Abstract
Candida albicans has been previously shown to stimulate the production of Pseudomonas aeruginosa phenazine toxins in dual-species colony biofilms. Here, we report that P. aeruginosa lasR mutants, which lack the master quorum sensing system regulator, regain the ability to produce quorum-sensing-regulated phenazines when cultured with C. albicans. Farnesol, a signalling molecule produced by C. albicans, was sufficient to stimulate phenazine production in LasR− laboratory strains and clinical isolates. P. aeruginosa ΔlasR mutants are defective in production of the Pseudomonas quinolone signal (PQS) due to their inability to properly induce pqsH, which encodes the enzyme necessary for the last step in PQS biosynthesis. We show that expression of pqsH in a ΔlasR strain was sufficient to restore PQS production, and that farnesol restored pqsH expression in ΔlasR mutants. The farnesol-mediated increase in pqsH required RhlR, a transcriptional regulator downstream of LasR, and farnesol led to higher levels of N-butyryl-homoserine lactone, the small molecule activator of RhlR. Farnesol promotes the production of reactive oxygen species (ROS) in a variety of species. Because the antioxidant N-acetylcysteine suppressed farnesol-induced RhlR activity in LasR− strains, and hydrogen peroxide was sufficient to restore PQS production in las mutants, we propose that ROS are responsible for the activation of downstream portions of this quorum sensing pathway. LasR mutants frequently arise in the lungs of patients chronically infected with P. aeruginosa. The finding that C. albicans, farnesol or ROS stimulate virulence factor production in lasR strains provides new insight into the virulence potential of these strains.
INTRODUCTION
Cystic fibrosis (CF) is a genetic disease in which one of the pathologies is the inability to effectively clear potential pathogens from the airways, leading to chronic colonization of the lungs by a number of bacterial and fungal strains. Over 75 % of CF patients over 18 years of age are chronically colonized with Pseudomonas aeruginosa, which often persists throughout the life of the patient (Rajan & Saiman, 2002). While P. aeruginosa is the predominant pathogen in the CF lung, fungi such as Candida albicans are also frequently detected (Bakare et al., 2003; Chotirmall et al., 2010; Haase et al., 1991; Navarro et al., 2001). Bacterial–fungal co-infections may alter the host response (Allard et al., 2009) or virulence factor expression in chronic biofilms (Cugini et al., 2007; Gibson et al., 2009; McAlester et al., 2008).
The complexity of CF lung infections is further increased by phenotypic diversification of P. aeruginosa strains within these chronic infections (Ernst et al., 2007; Li et al., 2005; Mahenthiralingam et al., 1994; Smith et al., 2006). LasR, a transcriptional regulator that directly and indirectly controls a large set of virulence-related genes, is frequently defective in CF isolates (D'Argenio et al., 2007; Hoffman et al., 2009; Smith et al., 2006; Tingpej et al., 2007), and these mutations confer a variety of advantages in the CF lung environment (D'Argenio et al., 2007). In a recent study by Hoffman et al. (2009), it was found that approximately 30 % of strains isolated from the airways of CF patients lacked LasR function, and that infection with LasR− strains correlates with decreased lung function and poor patient prognosis. While LasR− strains have a growth advantage in some environments and increased resistance to some antimicrobials (D'Argenio et al., 2007; Hoffman et al., 2010), the in vivo regulation of virulence factors in LasR-defective strains is not completely understood. Evidence suggests that LasR− strains can exist as social cheaters by residing in the presence of LasR+ counterparts that continue to produce quorum sensing signals and quorum-sensing-controlled virulence factors, which can cross-complement (Sandoz et al., 2007; Wilder et al., 2009). In addition, data indicate that alternate pathways for virulence factor production may exist in LasR loss-of-function mutants, as strains PA14ΔlasR and PAO1ΔlasR regain the ability to produce pyocyanin at late time points (Dekimpe & Deziel, 2009; Diggle et al., 2003).
LasR is at the top of the P. aeruginosa quorum sensing regulatory cascade. Upon binding the acyl homoserine lactone (AHL) 3-oxo-C12-homoserine lactone (3OC12HSL), LasR activates the transcription of RhlR, another transcription factor, and stimulates the production of N-butyryl-homoserine lactone (C4HSL) (Pesci et al., 1997). LasR also participates in the synthesis of the Pseudomonas quinolone signal (PQS; 2-heptyl-3-hydroxy-4-quinolone) (Pesci et al., 1999; Xiao et al., 2006b) by affecting the expression of PqsR (MvfR), which regulates the pqsA–D operon that encodes the enzymes to synthesize 2-heptyl-4-quinolone (HHQ), and by inducing the expression of pqsH, which catalyses the conversion of HHQ to PQS (Gallagher et al., 2002). PqsE, which is induced by PQS through PqsR, in concert with RhlR, controls the expression of phenazine biosynthetic genes (Farrow et al., 2008).
Production of PQS by P. aeruginosa is inhibited by farnesol, a C. albicans signalling molecule that participates in inter- and intraspecies interactions (Langford et al., 2010; Peleg et al., 2010; Shirtliff et al., 2009b). Farnesol inhibits the PqsR-mediated transcription of the pqsA–E operon, probably due to direct interaction with PqsR, though the effects of farnesol on PqsR activity can be suppressed by higher levels of PQS (Cugini et al., 2007). In P. aeruginosa–C. albicans biofilms, where P. aeruginosa and PQS concentrations are high, the presence of the fungus leads to increased production of phenazines through an uncharacterized pathway, suggesting that the effects of C. albicans on P. aeruginosa are complex (Gibson et al., 2009). Here, we report that in colony biofilms formed by LasR-defective strains, the defects in PQS production that are normally associated with the ΔlasR genotype were no longer apparent in the presence of farnesol or farnesol-producing C. albicans. The increased PQS production translated into increased phenazine production. Farnesol led to an induction of pqsH transcription, and pqsH overexpression was sufficient to restore pyocyanin and PQS production in the absence of farnesol. The induction of pqsH by farnesol was dependent on RhlR, and we have demonstrated that farnesol stimulated RhlR activity and the production of C4HSL. Exogenous C4HSL was sufficient to stimulate PQS production in the ΔlasR strain. Clinical isolates of P. aeruginosa with LasR loss-of-function mutations also produced increased PQS in the presence of farnesol or farnesol-producing C. albicans. Because farnesol leads to the production of intracellular reactive oxygen species (ROS) in a variety of species (Machida et al., 1998; Semighini et al., 2006; Shirtliff et al., 2009a), farnesol-induced PQS production in PA14ΔlasR is decreased upon the addition of the antioxidant N-acetyl cysteine (NAC), and hydrogen peroxide is sufficient to stimulate PQS production in lasR but not lasRrhlR mutants, we suggest that the activation of RhlR signalling in PA14ΔlasR may occur in response to oxidative stress. We propose that the stimulation of PQS production by the presence of C. albicans or other sources of oxidative stress, such as those present in association with inflammation, can lead to high level expression of downstream elements in the LasR-controlled quorum sensing pathway and the reactivation of quorum-sensing-controlled genes in LasR-defective strains.
METHODS
Bacterial strains, media and culture conditions.
The bacterial and fungal strains used are listed in Table 1. Bacterial overnight cultures were grown in 5 ml LB at 37 °C on a roller drum and, when needed, antibiotics were provided at the following concentrations: gentamicin, 10 μg ml−1 for Escherichia coli and 75 μg ml−1 for P. aeruginosa; carbenicillin, 150 μg ml−1 for E. coli and 750 μg ml−1 for P. aeruginosa. C. albicans overnight cultures were grown in 5 ml YPD at 30 °C on a roller drum.
Table 1.
Strains and plasmids
| Strains | Lab DH no. | Description | Source |
|---|---|---|---|
| P. aeruginosa | |||
| PA14 | 122 | Wild-type | Rahme et al. (1995) |
| PA14ΔlasR | 164 | In-frame deletion of lasR | Hogan et al. (2004) |
| PA14ΔlasI | 132 | In-frame deletion of lasI | Hogan et al. (2004) |
| PA14 rhlR : : TetR | 6 | Gene replacement of rhlR | Hogan & Kolter (2002) |
| PA14ΔrhlI | 169 | In-frame deletion of rhlI | Hogan et al. (2004) |
| PA14ΔlasRrhlR : : TetR | 237 | In-frame deletion of lasR in DH6 | This study |
| PA14ΔlasRΔrhlI | 238 | In-frame deletion of lasR in DH169 | This study |
| PA14ΔpqsH | 1112 | In-frame deletion of pqsH | This study |
| PA14ΔlasRΔpqsH | 1113 | In-frame deletion of lasR in DH1112 | This study |
| PA14ΔpqsR | 1110 | In-frame deletion of pqsR | This study |
| PA14ΔlasRΔpqsR | 1111 | In-frame deletion of lasR in DH1110 | This study |
| CIA467G | 1097 | LasR mutation, 467 nt mutated, A→G, nonsynonomous D→G | Smith et al. (2006) |
| CIG691C | 1099 | LasR mutation, 691 nt mutated, G→C, nonsynonomous A→P | Smith et al. (2006) |
| CIT341C | 1100 | LasR mutation, 341 nt mutated, T→C, nonsynonomous L→P | Smith et al. (2006) |
| CIG608A | 1103 | LasR mutation, 608 nt mutated, G→A, nonsynonomous C→Y | Smith et al. (2006) |
| CIG179A | 1132 | LasR mutation, 179 nt mutated, G→A, nonsynonomous W→STOP | Smith et al. (2006) |
| CIC181T | 1134 | LasR mutation, 181 nt mutated, C→T, nonsynonomous R→C | Smith et al. (2006) |
| CIG455- | 1136 | LasR mutation, 455 nt deleted, G→ – | Smith et al. (2006) |
| E. coli | |||
| S17/λpir | 71 | Laboratory collection | |
| DH5α | – | F′/endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 (lacZYA-argF) U169 deoR [80 dlac (lacZ)M15 recA1] | Laboratory collection |
| JM109 | 1406 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15/Δ (lac-proAB) glnV44 e14− gyrA96 recA1 relA1 endA1 thi hsdR17 | Laboratory collection |
| C. albicans | |||
| SC5314 | 35 | Wild-type | Gillum et al. (1984) |
| S. cerevisiae | |||
| DC49-7.1C | – | MATα, leu2-3, 112 trp1-289 ura3-52 arg4− Δ57, RV | Shanks et al. (2006) |
| Plasmids | |||
| pUCP22 | 160 | GmR; Escherichia–Pseudomonas expression plasmid | West et al. (1994) |
| pEX18GM | GmR; oriTsacB, gene replacement vector with MCS from pUC18 | Hoang et al. (1998) | |
| pMQ30 | 962 | Deletion plasmid for yeast cloning | Shanks et al. (2006) |
| pLASR | 133 | PA14ΔlasR deletion construct | Hogan et al. (2004) |
| pPQSRdel | 949 | PA14ΔpqsR deletion construct | This study |
| pPQSHdel | 1109 | PA14ΔpqsH deletion construct | This study |
| pCR2.1TOPO | – | Cloning vector | Invitrogen |
| pPQSHFLAG | 1413 | pqsH promoter region and gene in pUCP22 | This study |
| pSB536 | 1407 | ahyR+ ahyI : : luxCDABE; AmpRcolE1 origin | Swift et al. (1997) |
P. aeruginosa and C. albicans co-cultures were formed in either 96-well plate wells with 150 μl YPD agar or in 180 mm petri dishes. Overnight cultures (10 μl for wells and 500 μl for plates) of C. albicans SC5314 were top-spread. C. albicans lawns were grown for 24 h at 30 °C prior to inoculation with P. aeruginosa. Co-cultures in 96-well plates were inoculated with 5 μl P. aeruginosa from cultures grown to OD600 0.2, and Petri plate co-cultures were spot-inoculated with 10 μl of LB-grown cultures.
For analysis of the effects of farnesol and hydrogen peroxide, P. aeruginosa was inoculated as described above onto LB agar containing DMSO (vehicle control), 250 μM farnesol, 5 or 10 mM NAC or 200 μM hydrogen peroxide, as specified, and visualized at time intervals described in the text. Farnesol (Sigma) was prepared as a 500 mM stock solution in DMSO prior to each experiment and added at a final concentration of 250 μM after autoclaving and cooling to pouring temperature; control plates received an equivalent amount of DMSO. C4HSL (Fluka) was prepared as a 1 mM stock solution in ethyl acetate and added at a final concentration of 25 μM to agar; the ethyl acetate solution was added to a glass bottle, and the solvent was allowed to evaporate prior to the addition of agar.
Construction of P. aeruginosa mutants.
To construct the pqsR deletion plasmid (pPQSRdel), flanking sequences of the target gene were amplified by PCR from PA14 chromosomal DNA using primers corresponding to the PA14 genomic sequence. The deletion constructs were made by splicing by overlapping extension (SOEing) PCR generating a fusion of the 5′ and 3′ flanking regions of the target gene following two rounds of PCR (Horton et al., 1989). PCR was performed using the high fidelity platinum Taq master mix kit (Invitrogen) with an additional 2.5 mM MgCl2 and 5 % DMSO. The first round of PCR created two 1 kb products using the following primers: pqsR5′F (CCGAGCTCCATCTCCAGCGAATCGGATACGC) with pqsRSOE5′ (GGAGCGCCTTCGGGCCTGAGCGGCGCTGCACCGGATCCCGAACCGGAGGCGATGACCTGGAG), and pqsR3′R (GGAAGCTTGGCGAACTGGATGCGCTGCATGC) with pqsRSOE3′ (ATGTTCCTCCAGGTCATCGCCTCCGGTTCGGGATCCGGTGCAGCGCCGCTCAGGCCCGAAGGCGCTCC). The second round of PCR used 20 pmol μl−1 of pqsR5′F/pqsR3′R primers and 1 μl of each of the 1 kb amplified products from round 1 as template, to generate 5′ and 3′ flanking regions fusions. 5′ Primers were designed with SacI sites at the 5′ terminus and 3′ primers were designed with HindIII, to allow for cloning into pGEX18Gm digested with SacI and HindIII (New England Biolabs).
To construct the pqsH deletion plasmid (pPQSHdel), flanking sequences of the target gene were amplified by PCR from PA14 chromosomal DNA using primers corresponding to the PA14 genomic sequence containing tails that correspond to the plasmid sequence. PCR was performed using the high fidelity platinum Taq master mix kit (Invitrogen) with an additional 2.5 mM MgCl2 and 5 % DMSO. The primers used were: pqsH/yeast primer 1 (CCAAGCTTGCATGCCTGCAGGTCGACTCTAGAGGATCCCCGCGCCAGCCCGGCGACGCC) pqsH/yeast primer 2 (ACCGACACCGGTGAAAAGACGCTGGTGCAGGCGCCTGCGACggatccGCGCCAGCGCCAGGCCGGCGATCCCGGCCCC), pqsH/yeast primer 3 (GGGGCCGGGATCGCCGGCCTGGCGCTGGCGCggatccGTCGCAGGCGCCTGCACCAGCGTCTTTTCACCGGTGTCGGT) with pqsH/yeast primer 4 (AACAGCTATGACCATGATTACGAATTCGAGCTCGGTACCCCGTGGCGGACGGCGAGCAGC). Nucleotides in bold are those that overlap with either the PA14 sequence or the pMQ30 sequence. The plasmid pMQ30 (Shanks et al., 2006) was digested with XbaI in order to linearize the plasmid for transformation in to Saccharomyces cerevisiae (strain DC49-7.1C). An overnight culture of S. cerevisiae (0.5 ml) grown in YPD was pelleted, the supernatant was removed, and the pellet was washed with TE. Transformation buffer (0.5 ml; 40 % polyethylene glycol, 0.1 M lithium acetate, 10 mM Tris/HCl, pH 7.5, and 1 mM EDTA), 20 μl 2 mg single stranded salmon sperm DNA ml−1, digested plasmid DNA (10 μl) and 5′ and 3′ PCR products (20 μl each) were added to the pellet; this was mixed by vortexing for 1 min and incubated overnight at room temperature. Heat shock was applied to the cells by incubation for 10 min at 42 °C; cells were pelleted and washed with TE. The pellets were plated to SC drop-out medium that lacked uracil and were incubated at 30 °C. The resulting colonies were patched to a fresh plate and allowed to reach a high cell density. The plasmid was liberated by a ‘smash and grab’ technique (Elble, 1992; Hoffman & Winston, 1987). All deletion and disruption constructs were electroporated into E. coli S17/λpir, mated into P. aeruginosa PA14 and single mutants, and resolved with gentamicin and sucrose selections as described previously (Hoang et al., 1998). The lasR double mutants were constructed using the pLASR deletion construct described previously (Hogan et al., 2004).
To construct the pqsH fragment for the overexpression plasmid (pPQSHFLAG) the following primers were used: pqsH5′ (ccgagctcTAGAAGGAGCAACGGATGACCG) and pqsH3′FLAG (cttaagcttCTACTTGTCGTCGTCGTCCTTGTAGTCCTGTGCGGCCATCTCACCGACACCGGTG). The 5′ and 3′ primers contain SacI and HindIII sites, respectively, and the 3′ primer encodes a C-terminal FLAG tag. The pqsH gene was cloned into pUCP22, and transformed into PA14, PA14ΔlasR, PA14 rhlR : : TetR, PA14ΔlasRrhlR : : TetR, PA14ΔpqsH and PA14ΔlasRΔpqsH.
Pyocyanin quantification.
P. aeruginosa cells were grown as described above. Samples were obtained at the times indicated and were prepared as described previously with modifications (Gallagher et al., 2002). At each time point, one agar plug was added to 750 μl chloroform, samples were mixed by vortexing for 2 min, then allowed to incubate for 2 h at 4 °C. The chloroform extracts were acidifified by the addition of 750 μl 0.2 M HCl. The aqueous layer, containing pyocyanin, was removed and measured at 390 nm using a Spectra Max plate reader. Samples were collected at each time point in quadruplicate and each sample was assayed in triplicate for pyocyanin concentrations.
To determine pyocyanin production in the presence of hydrogen peroxide, P. aeruginosa strains were grown in 96-well plates as outlined above. After 48 h of growth, agar plugs were transferred to tubes containing chloroform (500 μl), mixed by vortexing for 2 min and then incubated for 2 h at 4 °C. The chloroform extracts were acidified by the addition of 500 μl 0.2 M HCl, and the aqueous layer, containing pyocyanin, was diluted 1 : 2 in 1 M Tris/HCl pH 8.0. Pyocyanin was measured by recording absorbance at 310 nm. For each strain under each condition, the pyocyanin levels were measured in five separate plugs. In all experiments, concentrations were determined using a standard curve of known concentrations of pyocyanin (Cayman Chemicals) dissolved in the appropriate buffer.
PQS quantification.
P. aeruginosa was grown as described above. Samples were obtained at the times indicated and were prepared as described previously with modifications (Gallagher et al., 2002). At each time point, one agar plug was added to 1 ml acidified ethyl acetate; samples were mixed by vortexing for 2 min, centrifuged for 2 min at 13 000 g, and 750 μl was removed to a clean glass vial. The extraction was repeated, removing 1.5 ml total volume. The extracts were allowed to dry at 37 °C or under nitrogen. Extracts were resuspended in 200 μl 1 : 1 acidified ethyl acetate:acetonitrile by vortexing for 10 min at low speed. For thin-layer chromatography (TLC) analysis, aluminium TLC sheets (EMD Chemicals; silica gel 60 F254) were activated in a 0.5 % KH2PO4 solution, air-dried, and baked at 100 °C for 1 h prior to use. Samples (1 or 2 μl) were applied to each plate using 1 μl glass capillaries. A mixture of 17 : 2:1 methylene chloride:acetonitrile:dioxane was used as a solvent, and plates were visualized using long-wave UV light. For each strain and treatment, PQS extracts for three biological replicates were each spotted in triplicate on a TLC plate. Spot intensities were determined with ImageJ software (Abramoff et al., 2004). A dilution series of an authentic PQS standard was included on each plate in order to generate a standard curve for PQS quantification. An anthranilic acid standard was also included in some analyses for comparison.
Transcript analysis.
To examine the timing of the effects of farnesol on pqsH expression, three biological replicates of P. aeruginosa cells grown on standard Petri plates were recovered from the agar surface at the times indicated and stored at −80 °C until RNA extractions were performed. To isolate P. aeruginosa RNA, 100 μl 3 mg ml−1 lysozyme solution was added to cells, and samples were allowed to stand at room temperature for 3 min. The RNeasy kit (Qiagen) was used for subsequent steps, with modifications. On-column DNase (Qiagen) and RQ1 (Promega) treatment was used to remove DNA. PCRs using the purified RNA as the template and rplU primer set (below) were carried out to ensure no DNA contamination of the RNA. The purified RNA was then used as a template to construct cDNA using SuperScript III (Invitrogen), according to the manufacturer's instructions, and random decamer primers [(NS)5]. The resulting cDNA was used as a template in PCRs with the following primers: rplU forward (5′-GCAGCACAAAGTCACCGAAGG-3′)), rplU reverse (5′-CCGTGGGAAACCACTTCAGC-3′), pqsH forward (5′-ACGACCTCGAGGAGTTGG-3′) and pqsH reverse (5′-GAACAGGATCAGCGTCTCG-3′). The PCR cycles were 10 min at 94 °C; 30 cycles of 30 s at 94 °C, 30 s at 54 °C, 1 min at 72 °C; 15 min at 72 °C.
Real-time PCR was conducted with an Applied Biosystems 7500 cycler. Conditions were identical to those used in the previous PCRs, with the following exceptions: for ppiD we used ppiD forward (5′-CGGGCACCGGTTTCG-3′) and ppiD reverse (5′-AAGTCGCGGGTCTGCTTCT-3′) in place of rplU primers, 40 cycles were used instead of 30, and the addition of a dissociation curve cycling was added to the end of the 15 min at 72 °C hold. The Power SYBR Green PCR kit (ABI) was used in the reactions. Three technical replicates were analysed for each biological replicate.
C4HSL bioassay.
P. aeruginosa cell extracts were obtained as described for PQS production at 10 or 14 h post-inoculation in biological triplicates as described in the text. Dried cell extracts were solubilized in 200 μl 1 : 1 acetonitrile:acidified ethyl acetate. Overnight cultures of E. coli JM109 harbouring pSB536 were grown overnight, diluted 1 : 100, then allowed to reach an OD600 of 0.3. C4HSL extracts (5 μl) were added in triplicate to each well of black-welled polypropylene plates (Costar), and the solvent was allowed to evaporate. An authentic C4HSL standard was included in each bioassay. One hundred microlitres of culture was added to each well, and the plate was statically incubated for 3 h at 37 °C. Luminescence was measured with a Victor2 fluorometer (Perkin Elmer). Cells were transferred to a clear 96-well plate and the OD600 was measured to use to normalize luminescence readings.
Analysis of growth on agar plugs.
P. aeruginosa microtitre dish cultures were grown as described above. Cells were grown at 37 °C for 6, 10 or 14 h, plugs were added to 1 ml PBS, mixed by vortexing, serially diluted, and plated in triplicate to enumerate colonies.
RESULTS
C. albicans-produced farnesol stimulates phenazine production in P. aeruginosa ΔlasR mutants
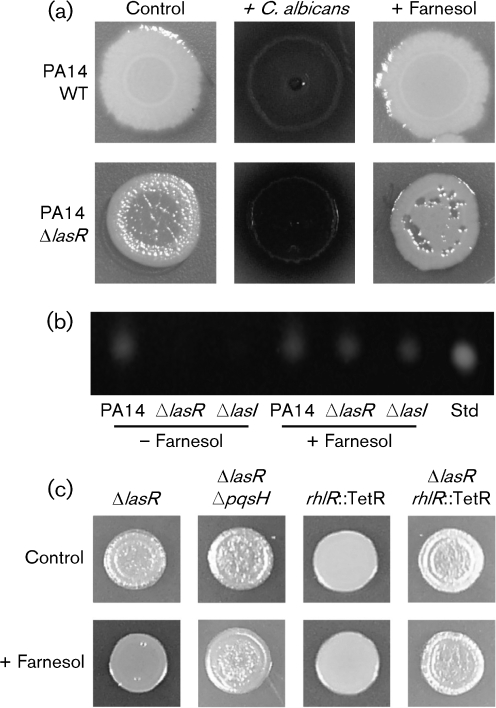
P. aeruginosa PA14 strains that lack lasR and lasI are defective in the production of quorum-sensing-controlled factors such as phenazines (Latifi et al., 1995). When inoculated onto lawns of C. albicans, however, ΔlasR (Fig. 1a) and ΔlasI (not shown) exhibited greatly enhanced phenazine production, as indicated by the red and blue colours conferred by 5-methylphenazium-1-carboxylate and pyocyanin, respectively (Gibson et al., 2009). C. albicans secretes farnesol, an auto-regulatory molecule, which participates in intraspecies (Hornby et al., 2001; Langford et al., 2009) and interspecies (Peleg et al., 2010; Shirtliff et al., 2009b) interactions. The addition of farnesol to the medium was sufficient to induce a striking increase in pyocyanin production in lasR and lasI mutant strains (Fig. 1a). Pyocyanin concentrations were more than threefold higher in PA14ΔlasR colonies grown with farnesol (0.22 μmol per plug) compared with control colonies (0.07 μmol per plug) (Supplementary Fig. S1, available with the online version of this paper). Farnesol induced only a modest increase in pyocyanin in PA14 wild-type (WT) colonies. In the presence of farnesol, the levels of pyocyanin in lasR colonies grown with farnesol were more than twofold higher than concentrations in WT colonies (Supplementary Fig. S1). Furthermore, pyocyanin appeared to be largely in its colourless reduced state in mature WT colonies, while the blue–green colour of oxidized pyocyanin was observed in lasR strains (Fig. 1a). In addition to the lack of pigmentation, PA14Δlas mutants appeared to accumulate HHQ, which gives colonies an iridescent sheen when grown on LB agar (Fig. 1a) (D'Argenio et al., 2007). The presence of farnesol suppressed the apparent HHQ accumulation (Fig. 1a). The increased phenazine production and apparent decrease in the accumulation of the PQS precursor, HHQ, upon growth with C. albicans or farnesol suggest that farnesol stimulates PQS production in the lasR and lasI mutant strains.
Fig. 1.
P. aeruginosa colony appearance and PQS production in the presence of C. albicans or farnesol. (a) P. aeruginosa strain PA14 WT and ΔlasR colonies after 24 h of growth on LB (control), in the presence of C. albicans or on LB plates containing 250 μM farnesol. (b) PQS analysis by TLC from cultures of PA14 WT, PA14ΔlasR and PA14ΔlasI grown in the absence or presence of 250 μM farnesol for 14 h. An authentic PQS standard (25 ng) was included for comparison. (c) P. aeruginosa strains PA14ΔlasR, PA14ΔlasRΔpqsH, PA14 rhlR : : TetR and PA14ΔlasRrhlR : : TetR grown in the absence (control) or presence of 250 μM farnesol. Colonies were photographed after 24 h of growth.
Farnesol stimulates PQS production in ΔlasR and ΔlasI strains and LasR-defective clinical strains
To test the hypothesis that farnesol was stimulating PQS production in lasR and lasI mutants, colonies were grown on agar in a 96-well plate format in either the presence or absence of farnesol, followed by extraction and analysis of PQS from the entire agar plug at different time points. By 6 h, P. aeruginosa strain PA14 WT colonies produced detectable levels of PQS that continued to increase over 24 h. As expected, PQS levels were much lower in PA14ΔlasR and PA14ΔlasI colony cultures, with PQS at barely detectable levels at the 14 h time point (Fig. 1b). In contrast, on farnesol-containing medium, ΔlasR, ΔlasI and PA14 WT colonies all had comparable PQS levels at 14 h (Fig. 1b). We previously showed that farnesol significantly delays PQS production in P. aeruginosa PA14 WT cultures grown in liquid media (Cugini et al., 2007), and we found that farnesol also inhibits PQS production in PA14 colonies at early time points (4 and 5 h) but this repression was no longer apparent once the population size reached approximately 109 c.f.u. per colony biofilm (data not shown). Farnesol did not cause any changes in growth rate in either WT or ΔlasR colonies (Supplementary Fig. S2, available with the online version of this paper).
Growth in co-culture with farnesol-producing C. albicans also abolished defects in PQS production in lasR and lasI mutant strains. In co-culture with the fungus, PA14 WT produced 71±1 ng PQS per colony and PA14ΔlasR produced 135±3 ng PQS per co-culture colony. The higher levels of PQS observed in ΔlasR colonies, relative to WT colonies, was consistent with the higher levels of pyocyanin in ΔlasR colonies grown with C. albicans or farnesol as described above (Fig. 1 and Supplementary Fig S1). To confirm that the fungus did not produce a compound that co-migrated with PQS, PA14ΔpqsR and PA14ΔlasRΔpqsR strains were constructed and analysed using the TLC method. No PQS was detected in these backgrounds upon co-culture with C. albicans (Supplementary Fig. S3, available with the online version of this paper).
LasR-defective isolates are frequently isolated from the lungs of CF patients (Hoffman et al., 2009; Smith et al., 2006). To determine if LasR− isolates from CF patients also showed enhanced PQS production upon growth with either C. albicans or farnesol, previously characterized P. aeruginosa strains with different loss-of-function mutations in lasR were examined. All of the seven LasR-defective clinical isolates tested produced little to no PQS on LB agar, and all but one strain showed a strong stimulation of PQS production on LB agar amended with farnesol (Supplementary Fig. S4a, available with the online version of this paper). In four of the six strains that produced PQS in the presence of farnesol, co-culture with C. albicans led to a strong increase in phenazine production (Supplementary Fig. S4b).
Farnesol promotes pqsH expression via the RhlR/C4HSL signalling pathway
Because farnesol caused a visual decrease in the apparent accumulation of HHQ (Fig. 1a) and stimulated the production of PQS (Fig. 1b), we hypothesized that farnesol was causing increased expression of pqsH. PqsH catalyses the conversion of HHQ to PQS, and in P. aeruginosa WT strains, LasR directly activates the transcription of pqsH (Gallagher et al., 2002; Gilbert et al., 2009). A ΔlasRΔpqsH mutant was constructed, and its colony phenotype was comparable to that of the ΔlasR parent. However, while the lasR mutant lost its iridescent sheen and exhibited increased pyocyanin production upon growth with farnesol, the ΔlasRΔpqsH mutant did not show any changes in colony morphology on farnesol-containing medium relative to control colonies (Fig. 1c). Consistent with published reports, deletion of pqsH in a WT strain abolished PQS production (Table 2) and led to the appearance of the iridescent sheen attributed to HHQ accumulation (data not shown) (Gallagher et al., 2002). Furthermore, pqsH expression in ΔlasR strains was sufficient to restore the production of PQS and pyocyanin in the absence of C. albicans or farnesol. PA14ΔlasR colonies carrying pqsH on a plasmid produced 252±7 ng PQS per plug at a time when no PQS was detected in colonies of ΔlasR with the empty vector (Table 2). Expression of pqsH also rescued PQS production in ΔlasRΔpqsH and ΔpqsH strains (Table 2). Together, these data show that pqsH was necessary for farnesol-induced PQS production, and that the expression of pqsH was sufficient to induce robust PQS production in ΔlasR and ΔlasI strains.
Table 2.
PQS production in lasR strains
nd, Not detected. Data shown are mean±sd.
| Strain | PQS (ng per plug) |
|---|---|
| PA14ΔlasR+pUCP22 | nd |
| PA14ΔlasR+pPQSH | 252±7 |
| PA14ΔpqsH+pUCP22 | nd |
| PA14ΔpqsH+pPQSH | 73±3 |
| PA14ΔlasRΔpqsH+pUCP22 | nd |
| PA14ΔlasRΔpqsH+pPQSH | 226±8 |
| PA14ΔlasR | nd |
| PA14ΔlasR+25 μM C4HSL | 106±7 |
| PA14ΔlasRrhlR : : TetR | nd |
| PA14ΔlasRrhlR : : TetR+25 μM C4HSL | nd |
| PA14ΔlasRΔrhlI | nd |
| PA14ΔlasRΔrhlI+25 μM C4HSL | 11±1 |
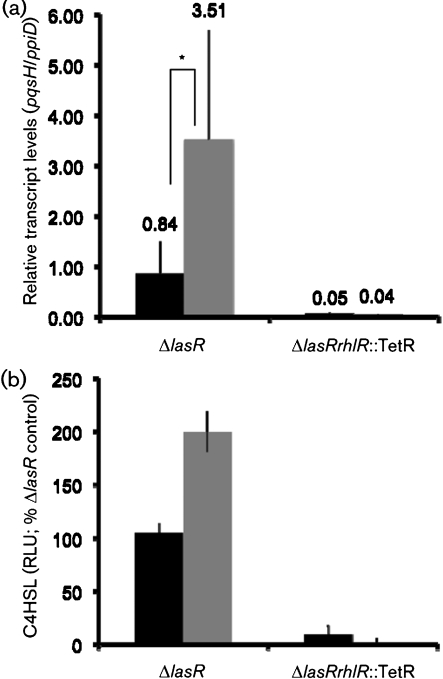
To determine whether farnesol led to increased pqsH transcript levels, ΔlasR colonies were grown in the presence and absence of farnesol, and RNA was extracted for analysis. PA14ΔlasR colonies grown in the presence of farnesol for 12 h had a significant increase in pqsH mRNA in comparison with those cells that received vehicle alone (Fig. 2a). As predicted by the model that PqsR positively regulates the expression of the pqsA–E operon in response to PQS, pqsA levels were several-fold higher in ΔlasR colonies that were grown in the presence of farnesol (data not shown).
Fig. 2.
Effects of farnesol on the levels of pqsH transcript and C4HSL. (a) RT-PCR analysis of strains PA14ΔlasR and PA14ΔlasRrhlR : : TetR colonies grown in the absence (black bars) and presence (grey bars) of 250 μM farnesol. The levels of pqsH and ppiD were measured in control colonies and colonies grown in the presence of farnesol for 12 h. While there is variability in the biological replicates within an experiment, likely due to the fact that quorum-sensing-controlled genes are rapidly changing in colonies at these time points, statistically significant increases in pqsH in the presence of farnesol were observed in four independent experiments. Data presented represent three biological replicates from one independent experiment (mean±sd). *P<0.1 based on Student's t-test. (b) C4HSL levels were measured from PA14ΔlasR and PA14ΔlasRrhlR : : TetR colonies grown for 10 h in the absence (black bars) and presence (grey bars) of 250 μM farnesol. Levels were determined by luciferase activity generated by the reporter plasmid pSB536, which detects short-chain AHLs, verified by comparison with a titrated authentic C4HSL standard and reported as relative luminescence units (RLU) after normalization to levels in control cultures. Error bars, sd.
In contrast with the stimulation of PQS production by farnesol in a lasR mutant, a ΔlasRrhlR : : TetR double mutant still exhibited the HHQ-associated iridescent sheen, and pyocyanin was not observed in colonies grown on medium with farnesol (Fig. 1c), suggesting that RhlR may play a role in the farnesol response. Phenazine production was also not observed upon co-culture of the ΔlasRrhlR : : TetR strain with the fungus (data not shown). Consistent with the phenotypic data, the increase in pqsH mRNA in lasR cells upon growth on farnesol was not seen in PA14ΔlasRrhlR : : TetR cells (Fig. 2a). Even in control conditions, levels of pqsH mRNA were lower in ΔlasRrhlR : : TetR compared with a ΔlasR mutant (Fig. 2a). Constitutive expression of pqsH restored PQS production in PA14ΔlasRrhlR : : TetR (Supplementary Fig. S5a, available with the online version of this paper) and PA14ΔlasRΔrhlI (data not shown). Evidence for RhlR-mediated regulation of pqsH expression in PA14ΔlasR in medium with farnesol is consistent with recently published data that show that RhlR is required for late (after 24 h) PQS production in the PA14ΔlasR liquid cultures (Dekimpe & Deziel, 2009).
Farnesol increases C4HSL production and C4HSL is sufficient to induce PQS production in ΔlasR
RhlR activity requires binding of C4HSL, which is synthesized by RhlI (Latifi et al., 1995). To determine if farnesol stimulated RhlR activity in a lasR mutant background by acting as an alternative ligand to C4HSL for RhlR activation, we determined if RhlI was necessary for PQS production in lasR mutants grown on farnesol. A PA14ΔlasRΔrhlI mutant was constructed, and found not to support PQS production on LB with farnesol, indicating that farnesol does not act as a surrogate for C4HSL and that it cannot stimulate RhlR activity in a C4HSL-independent manner. While RhlR and RhlI were required for PQS production in ΔlasR (Table 2 and Supplementary Fig. S5a), single rhlR : : TetR or ΔrhlI mutants were not defective in PQS production and, in fact, produced higher levels of PQS relative to WT control strains (Supplementary Fig. S5b), as has been previously reported (McGrath et al., 2004).
To determine if RhlR–C4HSL regulation was induced by farnesol, we determined if levels of C4HSL were higher in farnesol-grown cultures compared with controls. PA14ΔlasR grown with farnesol had a nearly twofold increase in C4HSL over ΔlasR control cultures as determined by the E. coli Lux-based biosensor assay that detects short chain AHLs (C4- and C6HSL) (Fig. 2b). As expected, PA14ΔlasRrhlR : : TetR did not produce detectable levels of C4HSL in either the presence or absence of farnesol, further supporting the hypothesis that the increase in C4HSL induced by farnesol is dependent on the activity of RhlR.
Given the elevated C4HSL levels, we sought to determine if the addition of exogenous C4HSL was sufficient to restore RhlR-dependent production of PQS in the lasR mutant. In the presence of 25 μM C4HSL, PA14ΔlasR colonies produced 106±7 ng PQS per plug at 14 h, whereas in control cultures without C4HSL, PQS was undetectable (Table 2). Furthermore, PA14ΔlasR colonies grown in the presence of C4HSL had a smooth colony morphology consistent with decreased accumulation of HHQ (data not shown). C4HSL addition to a PA14ΔlasRrhlR : : TetR did not rescue PQS production (Table 2). As expected based on the results shown above, the ΔlasRΔrhlI mutant regained the ability to produce PQS (Table 2) and pyocyanin (not shown) upon growth with C4HSL.
Farnesol-induced oxidative stress leads to increased C4HSL signalling
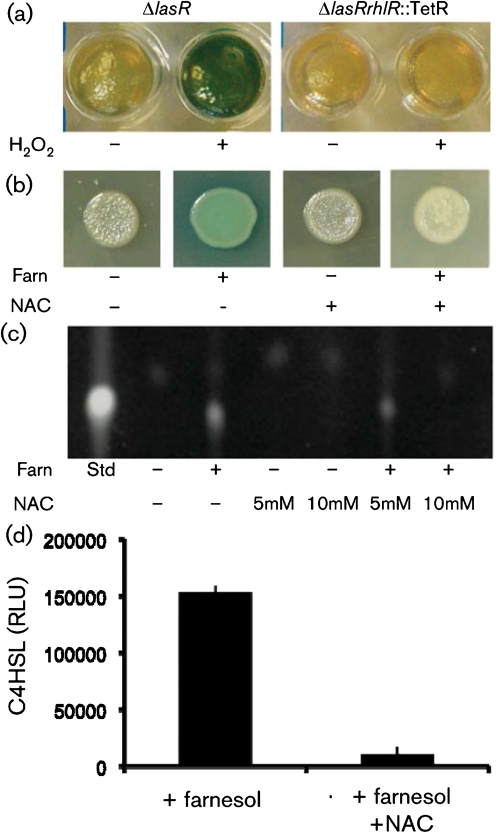
Previously, farnesol has been reported to induce oxidative stress in multiple microbes (Semighini et al., 2006; Shirtliff et al., 2009a), potentially through interactions with electron transfer reactions during respiration (Machida & Tanaka, 1999). To determine if oxidative stress induced by farnesol mediated the stimulation of RhlR–C4HSL signalling, we determined if hydrogen peroxide was sufficient to stimulate RhlR-dependent PQS and pyocyanin production in PA14ΔlasR strains. Hydrogen peroxide (200 μM) was added to the agar medium prior to inoculation, and the colonies were grown for 48 h. As shown in Fig. 3(a), hydrogen peroxide stimulated pyocyanin production in PA14ΔlasR, but not PA14ΔlasRrhlR : : TetR strains. Quantification of pyocyanin levels in these cultures is presented in Supplementary Fig. S6(a). Addition of NAC, an antioxidant, to the medium abolished the stimulation of pyocyanin production by hydrogen peroxide in lasR mutant strains. NAC did not affect pyocyanin levels in aerated WT cultures, indicating that it does not have an independent effect on phenazine production. NAC also repressed the farnesol-mediated stimulation of pyocyanin (Fig. 3b) and PQS (Fig. 3c) in PA14ΔlasR strains suggesting that ROS were mediating farnesol's effects on quorum sensing. The number of c.f.u. in colonies grown with farnesol alone or farnesol with NAC were comparable (Supplementary Fig. S6b). C4HSL levels in ΔlasR colonies were also lower upon the addition of 10 mM NAC to medium with farnesol (Fig. 3d).
Fig. 3.
P. aeruginosa colony appearance and PQS and C4HSL production by PA14ΔlasR grown in the presence of farnesol, hydrogen peroxide and the oxidative stress protectant NAC. (a) PA14 ΔlasR and ΔlasRrhlR : : TetR colonies were grown in the absence or presence of hydrogen peroxide and photographed after 48 h. Colonies were grown on agar plugs in a 96-well microtitre dish. (b) PA14ΔlasR grown as spotted colonies on agar plates containing vehicle alone, 250 μM farnesol, 10 mM NAC or the combination of 10 mM NAC and 250 μM farnesol. (c) PQS production by ΔlasR colonies grown for 14 h in the absence and presence of 5 and 10 mM NAC, 250 μM farnesol or the combination of 5 and 10 mM NAC and 250 μM farnesol. PQS production was determined by TLC analysis and PQS was identified by comparison with an authentic PQS standard (25 ng). (d) C4HSL levels were measured from ΔlasR colonies grown for 14 h in the absence and presence of 250 μM farnesol or farnesol with 10 mM NAC. Levels were determined by luciferase activity (RLU) generated by the reporter plasmid pSB536, which detects short-chain AHLs. The assay was validated using a titrated authentic C4HSL standard.
As has been reported previously, ΔlasR strains can produce PQS after prolonged incubation (Dekimpe & Deziel, 2009; Diggle et al., 2003), and we observed low levels of PQS in ΔlasR colonies after entry into stationary phase. The slight production of PQS and pyocyanin by PA14ΔlasR in late stationary phase cultures or colonies was also suppressed by the addition of NAC to the medium, suggesting that late PQS production in LasR− strains is due to the effects of oxidative stress (data not shown). The PA14ΔlasRΔpqsH and ΔlasRrhlR : : TetR mutants, which no longer produced PQS on medium with farnesol (Fig. 1c), also lacked PQS production in cultures even when examined at very late time points (data not shown).
DISCUSSION
Numerous studies in multiple models of acute infection have demonstrated a marked decrease in virulence upon loss of LasR function, and this attenuation is likely due to the inability to induce important virulence factors (Lesprit et al., 2003; Preston et al., 1997; Rumbaugh et al., 1999; Tang et al., 1996). In contrast, the emergence of lasR mutants in chronic CF-related infections correlates with decreased lung function, suggesting that these strains may retain some virulence properties to complement their growth advantages in the CF lung environment (Hoffman et al., 2009). Recent reports indicate that virulence factors, such as pyocyanin and elastase, which normally require LasR for production, are made by ΔlasR mutants in stationary phase cultures grown in TSB medium (Dekimpe & Deziel, 2009). The late production of these virulence factors is dependent on RhlR and C4HSL (Dekimpe & Deziel, 2009).
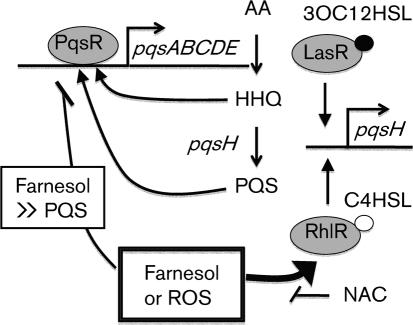
Here, we report that C. albicans and its secreted factor farnesol caused an increase in the levels of two quorum sensing molecules, PQS (Fig. 1b and Table 2) and C4HSL (Figs 2b and 3d), in the ΔlasR strain (see Fig. 4 for model). The increases in these signalling molecules translated into the increased production of the quorum-sensing-regulated phenazine virulence factors (Figs 1 and 3, and Supplementary Fig. S4). The increased production of PQS was dependent on rhlR, rhlI and pqsH, and RhlR was required for increased pqsH transcript levels in ΔlasR strains in the presence of farnesol (Fig. 2a). Similarly, the addition of C4HSL rescued PQS production in ΔlasR (Table 2) and ΔlasI strains (data not shown). While RhlR-dependent control of pqsH in a lasR mutant background was proposed previously (Dekimpe & Deziel, 2009), it had not previously been directly tested. We demonstrated that pqsH expression was sufficient to restore pyocyanin and PQS production in lasR and lasI mutants (Table 2 and data not shown). Farnesol induced C4HSL levels (Fig. 3b) and, as predicted by the positive feedback loop that controls rhlR and rhlI expression, rhlR and rhlI transcript levels increased by 11- and 7.3-fold, respectively, in the presence of farnesol (data not shown). Because RhlR is not required for the expression of pqsH in LasR+ backgrounds, and, in fact, PQS levels are higher in rhlI and rhlR single mutants (Supplementary Fig. S5b) (McGrath et al., 2004; Xiao et al., 2006a), this activity of RhlR in LasR− strains represents an alternate mechanism for the regulation of the downstream portion of the quorum sensing regulatory pathway when the LasR function is absent.
Fig. 4.
Model for the effects of farnesol on PQS production in P. aeruginosa. Anthranilic acid (AA) is converted into HHQ by genes encoded by pqsA–D. HHQ is converted into PQS by PqsH. HHQ and PQS contribute to the positive feedback regulation of the pqsA–D genes via PqsR. Farnesol inhibits PqsR activity when PQS is at low levels. Inhibition of PqsR activity by farnesol is not complete, and as HHQ or PQS levels increase, farnesol no longer competes with natural PqsR ligands. Expression of pqsH is controlled by LasR when it is complexed with 3OC12HSL, and the RhlR–C4HSL complex is not needed for induction of pqsH in wild-type cells. In lasR or lasI mutants, RhlR–C4HSL can activate pqsH. While LasR–3OC12HSL is normally required for induction of rhlR and rhlI, farnesol or hydrogen peroxide can stimulate RhlR–C4HSL activity in the absence of LasR–3OC12HSL.
Our data suggest that RhlR is activated in ΔlasR strains by ROS or oxidative stress. Farnesol perturbs electron transport in yeast mitochondria leading to the generation of ROS (Machida et al., 1998) and promotes the generation of ROS in other fungi (Semighini et al., 2006; Shirtliff et al., 2009a). Research suggests that the same is true in bacteria (Gomes et al., 2009). Because hydrogen peroxide is sufficient to induce PQS and pyocyanin production in ΔlasR, and NAC suppresses the induction of PQS production in ΔlasR strains grown with farnesol or hydrogen peroxide, we predict that farnesol-induced oxygen radicals are the key factor that triggers activation of the lower portion of the quorum sensing pathway (for a model see Fig. 4). While the mechanism by which ROS influence RhlR and RhlI remains the subject of future work, it is interesting to consider a potential role for the Lon protease. Lon degrades both short-lived regulators and damaged proteins (Van Melderen & Aertsen, 2009), and P. aeruginosa Lon has been shown previously to negatively regulate the RhlI synthase, which produces C4HSL (Takaya et al., 2008). Perhaps ROS exposure somehow alters Lon-mediated degradation of RhlI allowing for the accumulation of higher levels of the C4HSL synthase, activation of the RhlR autoregulatory loop and, ultimately, pqsH expression. NAC also affects PQS production in PA14ΔlasR cultures in the absence of farnesol. The production of PQS and pyocyanin that is observed in late stationary phase ΔlasR and ΔlasI liquid cultures was suppressed by NAC (data not shown). It is not yet clear whether farnesol affects P. aeruginosa WT and Δlas strains differently, though there is a clear change in respiration upon the loss of LasR (Hoffman et al., 2010), and these differences may impact how farnesol affects cells. It is interesting to note that NAC is currently in phase II clinical trials as a useful alternative therapy for treatment of CF (Cystic Fibrosis Foundation, 2006). In addition to the direct benefits of reducing ROS in the lungs, NAC might have the added benefit of decreasing P. aeruginosa virulence by preventing activation of this alternative regulatory pathway.
Published evidence supports the idea that there is a connection between oxidative stress and quorum sensing regulation. PQS not only induces genes involved in oxidative stress (Bredenbruch et al., 2006; Häussler & Becker, 2008) but also confers the induction of a protective oxidative stress response (Häussler & Becker, 2008). Furthermore, an oxyR mutant, with decreased expression of genes that encode ROS scavenging enzymes, exhibits increased levels of pyocyanin (Vinckx et al., 2010). While no differences in PQS levels were observed at 24 h, it would be interesting to determine whether the oxyR mutant has accelerated PQS production relative to WT strains. While it was not the focus of the work, we did observe slightly increased PQS when P. aeruginosa WT was grown on medium with farnesol or hydrogen peroxide (data not shown). Activation of PQS production by ROS may indicate a role for PQS in defence. In the presence of polymorphonuclear neutrophils, P. aeruginosa had elevated levels of RhlR-regulated rhamnolipids and increased expression of the PQS biosynthetic operon (Alhede et al., 2009).
Farnesol has multiple effects on P. aeruginosa cells. Previously, we reported that C. albicans and farnesol led to decreased PQS and pyocyanin levels in P. aeruginosa by affecting the ability of the PqsR–PQS complex to activate pqsA transcription (Cugini et al., 2007). We found that either PQS or farnesol promotes PqsR binding to the pqsA promoter, but only PQS leads to transcriptional activation of the pqsA–E promoter. The inhibition of PqsR activity by farnesol only occurred at lower cell densities due to the fact that PqsR appears to have a greater affinity for PQS than for farnesol by several orders of magnitude (see Fig. 4 for a model). As in liquid cultures, farnesol also led to a delay in early PQS production in PA14 WT colony biofilms, but this repression was not observed as colony density increased. These data indicate that farnesol affects P. aeruginosa quorum sensing through both interaction with specific targets (Cugini et al., 2007) and the generation of ROS.
Both clinical and environmental strains commonly have mutations in lasR, and LasR loss-of-function mutants can be selected for under laboratory conditions (Cabrol et al., 2003; Dénervaud et al., 2004; Heurlier et al., 2005; Hoffman et al., 2009; Luján et al., 2007; Smith et al., 2006; Wilder et al., 2009). Diffusible signals such as C4HSL and PQS can stimulate virulence factor production in las mutants (Déziel et al., 2004; Sandoz et al., 2007). Thus, the presence of LasR+ P. aeruginosa strains may be sufficient to compensate for loss of LasR function (Sandoz et al., 2007). Here, we demonstrate that other microbes, such as the fungus C. albicans, and the environmental conditions created through the secretion of small molecules, also induce virulence factor production in lasR-defective laboratory and clinical mutants (Fig. 1a and Supplementary Fig S4b).
Acknowledgments
This publication was made possible by the Cystic Fibrosis Foundation, the Pew Biomedical Scholars Program, and National Institutes of Health Grant Number 1 P20 RR018787 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources (D. A. H.). Additional support was provided by the National Institutes of Health Molecular Pathogenesis Training Grant T32 AI07519 (C. C.). We would like to thank Malathy Krishnamurthy, currently at University of Michigan, for providing the PQS standard used. We would also like to that Sathish Rajamani for providing pSB536 for the C4HSL studies.
Abbreviations
3OC12HSL, 3-oxo-C12-homoserine lactone
AHL, acyl homoserine lactone
C4HSL, N-butyryl-homoserine lactone
CF, cystic fibrosis
HHQ, 2-heptyl-4-quinolone
NAC, N-acetyl cysteine
PQS, Pseudomonas quinolone signal
RLU, relative luminescence units
ROS, reactive oxygen species
TLC, thin-layer chromatography
WT, wild-type
Footnotes
Six supplementary figures are available with the online version of this paper.
References
- Abramoff, M. D., Magelhaes, P. J. & Ram, S. J. (2004). Image processing with ImageJ. Biophotonics International 11, 36–42. [Google Scholar]
- Alhede, M., Bjarnsholt, T., Jensen, P. Ø., Phipps, R. K., Moser, C., Christophersen, L., Christensen, L. D., van Gennip, M., Parsek, M. & other authors (2009). Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155, 3500–3508. [DOI] [PubMed] [Google Scholar]
- Allard, J. B., Rinaldi, L., Wargo, M. J., Allen, G., Akira, S., Uematsu, S., Poynter, M. E., Hogan, D. A., Rincon, M. & Whittaker, L. A. (2009). Th2 allergic immune response to inhaled fungal antigens is modulated by TLR-4-independent bacterial products. Eur J Immunol 39, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakare, N., Rickerts, V., Bargon, J. & Just-Nubling, G. (2003). Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 46, 19–23. [DOI] [PubMed] [Google Scholar]
- Bredenbruch, F., Geffers, R., Nimtz, M., Buer, J. & Haussler, S. (2006). The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol 8, 1318–1329. [DOI] [PubMed] [Google Scholar]
- Cabrol, S., Olliver, A., Pier, G. B., Andremont, A. & Ruimy, R. (2003). Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol 185, 7222–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotirmall, S. H., O'Donoghue, E., Bennett, K., Gunaratnam, C., O'Neill, S. J. & McElvaney, N. G. (2010). Sputum Candida albicans presages FEV1 decline and hospitalized exacerbations in cystic fibrosis. Chest (in press ). doi: 10.1378/chest.09-2996 [DOI] [PubMed]
- Cugini, C., Calfee, M. W., Farrow, J. M., III, Morales, D. K., Pesci, E. C. & Hogan, D. A. (2007). Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol 65, 896–906. [DOI] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation. (2006). http://www.cff.org/treatments/Therapies/AlternativeTherapies/Antioxidants/.
- D'Argenio, D. A., Wu, M., Hoffman, L. R., Kulasekara, H. D., Déziel, E., Smith, E. E., Nguyen, H., Ernst, R. K., Larson Freeman, T. J. & other authors (2007). Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64, 512–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe, V. & Deziel, E. (2009). Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155, 712–723. [DOI] [PubMed] [Google Scholar]
- Dénervaud, V., TuQuoc, P., Blanc, D., Favre-Bonte, S., Krishnapillai, V., Reimmann, C., Haas, D. & van Delden, C. (2004). Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol 42, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel, E., Lepine, F., Milot, S., He, J., Mindrinos, M. N., Tompkins, R. G. & Rahme, L. G. (2004). Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101, 1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle, S. P., Winzer, K., Chhabra, S. R., Worrall, K. E., Camara, M. & Williams, P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 50, 29–43. [DOI] [PubMed] [Google Scholar]
- Elble, R. (1992). A simple and efficient procedure for transformation of yeasts. Biotechniques 13, 18–20. [PubMed] [Google Scholar]
- Ernst, R. K., Moskowitz, S. M., Emerson, J. C., Kraig, G. M., Adams, K. N., Harvey, M. D., Ramsey, B., Speert, D. P., Burns, J. L. & Miller, S. I. (2007). Unique lipid A modifications in Pseudomonas aeruginosa isolated from the airways of patients with cystic fibrosis. J Infect Dis 196, 1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow, J. M., Sund, Z. M., Ellison, M. L., Wade, D. S., Coleman, J. P. & Pesci, E. C. (2008). PqsE functions independently of PqsR–Pseudomonas Quinolone Signal and enhances the rhl quorum sensing system. J Bacteriol 190, 7043–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher, L. A., McKnight, S. L., Kuznetsova, M. S., Pesci, E. C. & Manoil, C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184, 6472–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, J., Sood, A. & Hogan, D. A. (2009). Pseudomonas aeruginosa–Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol 75, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, K. B., Kim, T. H., Gupta, R., Greenberg, E. P. & Schuster, M. (2009). Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol 73, 1072–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum, A. M., Tsay, E. Y. & Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Gomes, F. I. A., Teixeira, P., Azeredo, J. & Oliveira, R. (2009). Effect of farnesol on planktonic and biofilm cells of Staphylococcus epidermidis. Curr Microbiol 59, 118–122. [DOI] [PubMed] [Google Scholar]
- Haase, G., Skopnik, H., Groten, T., Kusenbach, G. & Posselt, H. G. (1991). Long-term fungal cultures from sputum of patients with cystic fibrosis. Mycoses 34, 373–376. [DOI] [PubMed] [Google Scholar]
- Häussler, S. & Becker, T. (2008). The Pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4, e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier, K., Denervaud, V., Haenni, M., Guy, L., Krishnapillai, V. & Haas, D. (2005). Quorum sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol 187, 4875–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J. & Schweizer, H. P. (1998). A broad-host-range Flp–FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Hoffman, C. S. & Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Hoffman, L. R., Kulasekara, H. D., Emerson, J., Houston, L. S., Burns, J. L., Ramsey, B. W. & Miller, S. I. (2009). Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8, 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. R., Richardson, A. R., Houston, L. S., Kulasekara, H. D., Martens-Habbena, W., Klausen, M., Burns, J. L., Stahl, D. A., Hassett, D. J. & other authors (2010). Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog 6, e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D. A. & Kolter, R. (2002). Pseudomonas–Candida interactions: an ecological role for virulence factors. Science 296, 2229–2232. [DOI] [PubMed] [Google Scholar]
- Hogan, D. A., Vik, A. & Kolter, R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol 54, 1212–1223. [DOI] [PubMed] [Google Scholar]
- Hornby, J. M., Jensen, E. C., Lisec, A. D., Tasto, J. J., Jahnke, B., Shoemaker, R., Dussault, P. & Nickerson, K. W. (2001). Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67, 2982–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K. & Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. [DOI] [PubMed] [Google Scholar]
- Langford, M. L., Atkin, A. L. & Nickerson, K. W. (2009). Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans. Future Microbiol 4, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Langford, M. L., Hasim, S., Nickerson, K. W. & Atkin, A. L. (2010). Activity and toxicity of farnesol towards Candida albicans are dependent on growth conditions. Antimicrob Agents Chemother 54, 940–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifi, A., Winson, M. K., Foglino, M., Bycroft, B. W., Stewart, G. S., Lazdunski, A. & Williams, P. (1995). Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol 17, 333–343. [DOI] [PubMed] [Google Scholar]
- Lesprit, P., Faurisson, F., Join-Lambert, O., Roudot-Thoraval, F., Foglino, M., Vissuzaine, C. & Carbon, C. (2003). Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. Am J Respir Crit Care Med 167, 1478–1482. [DOI] [PubMed] [Google Scholar]
- Li, Z. H., Kosorok, M. R., Farrell, P. M., Laxova, A., West, S. E. H., Green, C. G., Collins, J., Rock, M. J. & Splaingard, M. L. (2005). Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293, 581–588. [DOI] [PubMed] [Google Scholar]
- Luján, A. M., Moyano, A. J., Segura, I., Argarana, C. E. & Smania, A. M. (2007). Quorum-sensing-deficient (lasR) mutants emerge at high frequency from a Pseudomonas aeruginosa mutS strain. Microbiology 153, 225–237. [DOI] [PubMed] [Google Scholar]
- Machida, K. & Tanaka, T. (1999). Farnesol-induced generation of reactive oxygen species dependent on mitochondrial transmembrane potential hyperpolarization mediated by F(0)F(1)-ATPase in yeast. FEBS Lett 462, 108–112. [DOI] [PubMed] [Google Scholar]
- Machida, K., Tanaka, T., Fujita, K. & Taniguchi, M. (1998). Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J Bacteriol 180, 4460–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam, E., Campbell, M. E. & Speert, D. P. (1994). Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun 62, 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlester, G., O'Gara, F. & Morrissey, J. P. (2008). Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol 57, 563–569. [DOI] [PubMed] [Google Scholar]
- McGrath, S., Wade, D. S. & Pesci, E. C. (2004). Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol Lett 230, 27–34. [DOI] [PubMed] [Google Scholar]
- Navarro, J., Rainisio, M., Harms, H. K., Hodson, M. E., Koch, C., Mastella, G., Strandvik, B. & McKenzie, S. G. (2001). Factors associated with poor pulmonary function: cross-sectional analysis of data from the ERCF. European Epidemiologic Registry of Cystic Fibrosis. Eur Respir J 18, 298–305. [DOI] [PubMed] [Google Scholar]
- Peleg, A. Y., Hogan, D. A. & Mylonakis, E. (2010). Medically important bacterial-fungal interactions. Nat Rev Microbiol 8, 340–349. [DOI] [PubMed] [Google Scholar]
- Pesci, E. C., Pearson, J. P., Seed, P. C. & Iglewski, B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol 179, 3127–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci, E. C., Milbank, J., Pearson, J., McKnight, S., Kende, A., Greenberg, E. & Iglewski, B. (1999). Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 96, 11229–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, M. J., Seed, P. C., Toder, D. S., Iglewski, B. H., Ohman, D. E., Gustin, J. K., Goldberg, J. B. & Pier, G. B. (1997). Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect Immun 65, 3086–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. [DOI] [PubMed] [Google Scholar]
- Rajan, S. & Saiman, L. (2002). Pulmonary infections in patients with cystic fibrosis. Semin Respir Infect 17, 47–56. [DOI] [PubMed] [Google Scholar]
- Rumbaugh, K. P., Griswold, J. A., Iglewski, B. H. & Hamood, A. N. (1999). Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun 67, 5854–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz, K. M., Mitzimberg, S. M. & Schuster, M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A 104, 15876–15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semighini, C. P., Hornby, J., Dumitru, R., Nickerson, K. & Harris, S. (2006). Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol 59, 753–764. [DOI] [PubMed] [Google Scholar]
- Shanks, R. M., Caiazza, N. C., Hinsa, S. M., Toutain, C. M. & O'Toole, G. A. (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff, M. E., Krom, B. P., Meijering, R. A., Peters, B. M., Zhu, J., Scheper, M. A., Harris, M. L. & Jabra-Rizk, M. A. (2009a). Farnesol-induced apoptosis in Candida albicans. Antimicrob Agents Chemother 53, 2392–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirtliff, M. E., Peters, B. M. & Jabra-Rizk, M. A. (2009b). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D'Argenio, D. A., Miller, S. I., Ramsey, B. W., Speert, D. P. & other authors (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103, 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift, S., Karlyshev, A. V., Fish, L., Durant, E. L., Winson, M. K., Chhabra, S. R., Williams, P., Macintyre, S. & Stewart, G. S. (1997). Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J Bacteriol 179, 5271–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya, A., Tabuchi, F., Tsuchiya, H., Isogai, E. & Yamamoto, T. (2008). Negative regulation of quorum-sensing systems in Pseudomonas aeruginosa by ATP-dependent Lon protease. J Bacteriol 190, 4181–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H. B., DiMango, E., Bryan, R., Gambello, M., Iglewski, B. H., Goldberg, J. B. & Prince, A. (1996). Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun 64, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingpej, P., Smith, L., Rose, B., Zhu, H., Conibear, T., Al Nassafi, K., Manos, J., Elkins, M., Bye, P. & other authors (2007). Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J Clin Microbiol 45, 1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen, L. & Aertsen, A. (2009). Regulation and quality control by Lon-dependent proteolysis. Res Microbiol 160, 645–651. [DOI] [PubMed] [Google Scholar]
- Vinckx, T., Wei, Q., Matthijs, S. & Cornelis, P. (2010). The Pseudomonas aeruginosa oxidative stress regulator OxyR influences production of pyocyanin and rhamnolipids: protective role of pyocyanin. Microbiology 156, 678–686. [DOI] [PubMed] [Google Scholar]
- West, S. E., Schweizer, H. P., Dall, C., Sample, A. K. & Runyen-Janecky, L. J. (1994). Construction of improved Escherichia–Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148, 81–86. [DOI] [PubMed] [Google Scholar]
- Wilder, C. N., Allada, G. & Schuster, M. (2009). Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum sensing populations from cystic fibrosis lung infections. Infect Immun 77, 5631–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, G. P., Deziel, E., He, J. X., Lépine, F., Lesic, B., Castonguay, M. H., Milot, S., Tampakaki, A. P., Stachel, S. E. & Rahme, L. G. (2006a). MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62, 1689–1699. [DOI] [PubMed] [Google Scholar]
- Xiao, G. P., He, J. X. & Rahme, L. G. (2006b). Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152, 1679–1686. [DOI] [PubMed] [Google Scholar]