Abstract
The tight control of autolysis by Streptococcus mutans is critical for proper virulence gene expression and biofilm formation. A pair of dicistronic operons, SMU.575/574 (lrgAB) and SMU.1701/1700 (designated cidAB), encode putative membrane proteins that share structural features with the bacteriophage-encoded holin family of proteins, which modulate host cell lysis during lytic infection. Analysis of S. mutans lrg and cid mutants revealed a role for these operons in autolysis, biofilm formation, glucosyltransferase expression and oxidative stress tolerance. Expression of lrgAB was repressed during early exponential phase and was induced over 1000-fold as cells entered late exponential phase, whereas cidAB expression declined from early to late exponential phase. A two-component system encoded immediately upstream of lrgAB (LytST) was required for activation of lrgAB expression, but not for cid expression. In addition to availability of oxygen, glucose levels were revealed to affect lrg and cid transcription differentially and significantly, probably through CcpA (carbon catabolite protein A). Collectively, these findings demonstrate that the Cid/Lrg system can affect several virulence traits of S. mutans, and its expression is controlled by two major environmental signals, oxygen and glucose. Moreover, cid/lrg expression is tightly regulated by LytST and CcpA.
INTRODUCTION
The regulatory network for virulence factor expression by Streptococcus mutans, the principal aetiological agent of human dental caries (Loesche, 1986), is more complex than previously appreciated. Recent studies suggest that autolysins (Sturges & Rettger, 1922) may be central to modulation of virulence in many bacteria, including S. mutans (Ahn & Burne, 2006, 2007; Bayles, 2007; Rice et al., 2007; Rice & Bayles, 2008; Shibata et al., 2005). The autolytic process directly affects the biogenesis, stability and composition of the cell envelope, which is a dynamic and complex structure that expands as cells grow and is reshaped when cells divide or differentiate (Ghuysen et al., 1966; Höltje, 1995; Perkins, 1980; Shockman & Holfje, 1994; Ward & Williamson, 1984). Autolysins thus impact many cellular processes, including cell-wall turnover, cell growth, antibiotic resistance, cell-to-surface adhesion, genetic competence, protein secretion and pathogenicity (Berry et al., 1989; Blackman et al., 1998; Groicher et al., 2000; Heilmann et al., 1997; Mercier et al., 2002; Smith et al., 2000; Wuenscher et al., 1993).
Recently, autolysis has been proposed to be a form of programmed cell death (PCD) utilized by certain bacteria in an altruistic behaviour that enhances the survival of populations of organisms (Bayles, 2003, 2007; Engelberg-Kulka et al., 2006; Lewis, 2000; Rice & Bayles, 2003; Yarmolinsky, 1995). These studies are based on the observation that single-celled organisms can display complex social behaviours when organized in multicellular communities, such as biofilms. PCD by cell lysis would be beneficial to the persistence of multicellular biofilm structures by eliminating damaged cells from the population, while concurrently enhancing the survival of the remaining population via liberation of nutrients from dying cells. Programmed altruistic behaviours may also be connected to the ability of bacteria to cope with changes in the environment, previous studies having revealed that autolysis is strongly influenced by multiple environmental factors, including salt concentration, pH, glucose and oxygen (Ahn & Burne, 2007; Bowman & Redmond, 1956; Gilpin et al., 1972; Ochiai, 1999; Qoronfleh et al., 1998; Tobin et al., 1994; Wells & Russell, 1996; Yabu & Kaneda, 1995). Oxygen was recently demonstrated to have a profound impact on the regulatory pathways of AtlA, a major autolysin of S. mutans that modulates cell-surface biogenesis and composition in response to oxygen (Ahn & Burne, 2007; Ahn et al., 2007). Thus, the autolytic process of S. mutans is modulated by its genetic and physiological responses to environmental stimuli, particularly oxygen.
In our efforts to identify the regulatory mechanisms and pathways that control autolysis in S. mutans, we previously identified a pair of dicistronic operons, lrgAB (SMU.575/574) and cidAB (SMU.1701/1700), that were regulated in response to aerobic growth (Ahn et al., 2007). Here, we report that the lrg and cid operons are regulated in a complex manner and that their products have a significant impact on autolysis and virulence factor expression by S. mutans.
METHODS
Bacterial strains, plasmids, media and growth conditions.
Escherichia coli DH10B was grown in Luria broth and S. mutans UA159 and its derivatives were grown in brain heart infusion (BHI) broth (Difco). For selection of antibiotic-resistant colonies after genetic transformation, ampicillin (100 μg ml–1 for E. coli), erythromycin (300 μg ml–1 for E. coli and 10 μg ml–1 for S. mutans), kanamycin (50 μg ml–1 for E. coli and 1 mg ml–1 for S. mutans) and spectinomycin (50 μg ml–1 for E. coli and 1 mg ml–1 for S. mutans) were added to media, as required. For biofilm formation assays, S. mutans strains were grown in microtitre plates in the semi-defined medium BM (Loo et al., 2000) supplemented with glucose or sucrose at a final concentration of 20 mM.
Construction of mutant strains.
Standard DNA manipulation techniques were used to engineer plasmids and strains (Ahn et al., 2006; Sambrook et al., 1989). All mutants were created by using a PCR ligation mutagenesis approach (Lau et al., 2002), whereby genes were disrupted by replacing nearly all of the ORF with a non-polar resistance marker, such as a spectinomycin (SpR), erythromycin (Emr) or kanamycin (NPKm) marker, or a polar marker (ΩKm). Transformants were selected on BHI agar containing appropriate antibiotics, and double-crossover recombination into each gene was confirmed by PCR and sequencing to ensure that no mutations were introduced into flanking genes. The mutant strains of S. mutans constructed in this study are listed in Table 1.
Table 1.
S. mutans strains used in this study
| Strain | Description | Relevant genotype | Source or reference |
|---|---|---|---|
| UA159 | UA159 | Wild-type | |
| SAB113 | ΔlrgA : : NPSpr | lrgA− | This study |
| SAB119 | ΔlrgB : : NPEmr | lrgB− | This study |
| SAB115 | ΔlrgAB : : ΩKmr | lrgAB− | This study |
| SAB111 | ΔlytS : : NPKmr | lytS− | This study |
| SAB112 | ΔlytS : : ΩKmr | lytST− | This study |
| SAB117 | ΔcidA : : NPKmr | cidA− | This study |
| SAB118 | ΔcidA : : ΩKmr | cidAB− | This study |
| SAB121 | ΔcidB : : NPKmr | cidB− | This study |
| TW1 | ΔccpA : : ΩKmr | ccpA− | Wen & Burne (2002) |
Growth, biofilm and autolysis assays.
For growth rate comparisons, fresh medium was inoculated with 1 : 100 dilutions of overnight cultures of S. mutans. The optical density at 600 nm (OD600) was measured at 37 °C at fixed time intervals in a spectrophotometer or was monitored by using a Bioscreen C lab system (Ahn & Burne, 2007). To achieve anaerobic conditions, sterile mineral oil (50 μl per well) was placed on top of the cultures (Ahn & Burne, 2007; Ahn et al., 2007). To measure the growth rate under oxidative stresses, the strains were grown for 48 h in BHI containing hydrogen peroxide (0.001 %, v/v) or paraquat (10 mM). Cells were also observed by phase-contrast microscopy to record chain length. For growth on agar plates, the strains were streaked from mid-exponential phase cultures onto the surface of BHI agar. The plates were then incubated for 72 h in an aerobic chamber, and growth was compared from three independent experiments. The ability to form stable biofilms in microtitre plates and the autolysis assay were carried out as described by Ahn & Burne (2006, 2007) and Shibata et al. (2005).
Protein electrophoresis and Western blotting.
Protein extracts from S. mutans were prepared from cell pellets harvested from BHI cultures in mid-exponential phase (OD600=0.5), as previously described (Ahn & Burne, 2006; Chen et al., 1998). Proteins (10 μg) were separated by SDS-PAGE in a 10 % polyacrylamide gel with a 4.5 % stacking gel (Laemmli, 1970), or in a 3–8 % Tris/acetate gradient gel (Invitrogen). Proteins were transferred to Immobilon P membranes (Millipore) and were subjected to Western blot analysis according to standard protocols (Sambrook et al., 1989). Membranes were incubated with anti-GtfB (Ahn et al., 2007) polyclonal antisera. Peroxidase-labelled goat anti-rabbit IgG (KPL) and Sigma FAST (3,3′-diaminobenzidine tablets) were used to determine antibody reactivity. The protein concentration of samples was determined by a bicinchoninic acid assay (Sigma).
Transcriptional analysis.
The potential for co-transcription of two genes was examined by RT-PCR. Levels of mRNA were quantified by Northern blotting and real-time RT-PCR analyses. To measure the expression of genes using real-time RT-PCR, S. mutans wild-type (UA159) and its derivatives were grown in 7 ml BHI, and cells were harvested in early (OD600=0.2), mid- (OD600=0.5), late (OD600=0.9) exponential or stationary phase. Extraction of RNA, RT-PCR, real-time RT-PCR and data analysis were performed as described previously (Ahn et al., 2005). The primers used for reverse transcription reactions and real-time PCR are shown in Table 2. For Northern blotting, total RNA was isolated from S. mutans UA159 cultures grown for 6 h, corresponding to late exponential phase, in Todd–Hewitt broth supplemented with 11, 15, 20, 25, 30, 35, 40 or 45 mM glucose. S. mutans UA159 and TW1 (Table 1), the latter being an otherwise-isogenic ccpA mutant, were also grown in the same broth containing glucose at a final concentration of either 11 or 45 mM. Cells were removed for RNA isolation at 2, 6 and 12 h growth, corresponding to early, late exponential and late stationary phases, respectively. Total RNA was isolated from each sample by using an RNeasy Mini kit (Qiagen) and FASTPREP (MP Biomedicals) as described previously (Patton et al., 2005). Northern blotting was performed as described previously (Patton et al., 2005). DIG-labelled DNA probes specific for the cidB and lrgA genes were synthesized by using a DIG PCR probe synthesis kit (Roche Applied Science) and the primer pairs listed in Table 2. Hybridization and processing of the blots were performed by using the DIG system (Roche Applied Science) following the supplier's recommendations for Northern blot analysis. Transcript sizes were estimated by comparison with an RNA molecular mass ladder (Invitrogen).
Table 2.
Primers used in this study for construction of deletion mutants and real-time PCR
Italic type indicates the restriction site integrated into the primer.
| Primer purpose | 5′-end amplicon | Deleted gene/amplicon/probe* | 3′-end amplicon | ||
|---|---|---|---|---|---|
| Primer | Nucleotide sequence (5′→3′) | Primer | Nucleotide sequence (5′→3′) | ||
| General | |||||
| lrgA-A | GCAATTGGCACATCCTCCAC | lrgA | lrgA-SphI-C | CTATCAAAAGCATGCATGTGGCAAG | |
| lrgA-SphI-B | CTGAAATAAGCATGCAAACGAGCAG | lrgA-D | CCATGGCAGTGATGGCAGTA | ||
| lrgB-A | GCAATCGGGGACAGTTTTGA | lrgB | lrgB-EcoRI-C | GCAGCCTTTGAATTCGAATTAGGAG | |
| lrgB-SalI-B | GCAAAGAAAGTCGACTGTAAGAGAA | lrgB-D | ACAGACCGCTTTGAGGTTGC | ||
| lytS-A | ACTGAACAGCCAGTGCACCA | lytS | lytS-BamHI-C | GCAGTGCTAGGATCCTACACTTTGA | |
| lytS-BamHI | GCCAGAATCGGATCCATACCAAGTC | lytS-D | TCAAAACTGTCCCCGATTGC | ||
| cidA-A | TGCGGTCAGTTTTGCTGTGG | cidA | cidA-BamHI-C | GAGACATTAGGATCCAGACTTTCCA | |
| cidA-BamHI-B | CAAATTCGCGGATCCAAGAAAAGAG | cidA-D | TGAGACAAAAGTGTTCCCAACC | ||
| cidB-A | GCGCTTTTCAGGCAAGCAGA | cidB | cidB-BamHI-C | ACGGGTTTGGGATCCGTCTTTGTAT | |
| cidB-BamHI-B | TAGGCAAATGGATCCAGCCAAAGAC | cidB-D | TGGCGCCAAATCTCTTACGC | ||
| RT-PCR†or real-time RT-PCR | |||||
| lrgA-sense | TTGCCTAAAGCCTTACCGATTCC | lrgA | lrgA-antisense | GCCTGATGGGACAAACATAAAGC | |
| lrgB-sense | GGCAAAAGGATTGGGAACTGATG | lrgB | lrgB-antisense | TGGAACGGCAAAGGCAATGG | |
| lytS-sense | TTGTCAGTTCTGCTTTGGTAGG | lytS | lytS-antisense | CAATGACCTGCGAAGTAGATGG | |
| lytT-sense | CATCCTCCACTTGTCGTCTTTGC | lytT | lytS-antisense | CACACGCCCCTGCTCAAAAG | |
| cidA-sense | ATCCGTTTGCGTCATATCAATGC | cidA | cidA-antisense | CCATAATCCCCACTGCTGCTG | |
| DIG-probe synthesis | |||||
| lrgA-sense | CACAATCAAAATCAGCACCT | lrgA | lrgA-antisense | TCACCTTTTTGATAGACAGAA | |
| cidB-sense | TTTTTCGAATCCTCTTTTTG | cidB | cidB-antisense | CAACAACAACCAGTGTTACG | |
*Additional primers used for deletion of lrgAB were as follows: lrgA-BamHI-B, CTGAAATAAGGATCCAAACGAGCAG; lrgB-BamHI-C, GCAGCCTTTGGATCCGAATTAGGAG.
†Primer sets for RT-PCR were employed as follows: lrgA-sense/lrgB-antisense primers for lrgA–lrgB, and lrgB-sense/lytS-antisense primers for lrgB–lytS.
RESULTS
The cid/lrg operons of S. mutans
Previous studies revealed that oxygen modulates autolysis by S. mutans (Ahn & Burne, 2007). In a subsequent microarray analysis (Ahn et al., 2007) designed to identify oxygen-responsive genes, two paralogously related operons, SMU.575/574 and SMU.1701/1700, were strongly upregulated or downregulated in cells grown aerobically, respectively. SMU.575 shares 20 % amino acid sequence similarity with that of SMU.1701, whereas SMU.574 shares 27 % amino acid sequence similarity with that of SMU.1700, by using a local blast search. The SMU.575/574 and SMU.1701/1700 operons share substantial sequence similarity with the bacterial lrg and cid operons, respectively, a family of genes widely conserved across a range of bacterial species (Bayles, 2007) that are known to control murein hydrolase activity in Staphylococcus aureus (Groicher et al., 2000; Rice et al., 2003). Thus, we designated SMU.575/574 as lrgAB and SMU.1701/1700 as cidAB.
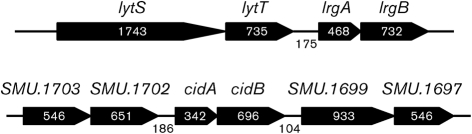
The genetic organization of the lrg and cid loci of S. mutans is shown in Fig. 1. Notably, a two-component signal transduction system (TCS), annotated as lytS (sensor kinse) and lytT (response regulator), is encoded immediately upstream of lrgA. The lytST operon is conserved in a subset of Gram-positive bacterial genera, including Bacillus and Staphylococcus (Bayles, 2007). However, apparent homologues of lyt–lrg are absent in some oral streptococci, including Streptococcus gordonii, Streptococcus mitis and Streptococcus sanguinis, but are present in Streptococcus agalactiae (http://www.oralgen.lanl.gov/). The S. mutans cidA and cidB genes overlap by 4 nt, but do not appear to be linked to regulatory genes. Instead, these genes lie between genes encoding conserved hypothetical proteins, including a putative membrane-associated phosphatase and an rRNA S-adenosylmethionine (SAM)-dependent methyltransferase (Fig. 1). It is also noteworthy that LrgA and, to a lesser extent, CidA each share structural features with the bacteriophage-encoded holins and antiholins that control bacteriophage-mediated cell death and lysis (Wang et al., 2000; Young & Blasi, 1995; Young, 2002). Similarities between LrgA and the prototypical holin, bacteriophage lamda S protein, include a relatively small size, two or more putative membrane-spanning domains, a polar N-terminal sequence and a charge-rich C-terminal domain (see Supplementary Fig. S1, available with the online version of this paper). Unlike LrgA, the CidA protein of S. mutans does not possess a polar N-terminal sequence and shows a shorter charged C-terminal sequence (Supplementary Fig. S1). LrgB and CidB, the presumed hydrophobic partners of LrgA and CidA, respectively, lack sequence similarity to holin proteins or known murein hydrolases.
Fig. 1.
Schematic diagram of the lrg and cid loci in the S. mutans UA159 genome. Gene assignments and gene numbers above the schematic diagram are based on the S. mutans UA159 genome (GenBank accession no. AE014133). Numbers inside the schematic diagram and between ORFs indicate the nucleotides (bp) in the ORFs and intergenic regions, respectively. Arrows indicate the direction of transcription. A dicistronic operon, lytST, is located immediately upstream of lrgAB. The lytS operon encodes the putative sensor kinase and lytT encodes the putative response regulator of a TCS. The lrgA and lrgB operons are annotated as a regulator and effector of murein hydroase, respectively. The SMU.1699 and SMU.1703 genes are annotated as conserved hypothetical proteins, and SMU.1697 and SMU.1702 are predicted to encode a possible rRNA methylase and uncharacterized phosphatase, respectively.
Role of the cid or lrg gene products in autolysis and biofilm formation of S. mutans cells
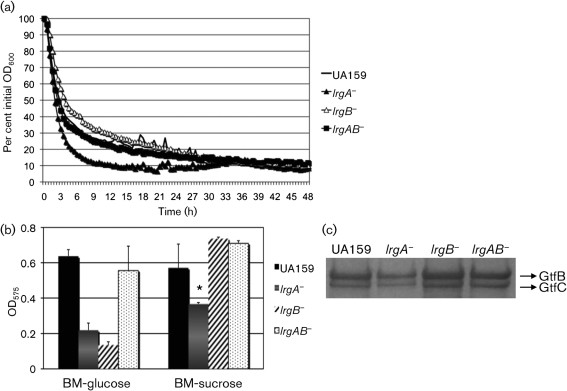
To begin to evaluate the role of LrgAB and CidAB in S. mutans, we constructed a series of mutant derivatives in the lrg and cid genes by replacing all or most of the structural genes with antibiotic resistance markers by using either a non-polar (NP) or polar (P) marker to inactivate a single gene or the entire operon (Table 1). A hallmark phenotype caused by the deficiency of lrg and cid in S. aureus is altered murein hydrolase activity (Bayles, 2000, 2003, 2007; Groicher et al., 2000; Rice et al., 2003; Rice & Bayles, 2008). To determine whether CidAB or LrgAB could impact lysis of S. mutans, autolysis assays were performed. Compared with the wild-type strain, the lrgA single mutant displayed enhanced autolysis and the lrgB single mutant displayed decreased autolysis (Fig. 2a). Within the first 6 h of the assay, the optical density (600 nm) of the cell suspension was reduced by 83 % in the lrgA mutant and 59 % in the lrgB mutant, whereas the wild-type cells showed a 70 % reduction. Inactivation of lrgAB (Fig. 2a) as well as cidAB genes (data not shown) did not have a discernible effect on autolysis.
Fig. 2.
Autolytic potential and biofilm-forming activity of S. mutans wild-type and its derivatives. (a) Autolysis assay. The autolytic activities of strains were monitored at 44 °C in a Bioscreen C system that was set to shake for 15 s before measurement every 30 min. Data are representative of three independent experiments and are presented as an average of triplicate samples. (b) Biofilm formation. Cultures were grown in BM medium supplemented with glucose or sucrose for 24 h. Data are representative of at least two separate experiments performed at least in triplicate. Error bars represent sd. *P<0.05; Student's t-test. (c) Production of GtfB and GtfC. Western blot analysis was performed by using bead-beaten, SDS-boiled extracts from S. mutans wild-type and its derivatives. Following SDS-PAGE, proteins were blotted onto an Immobilon P membrane and subjected to Western blotting with an anti-GtfB antiserum at a dilution of 1 : 500. Data are representative of at least three independent experiments. See text for details.
Given that autolysis has been implicated in the ability of bacteria to develop biofilms, we evaluated whether loss of the lrg or cid genes in S. mutans affected its capacity to form biofilms. Interestingly, the ability of both lrgA and lrgB mutants to form biofilms was dramatically reduced in BM–glucose medium, compared with that of the parental strain (Fig. 2b). In BM–sucrose medium, the lrgA mutant displayed a dramatic reduction in biofilm formation (P<0.05, Student's t-test), whereas the lrgB mutant formed biofilms as efficiently as the wild-type strains (Fig. 2b). Sucrose-dependent biofilm accumulation in S. mutans is mediated primarily by the production of glucan polymers from sucrose via specialized glycosyltransferase (Gtf) enzymes. Binding to these glucans is mediated by the Gtfs themselves, as well as by multiple glucan binding proteins (Yamashita et al., 1993). To examine whether the differences in the lrgA and lrgB mutants to form biofilms were due to altered levels of Gtfs, GtfB and GtfC production in these strains was monitored in mid-exponential phase BHI cultures via Western blot analysis by using an anti-GtfB serum (Ahn et al., 2007; Wunder & Bowen, 2000). The amount of GtfB and GtfC production under these growth conditions was decreased in the lrgA mutant, whereas the lrgB mutant displayed no apparent difference in the amount of these enzymes, compared with the wild-type strain (Fig. 2c). These results correlate with the impaired ability of the lrgA mutant (but not the lrgB mutant) to form biofilm in the presence of sucrose (Fig. 2b). In addition to a lack of effect on autolysis, no obvious differences in biofilm formation and GtfB or GtfC levels were noted in the cid mutants (data not shown). It is unclear from these results why glucose-dependent biofilm formation is more greatly affected compared with sucrose-dependent biofilm formation in the lrgA and lrgB mutants. The release of extracellular genomic DNA (eDNA) via cell lysis is important for biofilm attachment and development (Mann et al., 2009; Perry et al., 2009), and this process is influenced by the Cid/Lrg system. Therefore, it is possible that the lrgA and lrgB gene products are affecting cell lysis and/or eDNA release during glucose-dependent biofilm development. Although there is only limited evidence to support a role for eDNA release in S. mutans biofilm formation (Perry et al., 2009), we are currently investigating how lrg and cid might influence eDNA production and whether this could contribute to the observed phenotypes.
The Cid/Lrg system is required for efficient aerobic growth of S. mutans
Given that previously published microarray data showed that oxygenation profoundly regulates expression of lrg and cid in an opposing manner (Ahn et al., 2007), we evaluated whether oxygen affects the growth of the lrg- or cid-deficient mutants in this study. Surprisingly, when the strains were cultured in an aerobic incubator on BHI agar plates, growth of the ΔlrgAB, ΔcidAB or ΔcidB mutants was almost completely inhibited (Supplementary Fig. S2). These growth defects were also observed in liquid media (data not shown). However, when the strains were cultured on solid BHI medium in an anaerobic jar, growth of all mutants was as robust as that of the wild-type strain (data not shown). Notably, a deficiency of either lrgA or lrgB alone had no apparent effect on aerobic growth, compared with the wild-type strain (Supplementary Fig. S2). In contrast, deficiency of CidAB, or of CidB alone, critically impaired the growth of S. mutans in an aerobic environment, whereas the effect of a cidA mutation on aerobic growth of the organism was negligible (Supplementary Fig. S2). Therefore, the cid and lrg gene products, which are differentially regulated by aeration, appear to function in a way that augments the survival and growth of S. mutans in an oxidizing or oxygen-rich environment.
Deficiency of cid or lrg impacts oxidative stress tolerance by S. mutans
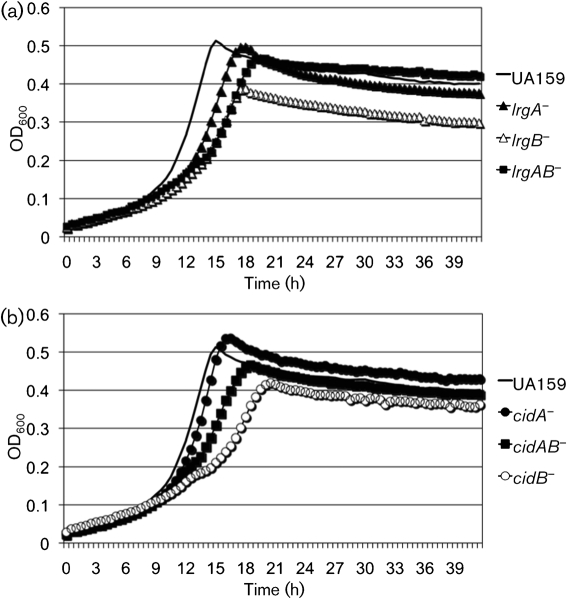
To determine whether the aerobic growth defects observed in the mutant strains were related to the capacity of cells to tolerate oxidative stress, the growth of each mutant was monitored under relatively anaerobic conditions (oil overlay) by using BHI medium containing hydrogen peroxide or paraquat. When the strains were grown in the presence of a low concentration of hydrogen peroxide (0.001 %, v/v), no obvious differences were observed in the growth of wild-type and all mutant strains (data not shown). In contrast, growth of the ΔlrgAB, ΔlrgB, ΔcidAB and ΔcidB mutants was profoundly affected by the presence of paraquat (10 mM), a superoxide anion-generating agent (Fig. 3). Although the lrgA mutant also displayed a growth defect (Fig. 3a), this was apparently not due to the presence of paraquat, as this mutant displayed similarly poor growth in the absence of paraquat (data not shown). These results suggest that the cid/lrg operons are required for growth of S. mutans during oxidative stress, and particularly that the lrgB and cidB gene products may be more important for stress tolerance compared with the lrgA and cidA gene products.
Fig. 3.
Growth curves of S. mutans wild-type and its lrg (a) and cid (b) derivatives under oxidative stress. Strains were grown in BHI medium containing 10 mM paraquat under anaerobic conditions. Growth was monitored in a Bioscreen C system that was set to shake for 15 s every 30 min. For anaerobic growth, sterile mineral oil (50 μl) was placed on top of the broth cultures. The results are representative of two independent experiments.
Acid tolerance is a major virulence attribute of S. mutans and we recently determined that growth under aerobic conditions significantly increased the susceptibility of S. mutans to acid stress (Ahn et al., 2009). Given the importance of cid and lrg for growth in the presence of oxidative stress, we investigated the impact of inactivation of these genes on acid tolerance. No differences were observed in the abilities of the strains to grow at pH 6.0, and all strains, including S. mutans UA159, did not grow well at pH 5.5 under the conditions tested (data not shown). Thus, the Cid/Lrg system, and in particular LrgB and CidB, may be important for tolerance of oxidative stress during aerobic growth of S. mutans, but these gene products do not appear to be required for growth at low pH under these experimental conditions (data not shown).
Expression of lrg and cid is growth-phase dependent
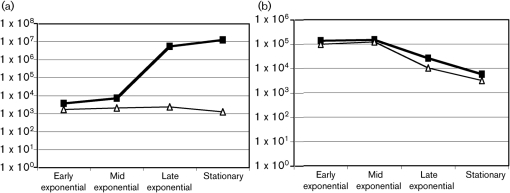
To begin to elucidate the factors influencing lrg and cid expression, we monitored lrg and cid mRNA levels during the growth cycle of planktonic cultures of UA159 by real-time PCR. Expression of cid and lrg was also evaluated in a lytS-deficient strain, as the LytSR TCS is known to regulate expression of lrgAB in Staphylococcus aureus (Groicher et al., 2000). Strikingly, the levels of lrgA mRNA increased over 1000-fold during growth of the wild-type strain, with maximal expression occurring in late exponential and stationary phases (Fig. 4). In contrast, cidA mRNA levels were most abundant in early exponential phase, and then decreased more than 10-fold as cells entered the late exponential phase of growth. Fig. 4 also shows that lack of LytST resulted in lower basal levels of lrgAB expression and a complete lack of induction of the lrg genes later in the growth cycle. In contrast, no significant effect on the expression of the cid genes was observed in the lytST mutant. Given that the lrgAB operon is located immediately downstream of the lytST genes (Fig. 1), the possibility that the lytST mutation may have had a polar effect on lrgAB expression was ruled out by using RT-PCR to show that the lytST and lrgAB operons were not co-transcribed (data not shown). Together, these results suggest that lrg and cid expression is growth-phase dependent and that lrgAB expression is tightly controlled by LytST.
Fig. 4.
Expression of lrgA and cidA with growth phase. The expression of lrgA (a) and cidA (b) genes was measured in UA159 (▪) and its isogenic lytST mutant (▵) in the early (OD600=0.2), mid (OD600=0.5), late (OD600=0.9) exponential and stationary phases of growth, using real-time RT-PCR. Results are the average of triplicate samples from three independent experiments.
Expression of lrg and cid is regulated by glucose
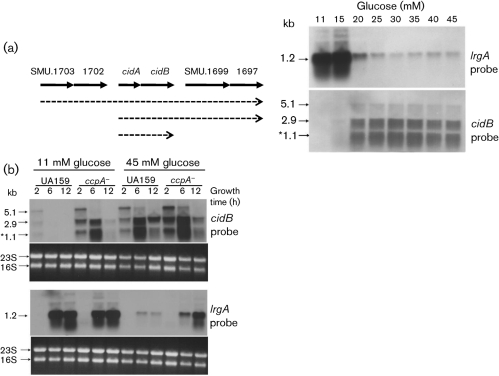
Based on the observation that the lrg and cid genes are differentially expressed throughout the growth cycle in BHI medium, which contains 0.3 % glucose (w/v), we hypothesized that depletion of glucose could stimulate changes in cid and lrg gene expression during the transition from early exponential phase to stationary phase. In this respect, it is noteworthy that the Bacillus subtilis ywbH and ywbG genes, predicted homologues of cidA and cidB, were induced when the organism was grown in the presence of 1 % (55.5 mM) glucose (Moreno et al., 2001), and that the cidABC operon of Staphylococcus aureus was induced in 35 mM glucose (Rice et al., 2005). We examined whether glucose concentration influences the expression of lrg and cid by Northern blot analysis of RNA samples isolated from UA159 cultures grown for 6 h, corresponding to late exponential phase, in the presence of increasing concentrations of glucose (Fig. 5a). Interestingly, the cidB probe hybridized to three distinct transcripts (1.1, 2.9 and 5.1 kb), of which the 1.1 kb transcript closely matched the predicted length of cidAB (1.045 kb). Subsequent Northern blot analysis with probes specific for cidA and the genes located immediately upstream and downstream of cidAB revealed that this locus comprises multiple overlapping transcripts: a 2.9 kb transcript contains cidA, cidB and two predicted ORFs (SMU.1697 and SMU.1699) located immediately downstream of cidAB, whereas the 5.1 kb transcript comprises these four genes and two predicted ORFs (SMU.1702 and SMU.1703) located immediately upstream of cidAB (data not shown). The amount of the transcript arising from the two dicistronic operons, cidAB and SMU.1697/1699, is similar to that of the cidAB transcript alone, indicating that transcription from the cidAB promoter contributes significantly to the expression of SMU.1697/1699.
Fig. 5.
Analysis of the effect of glucose concentration (a) and CcpA (b) on expression of the S. mutans cid and lrg operons by Northern blot. Total RNA was isolated from S. mutans UA159 cultures grown for 6 h in the presence of increasing concentrations of glucose (a), and UA159 and ccpA− (TW1) cultures grown in the presence of either 11 or 45 mM glucose, at 2 h (early exponential growth), 6 h (late exponential phase) or 12 h (late stationary phase) (b). RNA (5 μg) from each sample was subjected to Northern blotting with DIG-labelled DNA probes corresponding to either lrgA or cidB. The size of each transcript is indicated to the left of each blot, and is also shown in a schematic diagram of the cid locus by the dotted arrows. The 1.1 kb cidAB transcript is indicated by an asterisk. The corresponding ethidium-bromide-stained gel is presented beneath the blots with arrows highlighting the 23S and 16S RNA gene bands.
Expression of lrg and cid was regulated in an opposite manner in response to glucose levels (Fig. 5a). The lrgAB genes were highly induced in cultures containing lower levels of glucose (11 and 15 mM) and were dramatically repressed in cultures containing glucose at concentrations of 20 mM and higher, although detectable expression persisted at concentrations of up to 45 mM. In contrast, all three overlapping transcripts of the cid locus were nearly undetectable when cells were cultured in the presence of lower glucose concentrations (<20 mM), but increased dramatically at higher glucose concentrations (≥20 mM). Thus, expression of the lrg and cid genes is glucose-dependent in an opposite manner, and two dicistronic operons located immediately upstream and downstream of the cid locus are coordinately regulated and co-transcribed with the cid operon.
Expression of lrg and cid is under the control of CcpA
Given that cid and lrg expression is highly responsive to glucose levels in S. mutans, we investigated the involvement of CcpA (carbon catabolite protein A) in the regulation of lrg and cid genes. CcpA in S. mutans has been shown to serve as a major regulator of the expression of glycolytic and tricarboxylic acid (TCA) cycle enzymes, the pyruvate dehydrogenase complex, carbohydrate transporters and various catabolic pathways (Abranches et al., 2008). It was also noted in a previous microarray study from our group (Abranches et al., 2008) that expression of lrg and cid could be influenced by loss of CcpA and affected by growth under conditions that alleviate catabolite repression in S. mutans. Subsequently, we measured the expression of lrg and cid in the presence of the lower (11 mM) and higher (45 mM) levels of glucose by using Northern blot analysis (Fig. 5b). In medium containing the lower level of glucose, cid transcription was maximal in early exponential phase (2 h growth), whereas lrgAB was highly expressed in late exponential (6 h growth) and late stationary (12 h growth) phases, consistent with the results in Fig. 5, which were determined under similar conditions. Furthermore, the cidAB transcript (1.1 kb) as well as the other overlapping transcripts (2.9 and 5.1 kb) were differentially upregulated at all time points in media containing high levels (45 mM) of glucose (Fig. 5b, top).
In contrast with the observed effects of glucose on cid transcription, expression of the lrgAB operon was strongly repressed in stationary phase in the presence of 45 mM glucose (Fig. 5b, bottom). Interestingly, cid expression was upregulated in the ccpA mutant when grown in 11 mM glucose, relative to the parental strain UA159 (Fig. 5b, top), whereas lrgAB expression was similar between the two strains under these growth conditions. These results correlate well with recently published microarray data showing that cidB expression was upregulated threefold in the ccpA mutant of S. mutans (Abranches et al., 2008). Also of note, the cidAB transcript (1.1 kb) was markedly increased as the ccpA mutant culture entered the late exponential phase of growth (6 h), and decreased to nearly undetectable levels in late stationary phase (12 h growth). However, when these strains were grown in 45 mM glucose, lrgAB expression was increased in the ccpA mutant relative to UA159, whereas no apparent differences in cid expression were observed (Fig. 5b, bottom). It is not clear whether this CcpA-dependent regulation is due to direct interaction of CcpA with the lrg and cid promoters, or if this regulation is an indirect result of altered expression of other regulatory genes or due to an effect on glucose metabolism. In this respect, it is interesting to note that there appear to be two cre-like consensus elements, which are binding sites for CcpA (Miwa et al., 2000), in the DNA sequence immediately upstream of lrgAB (Fig. 6), suggesting that CcpA may directly regulate expression of lrgAB. However, identifiable cre elements were not apparent in the sequences upstream of cidAB and lytSR (data not shown). Collectively, these results demonstrate that growth phase, glucose metabolism and CcpA are important factors that regulate expression of the S. mutans cid and lrg genes.
Fig. 6.
Identification of cre sequences upstream of lrgAB. The B. subtilis cre consensus sequence derived by Miwa et al. (2000) is shown at the top, where W=A or T, and N=any nucleotide. The nucleotide sequence corresponding to 114 bp upstream of the ATG start codon (identified in bold italics) of the S. mutans lrgA gene is shown at the bottom. The putative cre elements are underlined and conserved nucleotides are shown in bold.
DISCUSSION
As part of an ongoing effort to identify the regulatory pathways controlling autolysis of S. mutans, we identified two dicistronic operons, lrgAB (SMU.575/574) and cidAB (SMU.1701/1700) (Ahn et al., 2007), encoding products with similarities to bacteriophage holin : antiholin proteins that modulate cell lysis (Bayles, 2000, 2003; Rice & Bayles, 2003; Young, 1992; Young & Blasi, 1995). The present study shows that the S. mutans CidAB and LrgAB proteins play important roles in biofilm formation, oxidative stress tolerance and regulation of autolysis. Also of note, expression of these two operons is tightly regulated by multiple systems, including the LytST signal-transduction complex and CcpA. Furthermore, cid/lrg transcription is highly responsive to environmental stimuli, particularly oxygen and glucose. Given that oxygen levels have a profound effect on the phenotypic properties of S. mutans through metabolic changes (Ahn et al., 2009), the exact mechanism by which cid and lrg gene products impact the virulence of this organism in the presence of oxygen and glucose remains unclear. However, the results presented herein show that the lrgAB and cidAB gene products probably interact to promote survival in a non-preferred oxidative environment (Fig. 3 and Supplementary Fig. S2). Additionally, the lrgAB gene products appear to play an important role in modulating virulence behaviour, including biofilm formation and autolysis, of S. mutans (Fig. 2). The results presented in this study highlight some fundamental differences between the Lrg/Cid systems of S. mutans and Staphylococcus aureus, the latter having been more intensively studied to date (Groicher et al., 2000; Rice et al., 2003). Namely, a role for the cid gene products in regulating cell autolysis and biofilm formation was not demonstrated for S. mutans in the present study. However, recent analysis of cid and lrg homologues in Bacillus anthracis found that inactivation of either lrgAB or clhAB did not appear to affect murein hydrolase activity or autolysis, but instead altered sporulation efficiency (Chandramohan et al., 2009). Collectively, these studies suggest that the Cid/Lrg systems may have evolved to respond to environmental, metabolic and developmental signals depending on the lifestyle of a particular organism.
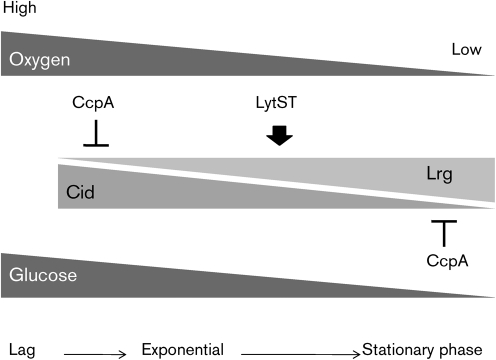
Based on the results of the current study, we provide a working model for the role and regulation of the S. mutans cid/lrg systems (Fig. 7). Primarily, Lrg and Cid levels are regulated at the transcriptional level in a growth-phase-dependent fashion that is integrated with availability of oxygen and glucose. In early exponential phase, where levels of oxygen and glucose are elevated, cid expression is dominant and lrg is repressed. As cells enter stationary phase, or in oxygen/glucose-limited cells, lrg seems to be de-repressed and activated through the LytST complex, whereas cid expression diminishes under these conditions. In this way, the expression levels of lrg and cid are counter-balanced throughout the growth cycle. Given that Staphylococcus aureus cid and lrg expression is regulated by acetic acid production during metabolism of excess glucose (Rice et al., 2005), it is possible that metabolic acids and/or low pH produced by S. mutans glucose fermentation act as the actual signals that regulate cid/lrg expression. We are currently investigating the effect and role of these acidic metabolites (i.e. lactate, acetate and formate) on S. mutans cid/lrg expression. However, it should be noted that the S. mutans lrg and cid mutant strains did not differ in their ability to grow at low pH, suggesting that the function of these gene products does not appear to be influenced by low pH.
Fig. 7.
Working model for glucose and oxygen-dependent regulation of the S. mutans Cid/Lrg system. Lrg and Cid activities are regulated in a growth-phase-dependent fashion by oxygen and glucose levels, which would change dramatically as cells progress from early exponential to stationary phase. As cells actively grow (high levels of oxygen and glucose available), cid expression is dominant but lrg is minimal and probably under the control of CcpA. As cells enter stationary phase (oxygen/glucose-limited cells), lrg expression is dramatically increased by the LytST TCS. In contrast, cid expression is gradually diminished, in part due to CcpA-dependent regulation. The expression level of lrg and cid seems to be tightly balanced throughout growth, and direct interactions between Lrg and Cid proteins may be involved in achieving this balance. Therefore, by sensing multiple environmental inputs and modulating the balance between Lrg and Cid, S. mutans is able to modulate growth and biofilm formation, which are critical for virulence expression by this pathogen.
Although certain aspects of this working model have yet to be tested directly, we propose that this regulatory pathway integrates environmental signals to control whether S. mutans is destined for active growth or for cell death/lysis in response to multiple environmental inputs, a process that is critical for the development of pathogenic biofilms. Although the cid and lrg genes were found to be regulated by CcpA in this study, the basis for this observation is still under investigation. Given that putative cre sites were identified in the lrg promoter region but not in the cid promoter region, it is likely that CcpA directly regulates lrgAB expression and indirectly regulates cid expression. As CcpA in S. mutans has been shown to serve as an important regulator of the expression of glycolytic and TCA cycle enzymes, carbohydrate transporters and catabolic pathways (Abranches et al., 2008), there are a number of possible ways by which CcpA deficiency could indirectly affect cid gene expression. For example, the CcpA-deficient strain may display alterations in lrgA expression that would affect cid transcription, or cause changes in metabolic end products, for example acetate, that could affect expression of cid genes. Alternatively, loss of CcpA may also affect the expression of transcriptional regulators that influence cid expression.
Clearly, lrg expression, but not that of cid, is under the tight control of LytST TCS. The primary regulatory role of LytST appears to be to activate lrg transcription in late exponential or stationary phase (Fig. 4), most likely by binding the lrg promoter region. Recently, by using a generic phylogenic footprinting/shadowing approach, the LytTR-family of response regulators was found to be associated with a specific operator motif consisting of direct repeats separated by 10–11 nt (de Been et al., 2008; Francke et al., 2008). A conserved motif (TGCAATTCAG-N11-TGCAATCGGG) was identified in the promoter region of lrgAB in S. mutans. By analysing the operator sequence identified in low-G+C Gram-positive bacteria, LytSR was also found to be involved in affecting or regulating transport of carbohydrates, peptides and/or amino acids (de Been et al., 2008), supporting our idea that activation of lrg genes by LytST is controlled by carbohydrate levels or metabolism. Given our previous microarray data that lytST is highly responsive to oxygen, the Cid/Lrg system appears to be hierarchically regulated in a signalling network involving carbohydrate availability and/or oxygen.
Additional interesting findings presented here are that Northern blot analysis with cid probes revealed three overlapping transcripts, and that the levels of these transcripts were coordinately regulated in response to the concentration of glucose in the growth medium. In particular, the amount of the 1.1 kb (cidAB) and 2.9 kb (cidAB–SMU.1699/1697) transcripts was very similar, which suggests that the cid operon may be regulated by, or functionally connected to, the dicistronic operon (SMU.1699/1697) located immediately downstream of cid. Interestingly, SMU.1697 is predicted to encode a putative rRNA SAM-dependent methyltransferase. These enzymes catalyse many diverse reactions, including methylation, isomerization, sulfur insertion, ring formation, anaerobic oxidation and protein radical formation (Sofia et al., 2001). As cid expression appears to be regulated by growth phase, oxidative stress and glucose metabolism, it is possible that these downstream ORFs participate in redox reactions that provide some protection to the cells. Importantly, however, the expression of these genes was not altered in the cidA or cidB mutants examined in these studies (data not shown), so the phenotypes described for these mutants are attributable to loss of CidA or CidB and not to polar effects on other genes. Studies are ongoing to determine whether the genes in this larger operon contribute to the function of the cid gene products and stress tolerance.
Based on the results presented here, it is not clear whether the Cid and Lrg systems of S. mutans are true holin : antiholin pairs. Specifically, inactivation of the cid genes did not have an opposing effect on the phenotypes measured in this study (growth rate, autolysis and biofilm formation) relative to inactivation of the lrg genes. Nevertheless, the fact that these genes display opposite patterns of expression in response to growth phase, glucose concentration, oxygenation (Ahn et al. 2007) and regulation by CcpA suggests that the functions of these gene products are interrelated and potentially antagonistic in S. mutans. Interestingly, a computational analysis via STRING (http://string.embl.de/), a database of known and predicted protein interactions, presents possible interactions of LrgA with LrgB or CidB (data not shown). In fact, loss of cidB was shown to reverse the effects of a cidA mutation (Fig. 3b). Moreover, autolysis is enhanced in an lrgA mutant and diminished in an LrgB-deficient strain. Thus, various possibilities exist for how LrgAB and CidAB may interact to regulate their biochemical activities and expression of these genes. Nonetheless, the data suggest that Lrg/Cid interations are critical for homeostasis in S. mutans. Indeed, a cidB mutation does not seem to be complemented by simply producing CidB in trans, as the cidB mutant strain harbouring a shuttle plasmid pMSP3535–CidB+ was still not able to grow on BHI agar plates under aerobic conditions (data not shown). It is also noteworthy that lrg expression was not induced in late exponential phase (when grown in BHI) when expression of cidAB genes was constitutively driven by the ldh promoter of S. mutans (unpublished data). Given that the lrg and cid mutants displayed very different phenotypes in the assays described above, and that they are regulated in an opposite manner in response to environmental conditions, the levels of cid and lrg gene products are critical and they may interact in as yet unknown ways. This may result in cross-regulation of these two operons, possibly explaining why complementation was not achieved in the experiment described above. Thus, the stoichiometry of these proteins in the cell appears to be critical. We are currently investigating whether there are any direct interactions between Lrg and Cid proteins and probing in more detail the potential for cross-regulation of expression of the lrg and cid operons.
Acknowledgments
This work was supported by a National Institute of Dental and Craniofacial Research (NIDCR) grant DE13239 to R. A. B., an NIDCR grant R03-DE019179 to K. C. R. and a National Institute of Allergies and Infectious Diseases grant R01 AI038901 to K. W. B.
Abbreviations
BHI, brain heart infusion
eDNA, extracellular genomic DNA
Gtf, glycosyltransferase
PCD, programmed cell death
TCA, tricarboxylic acid
TCS, two-component system
Footnotes
Supplementary figures showing structural similarities among LrgA, CidA and the prototypical bacteriophage lambda S holin, and aerobic growth of Streptococcus mutans wild-type and its derivatives are available with the online version of this paper.
References
- Abranches, J., Nascimento, M. M., Zeng, L., Browngardt, C. M., Wen, Z. T., Rivera, M. F. & Burne, R. A. (2008). CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol 190, 2340–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J. & Burne, R. A. (2006). The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J Bacteriol 188, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J. & Burne, R. A. (2007). Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol 189, 6293–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J., Lemos, J. A. & Burne, R. A. (2005). Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol 187, 3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J., Wen, Z. T. & Burne, R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74, 1631–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J., Wen, Z. T. & Burne, R. A. (2007). Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol 189, 8519–8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. J., Ahn, S. J., Browngardt, C. M. & Burne, R. A. (2009). Changes in biochemical and phenotypic properties of Streptococcus mutans during growth with aeration. Appl Environ Microbiol 75, 2517–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles, K. W. (2000). The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol 8, 274–278. [DOI] [PubMed] [Google Scholar]
- Bayles, K. W. (2003). Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol 11, 306–311. [DOI] [PubMed] [Google Scholar]
- Bayles, K. W. (2007). The biological role of death and lysis in biofilm development. Nat Rev Microbiol 5, 721–726. [DOI] [PubMed] [Google Scholar]
- Berry, A. M., Lock, R. A., Hansman, D. & Paton, J. C. (1989). Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun 57, 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman, S. A., Smith, T. J. & Foster, S. J. (1998). The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144, 73–82. [DOI] [PubMed] [Google Scholar]
- Bowman, B. U., Jr & Redmond, W. B. (1956). The effects of glucose and of oxygen on autolysis of Mycobacterium tuberculosis. Am Rev Tuberc 73, 907–916. [DOI] [PubMed] [Google Scholar]
- Chandramohan, L., Ahn, J. S., Weaver, K. E. & Bayles, K. W. (2009). An overlap between the control of programmed cell death in Bacillus anthracis and sporulation. J Bacteriol 191, 4103–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. Y., Weaver, C. A., Mendelsohn, D. R. & Burne, R. A. (1998). Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol 180, 5769–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Been, M., Bart, M. J., Abee, T., Siezen, R. J. & Francke, C. (2008). The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ Microbiol 10, 2796–2809. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I. & Hazan, R. (2006). Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke, C., Kerkhoven, R., Wels, M. & Siezen, R. J. (2008). A generic approach to identify transcription Factor-specific operator motifs; inferences for LacI-family mediated regulation in Lactobacillus plantarum WCFS1. BMC Genomics 9, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen, J.-M., Tipper, D. J. & Strominger, J. L. (1966). Enzymes that degrade bacterial cell walls. Methods Enzymol 8, 685–699. [Google Scholar]
- Gilpin, R. W., Chatterjee, A. N. & Young, F. E. (1972). Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J Bacteriol 111, 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groicher, K. H., Firek, B. A., Fujimoto, D. F. & Bayles, K. W. (2000). The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J Bacteriol 182, 1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann, C., Hussain, M., Peters, G. & Gotz, F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Höltje, J. V. (1995). From growth to autolysis: the murein hydrolases in Escherichia coli. Arch Microbiol 164, 243–254. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lau, P. C., Sung, C. K., Lee, J. H., Morrison, D. A. & Cvitkovitch, D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49, 193–205. [DOI] [PubMed] [Google Scholar]
- Lewis, K. (2000). Programmed death in bacteria. Microbiol Mol Biol Rev 64, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche, W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiol Rev 50, 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, C. Y., Corliss, D. A. & Ganeshkumar, N. (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182, 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., Tsang, L. H., Smeltzer, M. S., Horswill, A. R. & Bayles, K. W. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4, e5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, C., Durrieu, C., Briandet, R., Domakova, E., Tremblay, J., Buist, G. & Kulakauskas, S. (2002). Positive role of peptidoglycan breaks in lactococcal biofilm formation. Mol Microbiol 46, 235–243. [DOI] [PubMed] [Google Scholar]
- Miwa, Y., Nakata, A., Ogiwara, A., Yamamoto, M. & Fujita, Y. (2000). Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 28, 1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, M. S., Schneider, B. L., Maile, R. R., Weyler, W. & Saier, M. H., Jr (2001). Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol 39, 1366–1381. [DOI] [PubMed] [Google Scholar]
- Ochiai, T. (1999). Salt-sensitive growth of Staphylococcus aureus: stimulation of salt-induced autolysis by multiple environmental factors. Microbiol Immunol 43, 705–709. [DOI] [PubMed] [Google Scholar]
- Patton, T. G., Rice, K. C., Foster, M. K. & Bayles, K. W. (2005). The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol Microbiol 56, 1664–1674. [DOI] [PubMed] [Google Scholar]
- Perkins, H. R. (1980). The Bacterial Autolysins. London: Chapman & Hall.
- Perry, J. A., Cvitkovitch, D. G. & Levesque, C. M. (2009). Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qoronfleh, M. W., Gustafson, J. E. & Wilkinson, B. J. (1998). Conditions that induce Staphylococcus aureus heat shock proteins also inhibit autolysis. FEMS Microbiol Lett 166, 103–107. [DOI] [PubMed] [Google Scholar]
- Rice, K. C. & Bayles, K. W. (2003). Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol Microbiol 50, 729–738. [DOI] [PubMed] [Google Scholar]
- Rice, K. C. & Bayles, K. W. (2008). Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev 72, 85–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C., Firek, B. A., Nelson, J. B., Yang, S. J., Patton, T. G. & Bayles, K. W. (2003). The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J Bacteriol 185, 2635–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C., Nelson, J. B., Patton, T. G., Yang, S. J. & Bayles, K. W. (2005). Acetic acid induces expression of the Staphylococcus aureus cidABC and lrgAB murein hydrolase regulator operons. J Bacteriol 187, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, K. C., Mann, E. E., Endres, J. L., Weiss, E. C., Cassat, J. E., Smeltzer, M. S. & Bayles, K. W. (2007). The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A 104, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Shibata, Y., Kawada, M., Nakano, Y., Toyoshima, K. & Yamashita, Y. (2005). Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect Immun 73, 3512–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockman, G. D. & Holfje, J.-V. (1994). Microbial Peptidoglycan (Murein) Hydrolases. Amsterdam: Elsevier.
- Smith, T. J., Blackman, S. A. & Foster, S. J. (2000). Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146, 249–262. [DOI] [PubMed] [Google Scholar]
- Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. (2001). Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturges, W. S. & Rettger, L. F. (1922). Bacterial autolysis. J Bacteriol 7, 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin, P. J., Mani, N. & Jayaswal, R. K. (1994). Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie van Leeuwenhoek 65, 71–78. [DOI] [PubMed] [Google Scholar]
- Wang, I. N., Smith, D. L. & Young, R. (2000). Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol 54, 799–825. [DOI] [PubMed] [Google Scholar]
- Ward, J. B. & Williamson, R. (1984). Bacterial Autolysins: Specificity and Function. Amsterdam: Elsevier.
- Wells, J. E. & Russell, J. B. (1996). The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl Environ Microbiol 62, 1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z. T. & Burne, R. A. (2002). Analysis of cis- and trans-acting factors involved in regulation of the Streptococcus mutans fructanase gene (fruA). J Bacteriol 184, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenscher, M. D., Kohler, S., Bubert, A., Gerike, U. & Goebel, W. (1993). The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol 175, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder, D. & Bowen, W. H. (2000). Effects of antibodies to glucosyltransferase on soluble and insolubilized enzymes. Oral Dis 6, 289–296. [DOI] [PubMed] [Google Scholar]
- Yabu, K. & Kaneda, S. (1995). Salt-induced cell lysis of Staphylococcus aureus. Curr Microbiol 30, 299–303. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y., Bowen, W. H., Burne, R. A. & Kuramitsu, H. K. (1993). Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun 61, 3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarmolinsky, M. B. (1995). Programmed cell death in bacterial populations. Science 267, 836–837. [DOI] [PubMed] [Google Scholar]
- Young, R. (1992). Bacteriophage lysis: mechanism and regulation. Microbiol Rev 56, 430–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R. (2002). Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol 4, 21–36. [PubMed] [Google Scholar]
- Young, R. & Blasi, U. (1995). Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev 17, 191–205. [DOI] [PubMed] [Google Scholar]