Abstract
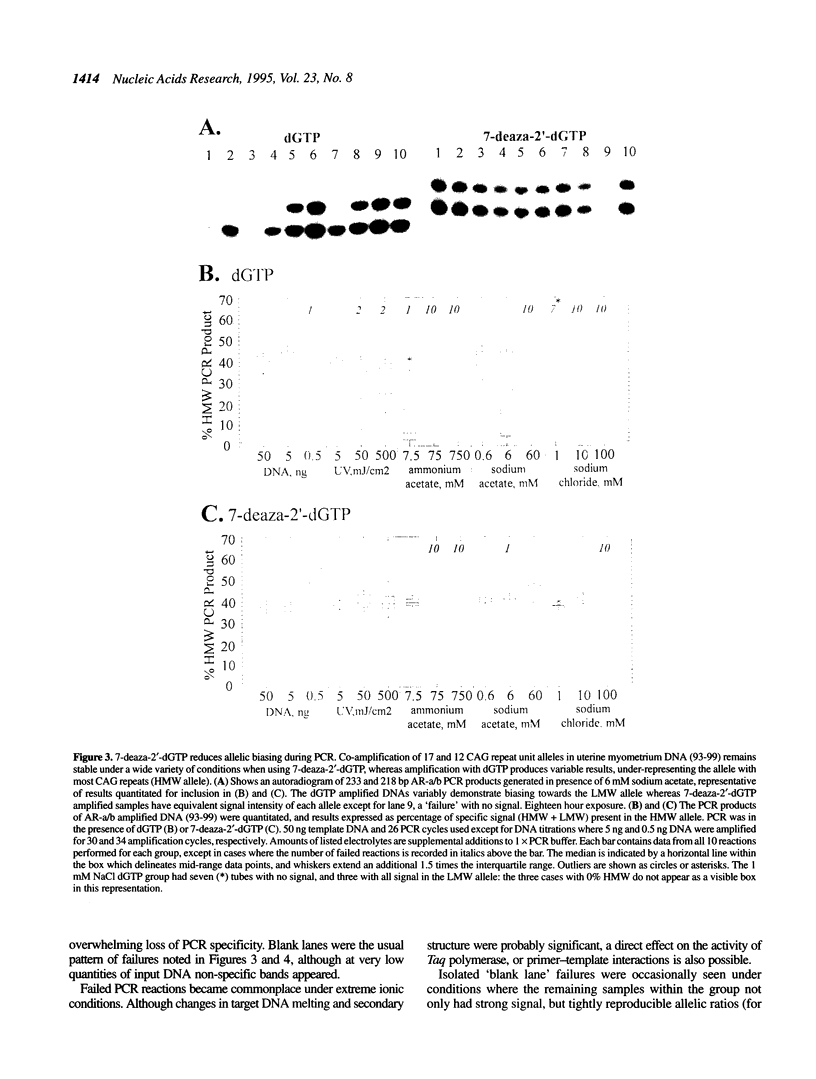
Trinucleotide CAG repeats in the X-linked human androgen receptor gene (HUMARA) have proved a useful means of determining X chromosome haplotypes, and when combined with methylation analysis of nearby cytosine residues permits identification of non-random X inactivation in tumors of women. Co-amplification of two alleles in a heterozygote generates PCR products which differ in the number of CAG units, and thus their melting and secondary structure characteristics. We have shown that under optimal conditions amplification efficiency of two HUMARA alleles is near-equivalent, generating PCR products in a ratio proportional to that of the genomic template. In contrast, reduction of template quantity, damage of template by ultraviolet irradiation or addition of monovalent salts (sodium chloride, sodium acetate or ammonium acetate) produces highly variable imbalances of allelic PCR products, with a strong tendency to preferentially amplify lower molecular weight alleles. Variability and biasing was diminished by substitution of 7-deaza-2'-dGTP for dGTP during amplification, an intervention which reduces stability of intramolecular and intermolecular GC base pairing. We conclude that DNA which is scanty, damaged or salt contaminated may display amplification bias of GC-rich PCR targets, potentially confounding accurate interpretation or reproducibility of assays which require co-amplification of alleles.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen R. C., Zoghbi H. Y., Moseley A. B., Rosenblatt H. M., Belmont J. W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992 Dec;51(6):1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Chou Q., Russell M., Birch D. E., Raymond J., Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992 Apr 11;20(7):1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fialkow P. J. Primordial cell pool size and lineage relationships of five human cell types. Ann Hum Genet. 1973 Jul;37(1):39–48. doi: 10.1111/j.1469-1809.1973.tb01813.x. [DOI] [PubMed] [Google Scholar]
- Fialkow P. J. Use of genetic markers to study cellular origin and development of tumors in human females. Adv Cancer Res. 1972;15:191–226. doi: 10.1016/s0065-230x(08)60375-9. [DOI] [PubMed] [Google Scholar]
- Gale R. E., Wheadon H., Linch D. C. X-chromosome inactivation patterns using HPRT and PGK polymorphisms in haematologically normal and post-chemotherapy females. Br J Haematol. 1991 Oct;79(2):193–197. doi: 10.1111/j.1365-2141.1991.tb04521.x. [DOI] [PubMed] [Google Scholar]
- Jackson D. P., Lewis F. A., Taylor G. R., Boylston A. W., Quirke P. Tissue extraction of DNA and RNA and analysis by the polymerase chain reaction. J Clin Pathol. 1990 Jun;43(6):499–504. doi: 10.1136/jcp.43.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada A. R., Wilson E. M., Lubahn D. B., Harding A. E., Fischbeck K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991 Jul 4;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- Lerman L. S., Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- Lobel S. M., Pomponio R. J., Mutter G. L. The sex ratio of normal and manipulated human sperm quantitated by the polymerase chain reaction. Fertil Steril. 1993 Feb;59(2):387–392. doi: 10.1016/s0015-0282(16)55682-9. [DOI] [PubMed] [Google Scholar]
- McConlogue L., Brow M. A., Innis M. A. Structure-independent DNA amplification by PCR using 7-deaza-2'-deoxyguanosine. Nucleic Acids Res. 1988 Oct 25;16(20):9869–9869. doi: 10.1093/nar/16.20.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutter G. L., Chaponot M. L., Fletcher J. A. A polymerase chain reaction assay for non-random X chromosome inactivation identifies monoclonal endometrial cancers and precancers. Am J Pathol. 1995 Feb;146(2):501–508. [PMC free article] [PubMed] [Google Scholar]
- Pergolizzi R. G., Erster S. H., Goonewardena P., Brown W. T. Detection of full fragile X mutation. Lancet. 1992 Feb 1;339(8788):271–272. doi: 10.1016/0140-6736(92)91334-5. [DOI] [PubMed] [Google Scholar]
- Richards R. I., Sutherland G. R. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992 Sep 4;70(5):709–712. doi: 10.1016/0092-8674(92)90302-s. [DOI] [PubMed] [Google Scholar]
- Sarkar G., Sommer S. S. Parameters affecting susceptibility of PCR contamination to UV inactivation. Biotechniques. 1991 May;10(5):590–594. [PubMed] [Google Scholar]
- Seela F., Driller H. Alternating d(G-C)3 and d(C-G)3 hexanucleotides containing 7-deaza-2'-deoxyguanosine or 8-aza-7-deaza-2'-deoxyguanosine in place of dG. Nucleic Acids Res. 1989 Feb 11;17(3):901–910. doi: 10.1093/nar/17.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata D., Hawes D., Li Z. H., Hernandez A. M., Spruck C. H., Nichols P. W. Specific genetic analysis of microscopic tissue after selective ultraviolet radiation fractionation and the polymerase chain reaction. Am J Pathol. 1992 Sep;141(3):539–543. [PMC free article] [PubMed] [Google Scholar]
- Shroyer K. R., Gudlaugsson E. G. Analysis of clonality in archival tissues by polymerase chain reaction amplification of PGK-1. Hum Pathol. 1994 Mar;25(3):287–292. doi: 10.1016/0046-8177(94)90201-1. [DOI] [PubMed] [Google Scholar]
- Singer-Sam J., LeBon J. M., Tanguay R. L., Riggs A. D. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 1990 Feb 11;18(3):687–687. doi: 10.1093/nar/18.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier Y., Biancalana V., Mandel J. L. Instability of CAG repeats in Huntington's disease: relation to parental transmission and age of onset. J Med Genet. 1994 May;31(5):377–382. doi: 10.1136/jmg.31.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Fearon E. R., Hamilton S. R., Preisinger A. C., Willard H. F., Michelson A. M., Riggs A. D., Orkin S. H. Clonal analysis using recombinant DNA probes from the X-chromosome. Cancer Res. 1987 Sep 15;47(18):4806–4813. [PubMed] [Google Scholar]
- Walsh P. S., Erlich H. A., Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992 May;1(4):241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- Warner J. P., Barron L. H., Brock D. J. A new polymerase chain reaction (PCR) assay for the trinucleotide repeat that is unstable and expanded on Huntington's disease chromosomes. Mol Cell Probes. 1993 Jun;7(3):235–239. doi: 10.1006/mcpr.1993.1034. [DOI] [PubMed] [Google Scholar]
- Zhang L., Leeflang E. P., Yu J., Arnheim N. Studying human mutations by sperm typing: instability of CAG trinucleotide repeats in the human androgen receptor gene. Nat Genet. 1994 Aug;7(4):531–535. doi: 10.1038/ng0894-531. [DOI] [PubMed] [Google Scholar]
- van Kamp H., Jansen R., Willemze R., Fibbe W. E., Landegent J. E. Studies on clonality by PCR analysis of the PGK-1 gene. Nucleic Acids Res. 1991 May 25;19(10):2794–2794. doi: 10.1093/nar/19.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]





