mice were unable to elicit induction of IL-12p40, TNF-α, IFN-γ and iNOS to levels comparable to those obtained in H37Rv-infected mice (Fig. 5). In fact, the levels of IL-12p40, IFN-γ and iNOS were significantly repressed, by 1.47-, 3.6- and 3.4-fold, respectively, in the tissues from LIX11-infected mice (Fig. 5, P<0.01). By week 3, the expression levels of all genes except TNF-α were still lower in the LIX11-infected than in the H37Rv-infected mice; however, these differences were not significant. We also analysed cytokine levels in LIX16-infected animals and observed complementation only at week 1 post-infection (data not shown).We attribute the lack of complementation at other time points to possible pleiotropic effects of PknK deletion. These data suggest an immunomodulatory function of PknK during establishment of acute infection in mice.
Fig. 5.
Role of PknK in modulation of host immunity. The IL-12p40, TNF-α, IFN-γ and iNOS mRNA levels in mice infected with LIX11 or H37Rv were determined by qRT-PCR over 3 weeks post-infection in RNA extracted from lung tissues pooled from four infected mice in each group. Normalization of expression was done with β-actin mRNA. qRT-PCR analysis was done in triplicate and data are presented as means±sd from at least two independent cDNA preparations. ** and *** denote P<0.01 and P<0.001, respectively, for the differences in the expression of IL-12p40, TNF-α, IFN-γ and iNOS in the mutant-infected animals versus the H37Rv-infected mice at a given time point.
PknK associates with bacterial cell wall/membrane components and may be secreted during infection
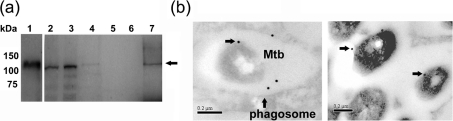
Based on our results implicating PknK in the regulation of M. tuberculosis growth and modulation of host immunity in vivo, we investigated the location of PknK in M. tuberculosis in vitro and within infected macrophages. Immunoblot analyses of subcellular fractions of M. tuberculosis revealed the presence of PknK in the cell wall and cell membrane fraction; however, the signal was very faint in the latter (Fig. 6a, lanes 3 and 4). We also detected the presence of PknK in the soluble cell wall fraction (Fig. 6a, lane 7), which contains soluble proteins typically associated with cell wall/membrane components. In vivo localization of PknK was observed by IEM. At day 1 post-infection, M. tuberculosis was visualized within phagocytic vacuoles inside infected macrophages. Although the overall labelling for PknK was low, gold particles were observed associated with cell walls of M. tuberculosis residing within the phagosomes inside the macrophage (Fig. 6b). Interestingly, we also observed PknK-specific labelling in the lumen of the phagosome. Because of the low signal, the experiment was repeated and observations over several sections revealed reproducible results. No labelling was noted either with uninfected macrophages or with infected macrophages not treated with anti-PknK antibody, indicating the specificity of the labelling (data not shown). These findings indicate that PknK is synthesized by M. tuberculosis in the phagosomal environment of human macrophages, is located in the bacterial cell wall and may also be secreted into the phagosomal lumen.
Fig. 6.
PknK localization in broth cultures and within infected macrophages. (a) Western blot analyses of the whole-cell lysate (lane 2), and the cell wall (lane 3), cell membrane (lane 4), culture filtrate (lane 5), cytosolic (lane 6) and soluble cell wall fractions (lane 7) of M. tuberculosis with anti-PknK antibody. Purified full-length, 6xHis-SUMO-tagged PknK recombinant protein (∼130 kDa) was used as a positive control (lane 1). The presence of native M. tuberculosis PknK (∼119 kDa) is indicated by an arrow. (b) Immunolocalization of PknK within infected human macrophages at day 1 post-infection (bars, 0.2 μm). Arrows indicate PknK-specific labelling on the cell wall of M. tuberculosis (Mtb) and also within the phagocytic vacuole (phagosome). Two representative pictures are presented.
DISCUSSION
Protein kinase K is one of the largest, multi-domain STPK proteins annotated as a transcriptional regulator (Av-Gay & Everett, 2000). Besides its recent association with the transcriptional regulation of the mycobacterial monooxygenase (mymA) operon of M. tuberculosis (Kumar et al., 2009), little is known about the function of PknK in mycobacterial growth, physiology and virulence. In the present report, we have established a role for PknK in the pathogenesis of M. tuberculosis. Our results implicate PknK in the regulatory mechanisms that modulate the growth of M. tuberculosis in culture and during persistent infection in mice. We also show that PknK functions as a modulator of host immunity that is accompanied by positive or negative effects on M. tuberculosis growth during early and late stages of infection, respectively, in immunocompetent mice.
The goal of this study was to identify candidate STPK virulence factors through a screening approach based on the differential expression of STPK genes in broth-grown cultures of H37Rv and H37Ra, and in H37Rv-infected human macrophages. Extending the rationale that has led to the successful identification of virulence factors in the past (Sharma & Tyagi, 2007), we reasoned that STPK genes having higher expression in H37Rv compared to H37Ra could be associated with functions critical to pathogenicity. In the second screen, we used SCOTS-generated cDNA mixtures to analyse STPK gene expression in H37Rv-infected human macrophages. Although quantitative estimations were not possible by this approach, it provided useful insights into the temporal expression of STPK genes during infection and correlated with the available data for most of the genes. For example, PknL has been grouped with PknA/PknB and is implicated in the regulation of cell shape and cell division (Narayan et al., 2007). The constitutive expression of pknL and pknB in H37Rv-infected human macrophages (Table 2) supports this hypothesis and exemplifies the robustness of the screen. Absence of pknI transcripts in our experiments could be due to the limit of detection since pknI expression decreases during infection in THP1 cells (Singh et al., 2006). Results from both screens highlighted pknK among STPK genes that were significantly expressed in H37Rv compared to H37Ra and during early time points of infection in human macrophages (Table 2). These data provided the impetus to investigate the role of PknK in mycobacterial pathobiology.
To address these questions, we constructed and characterized LIX11, a ΔpknK strain of H37Rv. Surprisingly, deletion of pknK affected physical attributes like cell size and the cell wall ultrastructure of the bacilli. These observations, particularly the perturbations in the cell wall, provide some insights into the target proteins of PknK. PknK is known to phosphorylate VirS and several members of the mymA operon of M. tuberculosis (Kumar et al., 2009). Interestingly, proteins encoded by the mymA operon are involved in the biosynthesis of mycolic acids, essential components of the mycobacterial cell wall that are also immunogenic (Korf et al., 2005). It is thus plausible that the aberrant synthesis of these key molecules or downstream modified lipids in LIX11 is responsible for the differential cell wall staining patterns compared to the wild-type. Our interpretations are supported by previous findings demonstrating a requirement of mymA operon in the maintenance of cell wall structure and in persistent infection in guinea pigs (Singh et al., 2003, 2005).
The LIX11 mutant is phenotypically different from the parent strain in terms of its growth and survival in vitro and in vivo. The most striking feature of the growth of LIX11 in broth cultures is its increased survival during stationary phase (Fig. 3a). In addition, increased resistance of the mutant to specific stresses, acidic pH, oxidative stress and hypoxia (Supplementary Fig. S2), provides evidence for PknK-mediated mechanisms that regulate growth under stress conditions and suggests a link between PknK function and energy status, catabolic activity, and/or the environment of the bacterial cell. Contrary to in vitro growth, the dynamics of M. tuberculosis growth in vivo are governed by combined effects of the pathogen and the host. We investigated the virulence of LIX11 in C57BL/6 mice using the low-dose aerosol model of infection, wherein unrestricted growth of M. tuberculosis occurred during the acute infection, after which the viable counts decreased and stabilized, characteristic of a chronic or persistent infection (Kelly et al., 1996). The transition from acute to chronic phase is marked by the reduced transcriptional activity of M. tuberculosis genes and decrease in growth (Shi et al., 2003; Talaat et al., 2007). The increase in the viable counts of the mutant during the chronic phase of infection (Fig. 4d, e) suggests involvement of PknK in such growth adaptive events. A common theme that is evident from our in vitro and in vivo studies is the role of PknK in slowing the growth of M. tuberculosis during stress conditions in culture and during chronic infection. It is of note that the deletion of pknH (Papavinasasundaram et al., 2005) and pknI (Gopalaswamy et al., 2009) also resulted in increased bacillary loads during persistent infection in mice. Although very little is known about the persistent phase, it is apparent that M. tuberculosis possesses distinct regulatory mechanisms that enable the switch between fast and slow growth rates (Beste et al., 2007, 2009) and, importantly, STPKs seem to be integral to these strategies.
PknK is the fourth STPK implicated in mycobacterial pathogenesis, but it is the first one shown to play a role in early infection events. Although the molecular events that lead to the early establishment of tuberculosis infection are not well understood, evidence suggests that M. tuberculosis alters macrophage signalling for the production of cytokines and effector molecules (Beltan et al., 2000; Falcone et al., 1994). We show that PknK modulates the host immune response as early as 1 week post-infection. Absence of PknK in LIX11 resulted in the upregulation of TNF-α, a key pro-inflammatory cytokine with anti-mycobacterial effects (Fig. 5). Most importantly, TNF-α induction coincided with the inhibition of growth of the mutant at 1 week post-infection (Fig. 4a) and correlated with the exaggerated inflammatory response in LIX11-infected mice (Fig. 4b, c). It is plausible that in the wild-type H37Rv, PknK plays a direct or indirect role in immune suppression, thus enabling exponential growth by providing a survival advantage to the wild-type during early stages of infection. It is established that IL-12p40 induces production of IFN-γ, a key Th1 cytokine (Magram et al., 1996); hence in view of the downregulation of IL-12p40 mRNA levels in LIX11-infected mice at 1 and 2 weeks post-infection, it is not surprising that the LIX11 mutant failed to induce IFN-γ at 2 weeks post-infection to the levels observed in H37Rv-infected mice (Fig. 5). Together, IL-12p40 (Cooper et al., 1997), IFN-γ (Flynn et al., 1993) and iNOS (MacMicking et al., 1997) are known to play protective roles in murine tuberculosis infections, typically at the onset of chronic infection (weeks three to four) (Shi et al., 2003). Thus their downregulation may be a prime factor contributing to the increased bacillary load of the mutant observed in the lungs and in spleens of mice during the persistent phase of infection. These results are indicative of distinct roles of PknK in responding to the intracellular environment. Specifically, the ability of PknK to subvert the immune response to facilitate exponential growth of mycobacteria during the acute phase of infection is intriguing and brings forth questions regarding the localization of PknK in vivo and possible cross-talk with eukaryotic signalling pathways.
Although the localization of PknK in subcellular fractions of M. tuberculosis has been recently reported (Kumar et al., 2009), we revisited this question, both in vitro and in vivo. Our results with broth-grown M. tuberculosis confirm their observations and add that PknK is a soluble protein associated with the proteins and/or lipids in the cell wall/membrane of M. tuberculosis (Fig. 6a). In view of the faint band in the cell membrane fraction (Fig. 6a; lane 4), it is likely that the association of PknK with the cell membrane is transient, but this result provides a clue to PknK function in signalling events or metabolite transport that may occur at these sites. Interaction of PknK with inner-membrane ABC transporters has been speculated through in silico studies (Cui et al., 2009). A similar localization of PknK in M. tuberculosis cells residing within the phagosome and, in addition, a consistent but low level of PknK-specific labelling in the phagosomal space, suggests that PknK is capable of sensing the intracellular environment cues and may also be secreted into the phagosomal compartment (Fig. 6b). One of the limitations of EM studies is the destruction of antigen due to chemical treatments; however, low levels of PknK cannot be excluded. Although PknK does not have a typical signal sequence, secretion could occur through alternative secretion pathways as seen in the case of PknG (Walburger et al., 2004). Nevertheless, in view of PknK's function in early infection and immune modulation, such a possibility warrants further exploration, and undoubtedly will have significant implications in host–pathogen interactions, particularly regarding the signalling pathways governing the early events of infection. These findings open exciting avenues for future research to address critical questions on strategies employed by mycobacteria to enable survival and the early establishment of infection in the host.
Acknowledgments
We gratefully acknowledge Richard F. Silver for providing the M. tuberculosis H37Ra strain and Shelley Haydel for providing control plasmids for hybridization with SCOTS-generated cDNA mixtures. The subcellular fractions of M. tuberculosis were obtained from Colorado State University as part of NIH, NIAID Contract no. HHSN266200400091C entitled ‘Tuberculosis Vaccine Testing and Research Materials’. We are thankful to the Electron Microscopy Laboratory in the School of Life Sciences, ASU, with special mention to David Lowry for his efforts, Daniel Clemens for assistance with macrophage experiments and James Megehee, Jr for assistance with animal experiments. We also thank Roy Curtiss III and Praveen Alamuri for valuable suggestions and critical review of the manuscript. This research was supported by Public Health Service grant AI46428 from the US National Institutes of Health and Arizona State University Start-up Funding, awarded to J. E. C.-C., and in part by funds from the Howard Hughes Medical Institute through the Undergraduate Science Education Program and from the ASU School of Life Sciences, awarded to G. C.
Abbreviations
ADS, albumin-dextrose-saline
BCG, Bacillus Calmette-Guérin
Ct, cycle threshold
HRP, horseradish peroxidase
IEM, immunoelectron microscopy
IFN, interferon
IL, interleukin
iNOS, inducible nitric oxide synthase
Mtb, Mycobacterium tuberculosis
qRT-PCR, quantitative RT-PCR
SCOTS, selective capture of transcribed sequences
SEM, scanning electron microscopy
STPK, serine/threonine protein kinase
TEM, transmission electron microscopy
TNF, tumour necrosis factor
Footnotes
A supplementary table of primers and two supplementary figures are available with the online version of this paper.
References
- Av-Gay, Y. & Everett, M. (2000). The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol 8, 238–244. [DOI] [PubMed] [Google Scholar]
- Av-Gay, Y., Jamil, S. & Drews, S. J. (1999). Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infect Immun 67, 5676–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, H., Wong, D. & Av-Gay, Y. (2009). Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem J 420, 155–160. [DOI] [PubMed] [Google Scholar]
- Bardarov, S., Bardarov, S., Jr, Pavelka, M. S., Jr, Sambandamurthy, V., Larsen, M., Tufariello, J., Chan, J., Hatfull, G. & Jacobs, W. R., Jr (2002). Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017. [DOI] [PubMed] [Google Scholar]
- Beltan, E., Horgen, L. & Rastogi, N. (2000). Secretion of cytokines by human macrophages upon infection by pathogenic and non-pathogenic mycobacteria. Microb Pathog 28, 313–318. [DOI] [PubMed] [Google Scholar]
- Beste, D. J., Laing, E., Bonde, B., Avignone-Rossa, C., Bushell, M. E. & McFadden, J. J. (2007). Transcriptomic analysis identifies growth rate modulation as a component of the adaptation of mycobacteria to survival inside the macrophage. J Bacteriol 189, 3969–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste, D. J., Espasa, M., Bonde, B., Kierzek, A. M., Stewart, G. R. & McFadden, J. (2009). The genetic requirements for fast and slow growth in mycobacteria. PLoS One 4, e5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaba, R., Raje, M. & Chakraborti, P. K. (2002). Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur J Biochem 269, 1078–1085. [DOI] [PubMed] [Google Scholar]
- Chao, J., Wong, D., Zheng, X., Poirier, V., Bach, H., Hmama, Z. & Av-Gay, Y. (2009). Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim Biophys Acta 1804, 620–627. [DOI] [PubMed] [Google Scholar]
- Cohen-Gonsaud, M., Barthe, P., Canova, M. J., Stagier-Simon, C., Kremer, L., Roumestand, C. & Molle, V. (2009). The Mycobacterium tuberculosis Ser/Thr kinase substrate Rv2175c is a DNA-binding protein regulated by phosphorylation. J Biol Chem 284, 19290–19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S. & other authors (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Cooper, A. M., Magram, J., Ferrante, J. & Orme, I. M. (1997). Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with Mycobacterium tuberculosis. J Exp Med 186, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley, S., Ko, M., Pick, N., Chow, R., Downing, K. J., Gordhan, B. G., Betts, J. C., Mizrahi, V., Smith, D. A. & other authors (2004). The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol 52, 1691–1702. [DOI] [PubMed] [Google Scholar]
- Cozzone, A. J. (2005). Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J Mol Microbiol Biotechnol 9, 198–213. [DOI] [PubMed] [Google Scholar]
- Cui, T., Zhang, L., Wang, X. & He, Z. G. (2009). Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Genomics 10, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, N., Kapur, V., Singh, K. K., Das, T. K., Sachdeva, S., Jyothisri, K. & Tyagi, J. S. (2000). Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis 80, 141–159. [DOI] [PubMed] [Google Scholar]
- Dasgupta, A., Datta, P., Kundu, M. & Basu, J. (2006). The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152, 493–504. [DOI] [PubMed] [Google Scholar]
- Deol, P., Vohra, R., Saini, A. K., Singh, A., Chandra, H., Chopra, P., Das, T. K., Tyagi, A. K. & Singh, Y. (2005). Role of Mycobacterium tuberculosis Ser/Thr kinase PknF: implications in glucose transport and cell division. J Bacteriol 187, 3415–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone, V., Bassey, E. B., Toniolo, A., Conaldi, P. G. & Collins, F. M. (1994). Differential release of tumor necrosis factor-alpha from murine peritoneal macrophages stimulated with virulent and avirulent species of mycobacteria. FEMS Immunol Med Microbiol 8, 225–232. [DOI] [PubMed] [Google Scholar]
- Flynn, J. L., Chan, J., Triebold, K. J., Dalton, D. K., Stewart, T. A. & Bloom, B. R. (1993). An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med 178, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, A. M. & Orlando, R. A. (2008). Curcumin and resveratrol inhibit nuclear factor-kappaB-mediated cytokine expression in adipocytes. Nutr Metab (Lond) 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalaswamy, R., Narayanan, S., Jacobs, W. R., Jr & Av-Gay, Y. (2008). Mycobacterium smegmatis biofilm formation and sliding motility are affected by the serine/threonine protein kinase PknF. FEMS Microbiol Lett 278, 121–127. [DOI] [PubMed] [Google Scholar]
- Gopalaswamy, R., Narayanan, S., Chen, B., Jacobs, W. R. & Av-Gay, Y. (2009). The serine/threonine protein kinase PknI controls the growth of Mycobacterium tuberculosis upon infection. FEMS Microbiol Lett 295, 23–29. [DOI] [PubMed] [Google Scholar]
- Graham, J. E. & Clark-Curtiss, J. E. (1999). Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci U S A 96, 11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein, A. E., Grundner, C., Echols, N., Gay, L. M., Lombana, T. N., Miecskowski, C. A., Pullen, K. E., Sung, P. Y. & Alber, T. (2005). Structure/function studies of Ser/Thr and Tyr protein phosphorylation in Mycobacterium tuberculosis. J Mol Microbiol Biotechnol 9, 167–181. [DOI] [PubMed] [Google Scholar]
- Greenstein, A. E., MacGurn, J. A., Baer, C. E., Falick, A. M., Cox, J. S. & Alber, T. (2007). M. tuberculosis Ser/Thr protein kinase D phosphorylates an anti-anti-sigma factor homolog. PLoS Pathog 3, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydel, S. E. & Clark-Curtiss, J. E. (2004). Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages. FEMS Microbiol Lett 236, 341–347. [DOI] [PubMed] [Google Scholar]
- Hou, J. Y., Graham, J. E. & Clark-Curtiss, J. E. (2002). Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect Immun 70, 3714–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J., Stella, A., Boudou, F., Levillain, F., Darthuy, E., Vaubourgeix, J., Wang, C., Bardou, F., Puzo, G. & other authors (2010). Functional characterization of the Mycobacterium tuberculosis serine/threonine kinase Pkn. Microbiology 156, 1619–1631. [DOI] [PubMed] [Google Scholar]
- Kang, C. M., Abbott, D. W., Park, S. T., Dascher, C. C., Cantley, L. C. & Husson, R. N. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev 19, 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, C. M., Nyayapathy, S., Lee, J. Y., Suh, J. W. & Husson, R. N. (2008). Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154, 725–735. [DOI] [PubMed] [Google Scholar]
- Kelly, B. P., Furney, S. K., Jessen, M. T. & Orme, I. M. (1996). Low-dose aerosol infection model for testing drugs for efficacy against Mycobacterium tuberculosis. Antimicrob Agents Chemother 40, 2809–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, J., Stoltz, A., Verschoor, J., De Baetselier, P. & Grooten, J. (2005). The Mycobacterium tuberculosis cell wall component mycolic acid elicits pathogen-associated host innate immune responses. Eur J Immunol 35, 890–900. [DOI] [PubMed] [Google Scholar]
- Kumar, P., Kumar, D., Parikh, A., Rananaware, D., Gupta, M., Singh, Y. & Nandicoori, V. K. (2009). The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J Biol Chem 284, 11090–11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking, J. D., North, R. J., LaCourse, R., Mudgett, J. S., Shah, S. K. & Nathan, C. F. (1997). Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A 94, 5243–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magram, J., Sfarra, J., Connaughton, S., Faherty, D., Warrier, R., Carvajal, D., Wu, C. Y., Stewart, C., Sarmiento, U. & Gately, M. K. (1996). IL-12-deficient mice are defective but not devoid of type 1 cytokine responses. Ann N Y Acad Sci 795, 60–70. [DOI] [PubMed] [Google Scholar]
- Malhotra, V., Tyagi, J. S. & Clark-Curtiss, J. E. (2009). DevR-mediated adaptive response in Mycobacterium tuberculosis H37Ra: links to asparagine metabolism. Tuberculosis (Edinb) 89, 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle, V., Brown, A. K., Besra, G. S., Cozzone, A. J. & Kremer, L. (2006). The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J Biol Chem 281, 30094–30103. [DOI] [PubMed] [Google Scholar]
- Narayan, A., Sachdeva, P., Sharma, K., Saini, A. K., Tyagi, A. K. & Singh, Y. (2007). Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol Genomics 29, 66–75. [DOI] [PubMed] [Google Scholar]
- O'Hare, H. M., Durán, R., Cerveñansky, C., Bellinzoni, M., Wehenkel, A. M., Pritsch, O., Obal, G., Baumgartner, J., Vialaret, J. & other authors (2008). Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol 70, 1408–1423. [DOI] [PubMed] [Google Scholar]
- Overbergh, L., Valckx, D., Waer, M. & Mathieu, C. (1999). Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 11, 305–312. [DOI] [PubMed] [Google Scholar]
- Papavinasasundaram, K. G., Chan, B., Chung, J. H., Colston, M. J., Davis, E. O. & Av-Gay, Y. (2005). Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J Bacteriol 187, 5751–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. T., Kang, C. M. & Husson, R. N. (2008). Regulation of the SigH stress response regulon by an essential protein kinase in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105, 13105–13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini, D. K., Malhotra, V., Dey, D., Pant, N., Das, T. K. & Tyagi, J. S. (2004). DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150, 865–875. [DOI] [PubMed] [Google Scholar]
- Scherr, N., Muller, P., Perisa, D., Combaluzier, B., Jeno, P. & Pieters, J. (2009). Survival of pathogenic mycobacteria in macrophages is mediated through autophosphorylation of protein kinase G. J Bacteriol 191, 4546–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, D. & Tyagi, J. S. (2007). The value of comparative genomics in understanding mycobacterial virulence: Mycobacterium tuberculosis H37Ra genome sequencing – a worthwhile endeavour. J Biosci 32, 185–189. [DOI] [PubMed] [Google Scholar]
- Sharma, K., Gupta, M., Krupa, A., Srinivasan, N. & Singh, Y. (2006). EmbR, a regulatory protein with ATPase activity, is a substrate of multiple serine/threonine kinases and phosphatase in Mycobacterium tuberculosis. FEBS J 273, 2711–2721. [DOI] [PubMed] [Google Scholar]
- Shi, L., Jung, Y. J., Tyagi, S., Gennaro, M. L. & North, R. J. (2003). Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc Natl Acad Sci U S A 100, 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A., Jain, S., Gupta, S., Das, T. & Tyagi, A. K. (2003). mymA operon of Mycobacterium tuberculosis: its regulation and importance in the cell envelope. FEMS Microbiol Lett 227, 53–63. [DOI] [PubMed] [Google Scholar]
- Singh, A., Gupta, R., Vishwakarma, R. A., Narayanan, P. R., Paramasivan, C. N., Ramanathan, V. D. & Tyagi, A. K. (2005). Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. J Bacteriol 187, 4173–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A., Singh, Y., Pine, R., Shi, L., Chandra, R. & Drlica, K. (2006). Protein kinase I of Mycobacterium tuberculosis: cellular localization and expression during infection of macrophage-like cells. Tuberculosis (Edinb) 86, 28–33. [DOI] [PubMed] [Google Scholar]
- Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T., Bansal, G. P., Young, J. F., Lee, M. H. & other authors (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. [DOI] [PubMed] [Google Scholar]
- Talaat, A. M., Ward, S. K., Wu, C. W., Rondon, E., Tavano, C., Bannantine, J. P., Lyons, R. & Johnston, S. A. (2007). Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J Bacteriol 189, 4265–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur, M. & Chakraborti, P. K. (2006). GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem 281, 40107–40113. [DOI] [PubMed] [Google Scholar]
- Thakur, M. & Chakraborti, P. K. (2008). Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochem J 415, 27–33. [DOI] [PubMed] [Google Scholar]
- Tiwari, D., Singh, R. K., Goswami, K., Verma, S. K., Prakash, B. & Nandicoori, V. K. (2009). Key residues in Mycobacterium tuberculosis protein kinase G play a role in regulating kinase activity and survival in the host. J Biol Chem 284, 27467–27479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyron-Churlet, R., Molle, V., Taylor, R. C., Brown, A. K., Besra, G. S., Zanella-Cleon, I., Futterer, K. & Kremer, L. (2009). The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J Biol Chem 284, 6414–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walburger, A., Koul, A., Ferrari, G., Nguyen, L., Prescianotto-Baschong, C., Huygen, K., Klebl, B., Thompson, C., Bacher, G. & Pieters, J. (2004). Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304, 1800–1804. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Munoz-Dorado, J., Inouye, M. & Inouye, S. (1992). Identification of a putative eukaryotic-like protein kinase family in the developmental bacterium Myxococcus xanthus. J Bacteriol 174, 5450–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]