Abstract
Despite an abundance of data describing expression of genes in the Candida albicans ALS (agglutinin-like sequence) gene family, little is known about the production of Als proteins on individual cells, their spatial localization or stability. Als proteins are most commonly discussed with respect to function in adhesion of C. albicans to host and abiotic surfaces. Development of a mAb specific for Als1, one of the eight large glycoproteins encoded by the ALS family, provided the opportunity to detect Als1 during growth of yeast and hyphae, both in vitro and in vivo, and to demonstrate the utility of the mAb in blocking C. albicans adhesion to host cells. Although most C. albicans yeast cells in a saturated culture are Als1-negative by indirect immunofluorescence, Als1 is detected on the surface of nearly all cells shortly after transfer into fresh growth medium. Als1 covers the yeast cell surface, with the exception of bud scars. Daughters of the inoculum cells, and sometimes granddaughters, also have detectable Als1, but Als1 is not detectable on cells from subsequent generations. On germ tubes and hyphae, most Als1 is localized proximal to the mother yeast. Once deposited on yeasts or hyphae, Als1 persists long after the culture has reached saturation. Growth stage-dependent production of Als1, coupled with its persistence on the cell surface, results in a heterogeneous population of cells within a C. albicans culture. Anti-Als1 immunolabelling patterns vary depending on the source of the C. albicans cells, with obvious differences between cells recovered from culture and those from a murine model of disseminated candidiasis. Results from this work highlight the temporal parallels for ALS1 expression and Als1 production in yeasts and germ tubes, the specialized spatial localization and persistence of Als1 on the C. albicans cell surface, and the differences in Als1 localization that occur in vitro and in vivo.
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen that causes oral and vaginal mucosal infections as well as systemic disease. C. albicans has several gene families that encode proteins involved in host–pathogen interactions (Jones et al., 2004). Among these is the Als (agglutinin-like sequence) family of large cell-surface glycoproteins (reviewed by Hoyer, 2001; Hoyer et al., 2008). Completion of the genome sequence of C. albicans strain SC5314 (Jones et al., 2004) has demonstrated that there are eight different ALS genes located on three of the eight C. albicans chromosomes (reviewed by Hoyer et al., 2008). ALS genes share a similar basic organization, consisting minimally of a relatively conserved 5′ domain, a central domain of tandemly repeated sequence units, and a 3′ domain of relatively variable length and sequence. Heavy glycosylation, primarily of the central and C-terminal Als domains, and localization of the mature Als proteins in the C. albicans cell wall, positions them optimally for contact with host and abiotic surfaces, where they function in adhesive processes (reviewed by Hoyer et al., 2008).
Since the discovery of the ALS family, efforts have focused on understanding the role of its multiple encoded proteins. Earlier approaches included extensive studies of ALS gene expression patterns in C. albicans cells from cultures, disease models and human clinical material (reviewed by Hoyer et al., 2008). Multiple possibilities could be envisioned, including transcription of a single ALS gene at one time, resulting in the domination of an individual Als protein on the C. albicans cell surface, or simultaneous expression of multiple ALS genes, resulting in the heterogeneous presence of similar quantities of Als proteins on the cell. Results from different studies have demonstrated simultaneous expression of ALS genes in various specimens, and found that, regardless of the source of the C. albicans cells, certain ALS genes can be expressed at high levels while others never rise above a low expression level. Some genes, such as ALS1, have distinct expression patterns (Zhao et al., 2004; Green et al., 2005b). In vitro studies using wild-type strains and also a PALS1–GFP reporter construct show a tremendous burst of ALS1 expression when cells from a saturated culture are placed into fresh growth medium. ALS1 expression levels trail off as culture growth progresses. In C. albicans cells recovered from disease models and human clinical specimens, ALS1 expression is detected readily without the temporal decrease in expression (Green et al., 2004, 2005a, 2006; Cheng et al., 2005), suggesting the potential for differences in ALS1 regulation in vitro and in vivo. Because gene expression data are derived from a population of cells, little is known about the production of Als1 on individual cells or its spatial localization or stability on the cell surface. The observed gene expression data could be consistent with numerous permutations of these variables.
To determine the cell-surface Als1 patterns that correspond to the gene expression data, we developed an anti-Als1 mAb. Development of an anti-Als1 mAb is part of a larger effort to raise a mAb specific for each protein in the Als family and study Als protein localization on the C. albicans cell surface. Characterization of an anti-Als3 mAb has been reported previously (Coleman et al., 2009). Indirect immunofluorescence of C. albicans cells showed the unique localization of Als1 on yeast and germ tubes/hyphae, and the stability of the protein, which resulted in a heterogeneous Als1 presence among cultured cells. Analysis of C. albicans cells recovered from a disease model revealed differences from cultured cells in Als1 localization, consistent with ALS1 regulatory differences in vitro and in vivo.
METHODS
Production of mAbs.
mAbs were raised against Pichia pastoris-produced, hexa-His-tagged fragments from the N-terminal domain of Als proteins, as described by Coleman et al. (2009). Briefly, Als N-terminal fragments were secreted into the culture supernatant and purified by His-Trap column chromatography according to the manufacturer's instructions (GE Healthcare). When necessary (for N-terminal domain fragments of Als2, Als6 and Als9), proteins were treated with endoglycosidase H (Roche) to remove N-linked carbohydrate. Proteins were analysed by MS to verify their predicted molecular mass, and visualized by SDS-PAGE and silver staining to confirm the presence of a single protein band. Als N-terminal domain fragments were used to immunize BALB/c mice, as described previously (Coleman et al., 2009). ELISA was used for initial specificity testing (Coleman et al., 2009). Absorbance values were calculated as A570 subtracted from A450. Typical results showed an absorbance reading of approximately 0.3 for the reaction between a specific anti-Als hybridoma culture supernatant and its corresponding immunogen, compared with background values of ≤0.1. ELISA analysis using protein G-purified (GE Healthcare) mAb typically produced absorbance readings of approximately 1.5 with background values of ≤0.1. Western blotting methods have also been described previously (Coleman et al., 2009). A mAb specific for Als1 (called 1-B2) was isotyped using the Monoclonal Antibody Isotyping kit (Pierce) and was IgG1 with a kappa light chain. mAb–antigen binding kinetics were measured on a Biacore 3000 Surface Plasmon Resonance system using an indirect capture method, as described previously (Coleman et al., 2009).
C. albicans strains.
Many of the C. albicans strains used in this work were derived from SC5314 (Gillum et al., 1984), such as CAI4 (iro1-Δura3 : : imm434/iro1-Δura3 : : imm434; Fonzi & Irwin, 1993) and CAI12, which is a CAI4 derivative (ura3/URA3; Porta et al., 1999). Strain 1467 (als1Δ/als1Δ; Zhao et al., 2004) was constructed from CAI4. The wild-type large ALS1 allele from strain SC5314 was reintegrated into 1467 to yield strain 2151 (Zhao et al., 2004). C. albicans strains of diverse origin and clade assignment (detailed in Coleman et al., 2009) were also used. These included strains WO-1, GC15, GC23, 1-28, 2309, SqF087, CrA038 and OKP77. A set of diverse Candida species was assembled, including Candida dubliniensis strains CD36 (from Derek Sullivan, Trinity College, Dublin, Ireland), CM1 and 16F (from Richard Barton, University of Leeds, UK), as well as isolates purchased from the American Type Culture Collection (Candida glabrata ATCC 2001, Candida krusei ATCC 14243, Candida parapsilosis ATCC 22109, Candida lusitaniae ATCC 42720, Candida tropicalis ATCC 201380 and Candida guilliermondii ATCC 6260).
ALS1 overexpression was accomplished using plasmid 1105 (Green et al., 2005a), which is a modified version of CIp10 (Murad et al., 2000). Plasmid 1105 encodes the C. albicans TPI1 promoter and terminator sequences separated by a polylinker that includes the restriction sites (5′→3′) XhoI-SmaI-NotI-BglII. The XhoI-BglII sites allow cloning for overexpression of any full-length ALS gene due to the lack of these restriction sites in any of the ALS gene coding regions. These sites are used in various vectors in the laboratory and allow interchangeable cloning of ALS genes between different constructs. The ALS1 large allele was amplified from genomic DNA of strain 1416, an als1Δ/ALS1 derivative of strain SC5314, using the primers ALS5-Xho (5′-CCC CTC GAG ATG ATT CAA CAA TTT ACA TTG TTA TTC C-3′) and ALS1-Bgl (5′-CCC AGA TCT TCA CTA AAT GAA CAA GGA CAA TAA TG-3′), and Pfu polymerase according to the manufacturer's instructions. The XhoI/BglII-digested fragment was ligated into XhoI/BglII-cut plasmid 1105. The PTPI1–ALS1 overexpression construct was linearized with HindIII, which cuts within the 3′ end of the ALS1 coding region to direct integration of the plasmid to the ALS1 locus in strain CAI4. The resulting strain, 2243, was verified by Southern blotting, which indicated that integration was directed to the large allele locus of strain CAI4 (data not shown). The growth rate of the overexpression strain was determined using published methods (Zhao et al., 2004). Briefly, doubling times were calculated from the linear portion of the growth curve, which was defined spectrophotometrically. Triplicate data points in triplicate independent experiments were analysed statistically and showed no difference between the strains (doubling time for CAI12=1.45±0.03 h and for 2243=1.47±0.03 h).
C. albicans culture conditions.
All C. albicans isolates were stored at –80 °C. Strains were streaked to YPD medium (per litre: 10 g yeast extract, 20 g peptone and 20 g glucose, with 20 g agar added for plates) and incubated for 24 h at 37 °C. Plates were transferred to 4 °C and kept for no longer than 1 week. A single colony was inoculated into 20 ml YPD liquid medium for each starter culture. The starter culture was incubated at 37 °C with 200 r.p.m. shaking for 16 h prior to experiments that utilized a 37 °C incubation temperature. Cells from the starter culture were washed in Dulbecco's PBS without calcium or magnesium (DPBS), counted and inoculated at 1×106 cells ml−1 into the growth medium appropriate for the experiment. Starter cultures for experiments that utilized a 30 °C incubation temperature were grown at 30 °C for 16 h with 200 r.p.m. shaking, and cells were handled similarly to those grown at 37 °C. Anti-Als1 immunolabelling experiments were conducted using cells grown at 37 or 30 °C with equivalent results. For growing germ tubes, C. albicans yeasts from the YPD starter culture were processed as detailed above and resuspended into germ tube-inducing medium at a density of 5×106 cells ml−1. Media included RPMI 1640 without l-glutamine, YPD with 10 % fetal bovine serum, DPBS with 10 % fetal bovine serum, and the medium described by Lee et al. (1975). Details regarding culture conditions are specified as needed for each experiment in Results (see below).
Immunolabelling of C. albicans cells.
Anti-Als mAbs were used to immunolabel C. albicans cells from culture, and from homogenized mouse kidney samples. Methods have been described previously (Coleman et al., 2009). Routinely, cultured cells were fixed in DPBS containing 3 % paraformaldehyde. Fixed cells were washed in DPBS, non-specific binding was blocked with normal goat serum, and the cells were incubated with the primary protein G-purified mAb and then with a secondary FITC-conjugated antibody. Fluorescence was detected either by microscopy (see below) or by flow cytometry (Coleman et al., 2009). For analysis of Als protein production in vivo, C. albicans cells were isolated from the kidney of a BALB/cByJ mouse at 28 h post-inoculation. Methods for electron microscopy detection of immunogold labelling have been described previously (Coleman et al., 2009).
Three different microscope systems were used to visualize immunolabelled cells. The first was an Olympus BX50 FluoView confocal microscope using Melles Griot argon (488 nm) and krypton (568 nm) lasers. The second was a Zeiss Axiovert 200M microscope equipped with a mercury X-Cite illuminator, an ApoTome Structured Illumination Optical Sectioning system with a VH ApoTome grid under a weak ApoTome filter setting, standard excitation and emission filters for FITC and rhodamine, a Plan-Apochromat ×63/1.40 numerical aperture oil objective lens, and an AxioCam MRc 5 colour camera. The third microscope was a Zeiss LSM 710 NLO confocal scanner, Axio Observer Z1 microscope and a Spectraphysics Mai Tai Ti : Sapphire laser. Some samples were observed as wet mounts, while others were embedded in ProLong Gold antifade reagent (Molecular Probes P36930) in an approximately 1 : 1 volume with cells in DPBS, placed on glass slides, coverslipped, and allowed to set for 24 h. The microscope used for each experiment is specified in the respective figure legend.
Covalent labelling of the C. albicans cell surface with Alexa Fluor 594.
C. albicans CAI12 cells from a 16 h overnight culture grown in YPD medium at 30 °C with 200 r.p.m. shaking were washed three times in 0.1 M sodium bicarbonate buffer (pH 8.3) and then incubated for 1 h at 25 °C in 1 ml bicarbonate buffer containing 1 mg Alexa Fluor 594 carboxylic acid, succinimidyl ester (Molecular Probes A-20004) dissolved in 0.1 ml DMSO. After incubation the cells were washed three times in DPBS, placed in YPD medium at a density of 1×106 cells ml−1, and incubated at 30 °C and 200 r.p.m. Aliquots of cells collected at multiple time points were washed, fixed and immunolabelled with anti-Als1, as described above. In certain experiments, C. albicans cells were stained with Calcofluor White (Sigma). A stock solution (1 mg ml−1) was prepared in 0.1 M Tris/HCl, pH 9.0, and diluted 1 : 10 in DPBS as a working solution. Anti-Als1-labelled C. albicans cells were resuspended in 100 μl Calcofluor White working solution and incubated at room temperature for 5 min. Cells were washed twice in DPBS prior to microscopic examination.
Adhesion assays.
The ability of the anti-Als1 mAb to inhibit adhesion of C. albicans to freshly collected human buccal epithelial cells (BECs) was tested. C. albicans yeasts were grown in YPD medium for 16 h at 30 °C. Yeasts were washed three times in DPBS, and inoculated either into YPD to grow yeasts or into RPMI 1640 medium to grow germ tubes. Germ tube adhesion assays have been described previously (Beucher et al., 2009). For yeast adhesion assays, 2×108 cells were inoculated into 20 ml fresh YPD and incubated for 1 h at 30 °C. Cells were collected, washed in DPBS and counted; 2×106 yeasts were added to a flask containing 4 ml DPBS. Anti-Als1 mAb or anti-Als6 mAb (isotype-matched negative control; Coleman et al., 2009) was added to the flask at a final concentration of 20 μg ml−1. Flasks were incubated for 30 min with 200 r.p.m. shaking and then 2×104 BECs were added. Following 30 min incubation, BECs were collected by filtration and processed as described previously (Beucher et al., 2009). Adhesion of yeasts to BECs was evaluated by microscopy and the mean number of yeasts adherent to 100 BECs calculated. Analysis of data from three separate days of experiments has been described previously (Beucher et al., 2009).
Real-time RT-PCR quantification of ALS1 transcription.
Yeasts from a YPD starter culture (described above) were inoculated into fresh medium and incubated with shaking at 200 r.p.m. At 1 h intervals, cells were removed from the culture and collected by filtration over a 0.45 μm pore-size membrane. Each membrane was transferred to a 50 ml conical Falcon tube, flash-frozen in a dry ice–ethanol bath, and stored at −80 °C until RNA was extracted using a hot phenol method (Hoyer et al., 1995). RNA was treated with DNase I and the samples were purified using an RNeasy Mini kit (Qiagen). PCR using primers against genomic DNA sequences was employed to verify that all detectable DNA was removed from the RNA preparation. The quality and integrity of the RNA were checked on a formaldehyde agarose gel (Hoyer et al., 1995). Primers QRTALS1F and QRTALS1R were used for real-time RT-PCR analysis of ALS1 gene expression, as described previously (Green et al., 2005b). Primers QRTTEF1F and QRTTEF1R, which amplify TEF1, were used as a control. Real-time RT-PCR analyses were run in triplicate for each time point, and the mean and sd of threshold values (Ct) graphed.
RESULTS
Validating the specificity of the anti-Als1 mAb
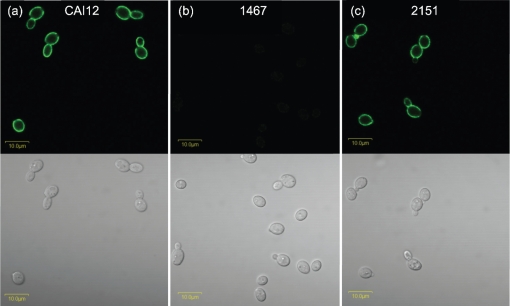
ELISA was used to select mAb 1-B2 from a panel of potential anti-Als1 mAb candidates (see Methods). mAb 1-B2 specificity for Als1 was tested further by immunolabelling of cultured C. albicans cells. Previous studies of ALS1 expression in cultured C. albicans cells showed a large peak of transcriptional activity after yeasts from a saturated culture were placed into fresh growth medium, and a gradual diminishing of measurable transcript abundance as the culture aged (Green et al., 2005b). Yeasts of control strain CAI12 were grown to saturation overnight at 37 °C (16 h) and then transferred to fresh YPD medium for 1 h. The 1 h yeast cells immunolabelled brightly with the anti-Als1 mAb, whereas no signal was detected for strain 1467, in which ALS1 is deleted (Fig. 1). The specificity of the anti-Als1 mAb was further demonstrated by immunolabelling of the ALS1 reintegrant strain 2151.
Fig. 1.
Immunolabelling of C. albicans yeasts with anti-Als1 mAb demonstrates cell-surface Als1 recognition and specificity of the mAb. Yeasts from saturated YPD cultures of C. albicans control strain CAI12 (a), 1467 (als1Δ/als1Δ) (b) and ALS1 reintegrant strain 2151 (als1Δ/als1Δ : : ALS1) (c) were transferred to fresh YPD medium for 1 h at 37 °C, immunolabelled with anti-Als1 and a FITC-conjugated secondary antibody, and viewed with an Olympus BX50 microscope. Upper panels with a dark background have laser illumination (488 nm); lower panels are illuminated with white light. Lack of Als1 on the mutant strain and detection of Als1 on the control and reintegrant strains supported the conclusion that the mAb recognized cell-surface Als1 and was specific for the protein. Bars, 10 μm.
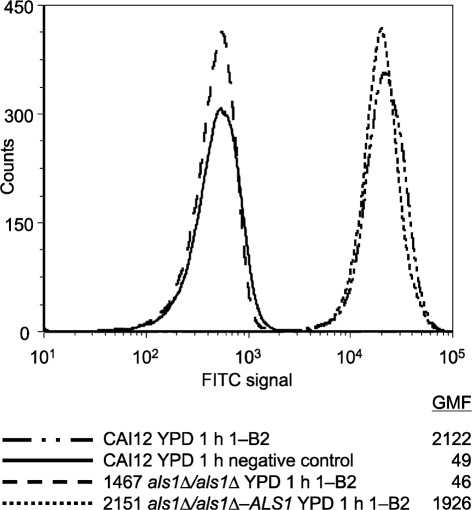
Flow cytometry analysis was also used to validate the specificity of the anti-Als1 mAb (Fig. 2). Similar to the results observed for immunofluorescence microscopy, flow cytometry showed a strong fluorescence on yeast cells of strain CAI12 grown under conditions known to promote high levels of ALS1 transcription. The als1Δ/als1Δ strain showed fluorescence at a level similar to that of the negative control (secondary antibody only). Reintegration of a wild-type ALS1 allele restored anti-Als1 recognition of the C. albicans cell surface.
Fig. 2.
Use of flow cytometry to demonstrate anti-Als1 specificity in populations of C. albicans yeast cells. C. albicans yeast cells of control strain CAI12, als1Δ/als1Δ mutant (1467) and ALS1 reintegrant (2151) strains were grown for 16 h in YPD and then transferred to fresh YPD for 1 h. Cells were immunolabelled with anti-Als1 and a FITC-conjugated secondary antibody. Geometric mean fluorescence (GMF) was measured as described previously (Coleman et al., 2009). The negative control sample was treated with the secondary antibody alone. An increased fluorescence intensity for strains that encoded ALS1 suggested that the mAb was specific for Als1.
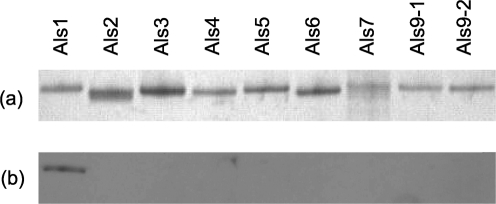
Fig. 3 shows a silver-stained SDS-PAGE gel of the N-terminal domain fragment from each protein in the Als family. Production and validation of these fragments have been detailed by Coleman et al. (2009) and are summarized in Methods. Western blotting with the anti-Als1 mAb detected the Als1 N-terminal domain fragment, but none of the fragments encoded by any other ALS genes, supporting the conclusions that mAb 1-B2 is specific for Als1 and that the mAb is useful for Western blotting. Surface plasmon resonance measurements of binding kinetics between mAb 1-B2 and the Als1 N-terminal domain fragment showed dissociation constants (KD) of 0.3 and 0.5 nM in duplicate experiments (mean=0.4 nM).
Fig. 3.
Western blotting of the N-terminal domain from each protein in the Als family to demonstrate specificity of the anti-Als1 mAb. N-terminal fragments of each Als protein were produced in P. pastoris, as described previously (Coleman et al., 2009). Each fragment had a hexa-histidine tag at its C terminus and was purified by nickel-affinity chromatography. (a) Approximately 0.5 mg of each purified protein was run on an SDS-PAGE gel and silver-stained. (b) A second gel with identical lanes, and just 50 ng of each protein, was blotted to a Hybond-P PVDF membrane and Western-blotted with anti-Als1 at a concentration of 4.8 μg ml−1. Recognition of only the Als1 immunogen with the anti-Als1 mAb further supported the conclusion that the mAb was specific for Als1.
Anti-Als1 labelling of C. albicans budding yeasts
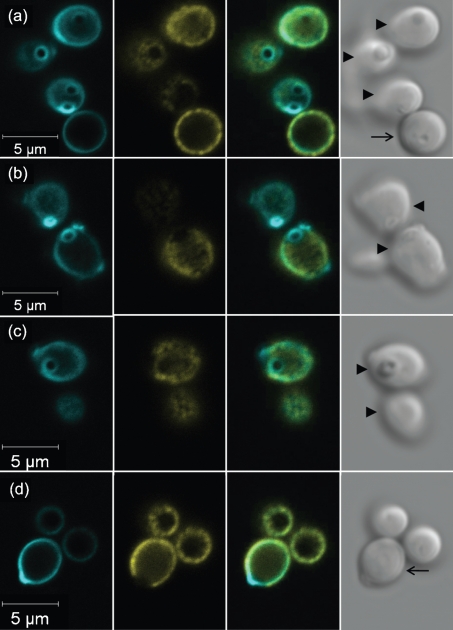
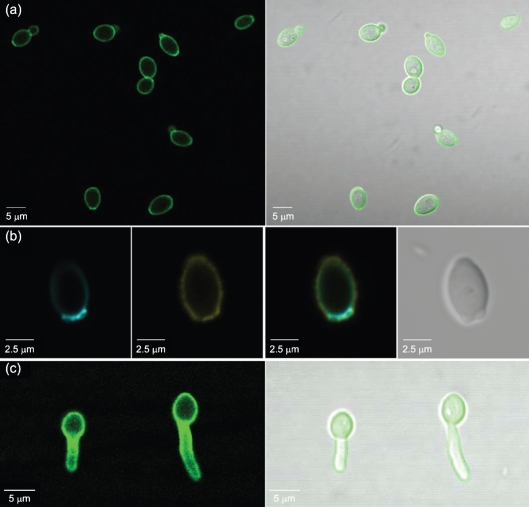
The cell-surface anti-Als1 labelling pattern appeared uniform, with the exception of gaps in signal at the poles of the cell (Fig. 1). The emergence of buds adjacent to the gaps suggested that Als1 was absent from bud scars. C. albicans yeasts were labelled with anti-Als1, counterstained with Calcofluor White and examined by microscopy (Fig. 4). Fig. 4(a) shows a lateral view of a yeast cell on which anti-Als1 labelling was missing from the pole. Intense Calcofluor White staining in this location was consistent with the presence of a bud scar. Images in Fig. 4(b–d) demonstrate that the Als1-negative portions of the yeast cell surface were stained intensely with Calcofluor and conformed to the shape of bud scars.
Fig. 4.
Anti-Als1 immunolabelling and Calcofluor White staining of C. albicans yeast cells to demonstrate the absence of Als1 localization to the bud scar. C. albicans strain CAI12 from a saturated culture was grown for 1 h in fresh YPD medium at 30 °C and fixed in paraformaldehyde. Fixed cells were immunolabelled with anti-Als1 and a FITC-conjugated secondary antibody. Cells were then stained with Calcofluor White, which binds to chitin, accentuating the fungal cell wall and bud scars. Cells were visualized with a Zeiss LSM 710 microscope. Rows (a–d) show images of Calcofluor-stained cells (left column, false coloured with cyan), FITC anti-Als1 labelling (second column from left, false coloured with yellow), a merged image (second column from right) and a bright-field image (right column). FITC labelling demonstrated Als1 on the cell surface with the exception of the bud scars, which were clearly stained with Calcofluor. Arrows within each bright-field image denote the plane of focus: an arrow with a tail shows cells with edges in focus, while arrowheads show cells where the cell surface was in focus so that the bud scar structure could be viewed more readily.
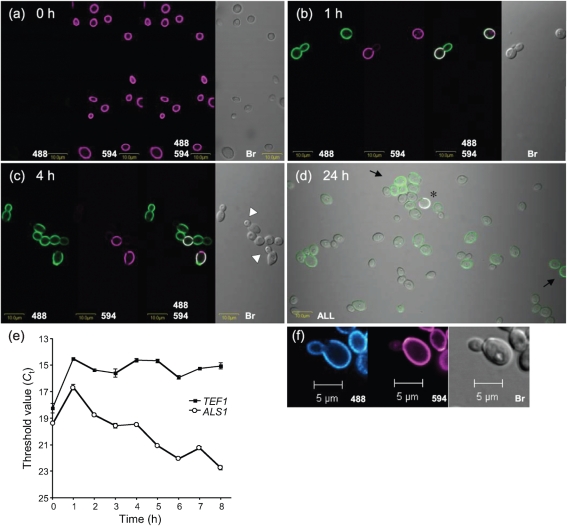
The persistence of Als1 on the C. albicans yeast cell was examined by covalently labelling the cell surface with Alexa Fluor 594 carboxylic acid, succinimidyl ester. CAI12 cells were grown for 16 h in YPD at 37 °C with 200 r.p.m. shaking. At this time point, ALS1 transcript levels are very low (Hoyer et al., 1995, 2008), and cells with detectable anti-Als1 surface labelling were rare in the culture (Fig. 5a). The Als1-negative, Alexa 594-positive cells (called ‘inoculum cells’) were placed into fresh YPD medium and followed over time. After 1 h, the inoculum cells, as well as their newly formed buds, were positive for Als1 (Fig. 5b). As culture growth progressed and new generations of buds formed, the Alexa 594-labelled inoculum cells became relatively less abundant in the culture. Als1-positive buds arose from Als1-positive mother yeasts until a time at which Als1-negative buds were observed [white arrowheads in Fig. 5(c), 4 h post-inoculation]. Four cells in Fig. 5(c) appeared connected, providing additional information regarding the generation that fell below the detection limit for the immunolabelling assay. The Alexa 594-positive inoculum cell in this group gave rise to two Als1-positive daughters. The progeny of one daughter was faintly Als1-positive, suggesting that the daughter of this cell (the inoculum cell's great-granddaughter) was likely to be Als1-negative. At the point when the culture reached saturation, Als1-positive cells were still visible, although rare, in the presence of many Als1-negative cells (Fig. 5d). The photomicrograph in Fig. 5(d) was difficult to obtain and suffers from obvious yeast autofluorescence that appears similar to that of Als1-positive cells; Als1-positive cells are indicated by black arrows. Also evident was a cell with signals of both Als1 and Alexa 594 (asterisk in Fig. 5d). The detection of this cell was evidence that Als1 deposited on the surface of an inoculum cell after it was placed into fresh YPD medium was still present 24 h after the culture was started. Alexa 594-positive, Als1-negative cells were not observed using this technique.
Fig. 5.
Photomicrographs of Alexa Fluor 594-labelled and anti-Als1-labelled C. albicans yeast cells to demonstrate decreasing cell-surface Als1 abundance on successive generations of yeast culture growth as well as stability of Als1 over time. Cells from a saturated YPD culture of strain CAI12 were transferred to fresh medium and grown at 30 °C for the indicated times [0 h, (a); 1 h, (b); 4 h, (c); 24 h, (d)]. In (a–c), replicate photomicrographs of the same image are shown illuminated with individual or combinations of wavelengths of light, as indicated. A Zeiss Axiovert 200M microscope was used to image cells. Anti-Als1 mAb was detected by a FITC-conjugated secondary antibody and 488 nm light (488). Alexa 594 covalently linked to the C. albicans cell surface was detected with 594 nm light (594). Bright-field microscopy was used to visualize all cells in a field (Br). Als1-negative buds are indicated by white arrowheads in (c). The photomicrograph in (d) was illuminated with all three light sources and shows many Als1-negative yeasts and a rare dual-labelled Als1/Alexa 594-positive cell (asterisk). Als1-positive cells in (d) are indicated by black arrows. Faint outlines of cells were the result of either autofluorescence (green) or bleeding of the FITC signal into the 594 channel (purple). Together, the images show that Als1-positive cells were rare in saturated cultures, that transfer of cells to fresh medium produced a coating of Als1, and that the protein coat persisted on the cell surface as the culture aged. (e) Real-time RT-PCR quantification of ALS1 transcriptional activity over the course of culture growth to demonstrate the burst in transcription that occurs following inoculation into fresh culture medium and the decreasing abundance of ALS1 RNA as culture growth progresses. Data from a representative biological replicate of the experiment are shown; triplicate data points were gathered for each time point. The threshold value (Ct) is graphed so that lower values indicate greater RNA abundance. (f) False-coloured image from a replicate time-course experiment viewed with a Zeiss LSM 710 NLO system. False colouring with a gradient of colour emphasizes the difference in anti-Als1 labelling intensity between the inoculum yeast (Alexa 594-positive, purple; Als1-positive, cyan gradient) and its daughter (Als1-positive, cyan gradient). Decreased anti-Als1 labelling intensity suggests that the transcriptional burst occurs in the inoculum cell and is diluted by cell division in each subsequent generation until anti-Als1 labelling intensity falls below the limit of detection for the assay.
The image in Fig. 5(c) suggested that the great-granddaughters of the inoculum cells were Als1-negative. Counting cells with respect to their Alexa 594 and anti-Als1 labelling status provided an additional estimate of the generation for which the anti-Als1 signal dropped below the antibody labelling detection limit. Table 1 shows results from two independent experiments. Counting the percentage of Alexa 594-positive cells in the culture over time provided an estimate of the number of cell divisions that had occurred at each time point of the experiment. For example, at 1 h post-inoculation, Alexa 594-positive cells were approximately 50 % of the total cells, suggesting that one doubling had occurred. By 3 h, Alexa 594-positive cells were 17 % of the population, indicating that inoculum cells were one of every six cells in the culture. Most simplistically, this figure can be interpreted to indicate that between two and three doublings occurred in the 3 h period. Over this same period of time, the percentage of Alexa 594-negative, Als1-positive yeasts (progeny of the inoculum cells) rose in the population to 66 % at 3 h. Alexa 594-negative, Als1-negative cells were roughly one in five cells present at 3 h. These figures were consistent with a culture population that contained inoculum cells and their daughters, granddaughters and possibly great-granddaughters. Counts of cells suggested that Als1-negative cells were the granddaughters or great-granddaughters of the inoculum cells. Micrographs suggested a trailing off of the anti-Als1 signal, as observed in the granddaughter cell in Fig. 5(c). Real-time RT-PCR quantification of ALS1 transcript levels provided the Ct values that correspond to the micrographs (Fig. 5e). The variable presence of Als1 on individual yeast cells in the culture provided an additional measure of heterogeneity in the population.
Table 1.
Evaluation of Alexa Fluor 594 and anti-Als1 cell-surface labelling patterns for cultured C. albicans yeasts to demonstrate reduced Als1 presence on offspring of the inoculum cells
| Labelling pattern | Time point (h) | Proportion of cells* | ||
|---|---|---|---|---|
| Expt. 1 | Expt. 2 | Mean | ||
| Alexa 594-positive, Als1-positive | 1 | 45 % | 57 % | 51 % |
| 2 | 31 % | 37 % | 34 % | |
| 3 | 16 % | 17 % | 17 % | |
| Alexa 594-negative, Als1-positive | 1 | 46 % | 41 % | 44 % |
| 2 | 54 % | 51 % | 53 % | |
| 3 | 62 % | 70 % | 66 % | |
| Alexa 594-negative, Als1-negative | 1 | 9 % | 2 % | 6 % |
| 2 | 15 % | 12 % | 14 % | |
| 3 | 22 % | 13 % | 18 % | |
*Inoculum cells were covalently labelled with Alexa 594. At the indicated time points, cells from the culture were harvested and immunolabelled with the anti-Als1 mAb and a FITC-conjugated secondary antibody. Cells were visualized using a Zeiss Axiovert 200M microscope and evaluated in digital images. The number of cells evaluated at each time point ranged from 54 to 265, with a mean of 129. Cells considered positive had a visible signal for Alexa 594 and/or anti-Als1, as tabulated above. The labelling intensity for Alexa 594 was more consistent than for anti-Als1, suggesting that Als1 protein abundance varied in an obvious manner on cultured yeast cells. The variability was likely due to the large increase in transcriptional activity when cells were placed into fresh growth medium, and the gradual diminishing of transcriptional activity as culture growth progressed.
The above experiment documented the trailing off of the presence of Als1 on the C. albicans cell surface as subsequent generations grew in a culture, and also the stability of Als1 on the cell surface over time. The Alexa 594 labelling experiment was repeated to gain a better understanding of which generation showed the first decrease in Als1 abundance. Careful examination of the images in Figs 1 and 5(b) suggested that daughter cells were labelled less intensely than the inoculum cells. The Alexa 594 labelling experiment was repeated, and this time, the slides were analysed using the Zeiss LSM 710 confocal system, which provides the option of false labelling images using gradations of colour that correspond to differences in signal strength. The image of cells from a 1 h culture showed that the daughter cell was labelled less intensely than the inoculum cell (Fig. 5f). This image suggested that the burst of ALS1 transcriptional activity occurred within the inoculum cell and was reduced by subsequent cell divisions until Als1 was no longer detectable on the cell surface of subsequent generations. This result has important implications for studying the mechanism that regulates ALS1 transcription in vitro.
Anti-Als1 labelling of C. albicans germ tubes and hyphae
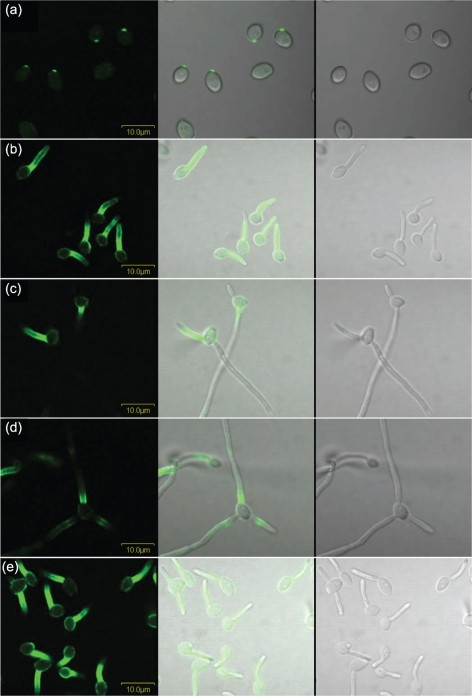
Surface localization of Als1 was also studied on germ tubes. The presence of Als1 on germ tubes has been reported elsewhere (Fu et al., 2002), and immunolabelling with mAb 1-B2 produced a similar result (Fig. 6). Analysis of germ tubes collected over a time-course of growth showed that Als1 was visible as early as 10 min following inoculation of YPD-grown yeasts into RPMI medium (Fig. 6a). On 1 h germ tubes, Als1 labelling was most intense proximal to the mother yeast (Fig. 6b). Similar to observations for yeast forms, Als1 persisted on germ tubes over time and was visible even after cells formed long hyphae (Fig. 6c, d). On longer hyphae, additional Als1-positive sections were observed further down the length of the hypha, but the signal intensity was considerably more faint than the signal proximal to the mother yeast. For 85 % of the cells observed, anti-Als1 labelling was detected only on the longer/longest germ tube to emerge from each mother yeast. However, other patterns were noted, including labelling only of the shorter germ tube on a mother yeast (Fig. 6c) and labelling of each germ tube present (Fig. 6d). Labelling of the mother yeast was visible, but faint, for C. albicans strains derived from SC5314 that were grown in YPD and then inoculated into RPMI medium.
Fig. 6.
Immunolabelling of C. albicans strain CAI12 with anti-Als1 over a time-course of germ tube growth. CAI12 cells were grown in YPD medium for 16 h at 37 °C, washed in DPBS, counted and transferred into pre-warmed RPMI medium at a cell density of 5×106 cells ml−1. The culture was sampled at various time points and cells were fixed in paraformaldehyde, as described previously (Coleman et al., 2009). Fixed cells were immunolabelled with anti-Als1 and a FITC-conjugated secondary antibody. Cells were imaged using an Olympus BX50 FluoView microscope. Cell-surface Als1 was visible as early as 10 min following inoculation of the cells into RPMI (a). Anti-Als1 labelling of germ tubes was most intense in the region proximal to the mother yeast (b). While the longer germ tube/hypha was Als1-positive for most mother yeasts, exceptions were noted, including labelling of the shorter germ tube/hypha (c), or labelling of each germ tube/hypha (d). In each case, Als1 persisted on the germ tube/hypha over time. Growth of germ tubes in different culture conditions, such as Lee medium (e), resulted in the same general pattern of anti-Als1 labelling, with differences in the intensity of labelling of the mother yeast and/or germ tubes (other patterns not shown). Bars, 10 μm.
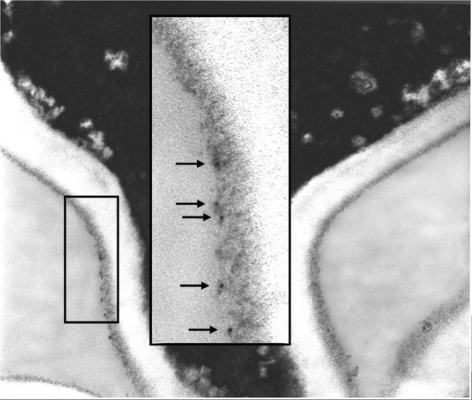
Growth of C. albicans germ tubes in media other than RPMI revealed growth medium-dependent effects in anti-Als1 labelling (Fig. 6e). In each experiment, yeast cells were grown in YPD for 16 h and then transferred to another medium to promote germ tube formation. Germ tubes were grown in YPD with 10 % fetal bovine serum, in DPBS with 10 % fetal bovine serum or in Lee medium, fixed in paraformaldehyde and immunolabelled with anti-Als1. Growth medium-dependent differences in anti-Als1 labelling were in intensity, rather than location of signal. While germ tubes grown in RPMI medium exhibited only faint labelling of the mother yeast (Fig. 6b), growth under serum-containing conditions or in Lee medium resulted in stronger anti-Als1 labelling of the mother yeast (Fig. 6e). Gaps in labelling appeared to correspond to bud scars. Labelling of the germ tube for Lee-grown cells was stronger than for the other growth media. Immunolabelling of germ tubes with anti-Als1, followed by treatment with an anti-mouse gold particle-linked antibody and processing for electron microscopy, visualized Als1 in the outer flocculant layer of the cell wall (Fig. 7).
Fig. 7.
Electron micrograph of a C. albicans mother yeast–germ tube junction immunolabelled with anti-Als1 to show Als1 localization in the outer flocculant layer of the germ tube surface. Strain CAI12 germ tubes were grown in RPMI medium at 37 °C, immunolabelled with anti-Als1 mAb and a gold-conjugated secondary antibody, embedded and sectioned for electron microscopy. Arrows in the higher-magnification inset highlight gold particles.
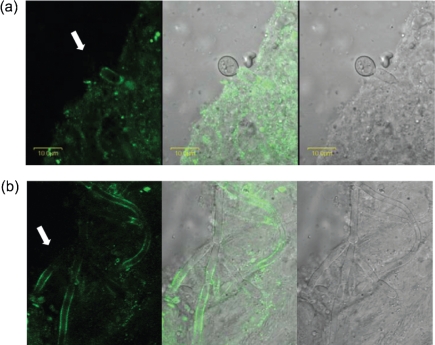
Anti-Als1 labelling was also examined on C. albicans cells from a murine model of disseminated candidiasis. C. albicans was injected into the mouse tail vein and infection proceeded for 28 h. Anti-Als1 immunolabelling of C. albicans cells dissected from homogenized mouse kidney showed instances of differential labelling between yeast and hyphae (Fig. 8a), as observed for RPMI-grown germ tubes. Unlike cultured cells, however, C. albicans hyphae from the mouse kidney were labelled on a more extensive length of the cell (Fig. 8b).
Fig. 8.
Anti-Als1 immunolabelling of C. albicans recovered from murine kidney tissue shows similarities and differences with respect to immunolabelling of cultured cells. A BALB/cByJ mouse was inoculated via the lateral tail vein with 5×105 cells of C. albicans strain CAI12. At 28 h post-inoculation, the kidneys were removed, minced with a razor blade, and then homogenized. A portion of the supernatant was treated with anti-Als1 mAb and an FITC-conjugated secondary antibody. Cells were imaged using an Olympus BX50 FluoView microscope. Images were illuminated with laser only (488 nm; left panel), white light (right panel) or both (centre panel). (a) Similar to cultured cells, hyphae were labelled with anti-Als1, but yeast cells were not (white arrow); however, yeast cells were difficult to find in the kidney tissues. (b) Although Als1-negative hypha segments were observed (white arrow), Als1 was detected on a much larger area of the surface of hyphae recovered from kidney tissue.
Anti-adhesive effects of the anti-Als1 mAb
Anti-Als1 mAb was tested for its ability to inhibit adhesion of C. albicans to freshly collected human BECs. Growth conditions that produced the greatest amount of anti-Als1 immunolabelling were examined. Treatment of 1 h yeast cells with anti-Als1 yielded significantly fewer adherent yeasts per buccal cell compared with treatment with an isotype-matched anti-Als6 (1.6±0.3 vs 5.0±0.3; P=0.02). Anti-Als6 was used as a negative control for adhesion assays because ALS6 expression levels are so low that little cell-surface protein is present (Coleman et al., 2009). Adhesion assays using 1 h C. albicans germ tubes produced similar results. Treatment with anti-Als1 yielded 7.0±0.9 adherent germ tubes per buccal cell compared with 10.9±0.9 for anti-Als6 (P=0.02). Inhibition of C. albicans adhesion using anti-Als1 is consistent with the role of Als1 in adhesion to host cells.
Anti-Als1 labelling of diverse Candida isolates
The anti-Als1 immunolabelling results presented above used derivatives of C. albicans strain SC5314. Anti-Als1 labelling patterns were examined on a set of eight C. albicans isolates of diverse clade assignment and collected from various body sites in either healthy or diseased humans or animals (Coleman et al., 2009). The collection included strains WO-1, GC15, GC23, 1-28, 2309, SqF087, CrA038 and OKP77. Prior to anti-Als1 immunolabelling, isolates were grown for 16 h in YPD, washed in DPBS and then transferred into YPD for 1 h or into RPMI for 1 h. For all isolates, immunolabelling of 1 h yeast cells produced results similar to those for SC5314-derived strains, with the exception of strain WO-1, for which a signal was not detected. Additional work demonstrated that the 5′ domain of the ALS1 coding region was missing from the WO-1 genome, providing additional support for the conclusion that the mAb was specific for Als1 (X. Zhao and others, unpublished results). Immunolabelling of germ tubes showed the most intense signal on the germ tube proximal to the mother yeast for each isolate, although the length of the labelled area varied between strains (data not shown). Sites of germ tube emergence were also labelled. Mother yeasts were labelled only faintly following growth in RPMI, although the labelling intensity varied between strains.
Isolates of various Candida species were tested to assess anti-Als1 immunolabelling. Coleman et al. (2009) have described the set of isolates, which included three strains of C. dubliniensis, and one isolate each of C. glabrata, C. krusei, C. parapsilosis, C. lusitaniae, C. tropicalis and C. guilliermondii. All Candida species were grown as starter cultures for 16 h in YPD at 37 °C and then transferred to fresh YPD for 1 h or to RPMI medium for 1 h at 37 °C. Many of these Candida species do not form germ tubes or hyphae, so the morphologies observed for C. albicans cells grown under these standard conditions were not present for all species tested. Immunolabelling was not detected for cells from any species or growth conditions using the anti-Als1 mAb.
Anti-Als1 immunolabelling of an ALS1 overexpression strain
Als1 that is synthesized as C. albicans yeast cells are placed into fresh culture medium was not incorporated into bud scars (Figs 1 and 4). This observation raises the question of whether Als1 can be placed into the cell wall at the location of a bud scar. This question was addressed by anti-Als1 immunolabelling of an ALS1 overexpression strain (strain 2243; Fig. 9). Constitutive ALS1 expression should result in Als1 presence on the surface of yeast cells, even at times in the growth curve when the majority of wild-type cells are Als1-negative. Fig. 9(a) shows yeast cells of strain 2243 grown in YPD medium for 8 h. Unlike wild-type C. albicans grown to this time point, each strain 2243 cell was labelled strongly with anti-Als1, including the poles (Fig. 9a). Calcofluor staining and higher-magnification imaging verified that Als1 was localized within the bud scars of strain 2243 (Fig. 9b). Germ tubes grown for 1 h in RPMI medium showed intense anti-Als1 labelling on both the mother yeast and germ tube (Fig. 9c).
Fig. 9.
C. albicans yeast and germ tubes overexpressing ALS1 have a uniform cell-surface coat of Als1, even within bud scars. Strain 2243, which overexpressed ALS1 under the control of the constitutive, high-activity TPI1 promoter, was grown in YPD for 16 h at 30 °C and transferred to fresh culture medium. (a) Yeast cells from 8 h growth at 30 °C in fresh YPD medium were fixed in paraformaldehyde and immunolabelled with anti-Als1 and a FITC-conjugated secondary antibody. In contrast to the labelling pattern of the control C. albicans strain CAI12 (Fig. 1), yeast forms of the ALS1 overexpression strain were labelled strongly at the poles of the cell, suggesting Als1 localization within bud scars. This conclusion was strengthened by observation of overlapping fluorescent signals from anti-Als1 immunolabelling and Calcofluor White staining (b). From left, a Calcofluor-stained cell false coloured with cyan, FITC anti-Als1 labelling false coloured yellow, the merged image, and a bright-field image. (c) YPD-grown 16 h yeast cells of strain 2243 were inoculated into RPMI medium for 1 h to produce germ tubes. Strong anti-Als1 immunolabelling was observed over the surface of the mother yeast and the entire germ tube. All images were produced using a Zeiss LSM 710 microscope.
DISCUSSION
Immunolabelling C. albicans cells with an anti-Als1 mAb visualized the pattern of Als1 cell-surface localization and stability that results from previously derived transcriptional data. Cell-surface Als1 protein abundance is directly related to ALS1 transcriptional abundance. The ALS1 transcriptional burst that occurs when cells are transferred to fresh medium results in a near-immediate cell-surface appearance of Als1. For cells that will grow as yeasts, the inoculum cell is coated in Als1, with the exception of the bud scars. Daughter cells and successive generations have a less-intense Als1 presence; this decrease continues as culture growth progresses. After the second or third generation of growth, Als1 is not detectable on the yeast cell surface by indirect immunofluorescence with the anti-Als1 mAb. Despite their apparent Als1-negative appearance, these cells also likely have some Als1 present. The persistence of Als1 on inoculum cells, even after that culture has reached saturation, demonstrates the stability of Als1 on the cell surface. Together, these data define a range of cell-surface Als1 abundance that varies from substantial to undetectable; the relative proportion of these cell types in the culture population varies over the stages of culture growth.
Intense anti-Als1 labelling was also noted as a germ tube emerged from the mother yeast. For the majority of cells, Als1 was found on the longest germ tube, although considerable variation was observed among multiple projections from the same mother yeast. It is open for debate whether anti-Als1-labelled subsequent projections from a mother yeast are germ tubes or pseudohyphae. Production of pseudohyphae from the mother yeast may trigger another burst of ALS1 transcription that is sufficient to produce Als1 on the new cell growth. Detection of a pseudohypha-specific cell-surface protein would clarify questions regarding morphology; however, recent work suggests that C. albicans may not produce a protein that uniquely identifies the pseudohyphal growth form (Carlisle et al., 2009).
One notable feature of Als1 on the yeast cell surface is its absence from bud scars. Most yeast cells in a saturated culture have divided at least once, leaving one, and for many cells, several, bud scars. Transfer of these cells into fresh medium, and the subsequent burst of ALS1 transcriptional activity, produces Als1 protein that is then distributed over the cell surface, with the exception of the bud scar(s). The bud scar structure is different from the remainder of the cell wall. Extensive investigations into Saccharomyces cerevisiae cell division processes, and conservation of major molecular elements between S. cerevisiae and C. albicans, allow S. cerevisiae to serve as a guide for C. albicans cell division (reviewed by Bi, 2001; Walther & Wendland, 2003; Sudbery et al., 2004). In S. cerevisiae, C. albicans and other yeasts, selection of the site of new bud production occurs in late G1 of the cell cycle with the deposition of septins that form a ring and act as a scaffold for many proteins involved in cytokinesis, including an actomyosin ring and localization of chitin synthase enzymes (DeMarini et al., 1997; Bi, 2001; Warenda & Konopka, 2002). Microscopically visible cell budding in C. albicans begins with a bulge in the cell wall due to the synthesis and deposition of new wall material (Rico et al., 1991). After cell division, the wall of C. albicans and S. cerevisiae cells contains bud scars consisting of thickened wall regions of increased chitin content (Mitchell & Soll, 1979). In C. albicans, the morphology of chitin microfibrils in bud scars and primary septa differs from that of microfibrils present in most of the cell wall. Bud scars and primary septa contain networks of longer interlaced microfibrils, while the majority of cell wall chitin microfibils are short microcrystalline rodlets (Lenardon et al., 2007). In yeast, newly synthesized cell wall material is deposited centripetally at the neck that separates mother and bud cell, forming a septum that splits down the middle after cytokinesis, allowing the cells to separate (Bi, 2001). In C. albicans, formation of an electron-lucent primary transverse septum is followed by electron-dense mother and bud secondary septa that form on either side of the primary septum (Shannon & Rothman, 1971).
After yeast cell division, the septin scaffold rings disassemble prior to formation of new rings for subsequent cell division events (Warenda & Konopka, 2002), suggesting transient localization of proteins associated with cell division in contrast to the permanent bud scars present in the wall. However, there is evidence that bud scar composition is more complex than simply differences in cell wall chitin microfibrils. Several studies have shown in both S. cerevisiae and C. albicans that new bud scars occur near but do not overlap previous bud scars (Chaffin, 1984; Chant & Pringle, 1995; Herrero et al., 1999), suggesting the presence of bud scar-associated intracellular signalling molecules that prevent septin localization. The significance of the lack of Als1 protein in bud scars is unknown, but may indicate spatial regulation of protein localization at these sites. Immunolabelling with an anti-Als4 mAb shows Als4 within bud scars, suggesting the possibility of specific localization signals for the various proteins in the Als family (D. A. Coleman and others, unpublished results). However, temporal regulation of Als1 protein production in the mother yeast late in the cell cycle could also account for Als1 absence from the bud scar. Overexpression of ALS1 under the control of the highly active, constitutive TPI1 promoter leads to Als1 localization in bud scars, demonstrating that bud scar localization is possible, although localization is differential when natural amounts of Als1 protein are produced. Although the function of Als1 is most commonly discussed in terms of adhesion, recent work suggests another phenotype evident in als1Δ/als1Δ strains: decreased cell size (X. Zhao and others, unpublished results). The small-cell-size phenotype suggests a role for Als1 in cell size homeostasis, an idea that is consistent with a role in cell division and perhaps differential localization on the yeast cell surface that excludes bud scars.
Other examples of differential localization with respect to bud scars are found in the yeast literature. Pir (proteins with internal repeats) proteins in S. cerevisiae are GPI-independent alkali-sensitive cell wall proteins that appear to function in cell wall stability (Sumita et al., 2005). Two of the four Pir proteins, Pir1 and Pir2, are localized only to S. cerevisiae bud scars, while Pir3 and Pir4 are found diffusely over the cell surface irrespective of bud scars. Signals in the C-terminal region of Pir1 are responsible for its localization to bud scars. C. albicans has only one Pir protein, Pir1 (Martínez et al., 2004). Pir1 in C. albicans is a cell wall protein similar in sequence to Pir4 in S. cerevisiae. The localization of Pir1 on the C. albicans cell surface is not known, and it may have a role in cell wall organization, based on observed increases in PIR1 expression during protoplast regeneration (Martínez et al., 2004).
For both the yeast and germ tube morphologies, the sharp increase in Als1 production immediately following transfer to fresh growth medium is a common link. The signal or signals initiating ALS1 transcription have been studied in far more detail in the context of germ tube growth, most likely due to a widespread misconception in the Candida literature that ALS1 transcription is specific for that growth form. Germ tube induction is a complex process. Multiple factors influence or regulate the process, including CPH1, CPH2, CZF1, EFG1, FLO8, NRG1, RAS1, RFG1, TEC1, TUP1 and UME6; UME6 is proposed as a central regulator of hypha formation downstream of the other factors (Banerjee et al., 2008; Zeidler et al., 2009). Several hypha-associated genes regulate ALS1 expression, including EFG1 (Braun & Johnson, 2000; Fu et al., 2002), BCR1 (Nobile & Mitchell, 2005) and TOR1, an upstream negative regulator of BCR1 and EFG1 (Bastidas et al., 2009). Deletion of TUP1 and CPH1, two genes related to filamentation, does not affect ALS1 expression (Braun & Johnson, 2000; Fu et al., 2002). YAK1 functions upstream of TUP1 and influences ALS1 transcription (Goyard et al., 2008). Investigations into ALS1 regulation in yeast will determine whether shared or unique regulatory proteins function in yeast and germ tubes.
The anti-Als1 immunolabelling patterns are different on cells cultured in vitro compared with those recovered from a disease model. Previously characterized ALS1 transcriptional patterns are consistent with this observation. Compared with the transcriptional burst and decline that is observed in vitro (Fig. 5e), ALS1 is detected consistently in C. albicans cells from disease models and human clinical material. The measurements were qualitative, rather than quantitative, but demonstrate ALS1 transcript in conditions where other ALS gene transcripts could not be detected. These conditions included oral scrapings from six HIV-positive patients with signs of oral candidiasis, oral tissues from a hyposalivatory rat model of oral candidiasis at 3 and 5 days post-inoculation, vaginal epithelium and discharge from women who had fungal elements present on direct microscopic examination, a murine model of vaginal candidiasis at day 4 and 7, vaginal, buccal and oesophageal reconstituted human epithelium at 12, 24 and 36 h of incubation with C. albicans, as well as model denture and catheter biofilms (Green et al., 2004, 2006; Cheng et al., 2005). ALS1 transcriptional activity was the most readily detected among genes in the ALS family in kidney tissue from the murine model of disseminated disease when assayed at 12, 24 and 48 h post-infection (Green et al., 2005a). Collectively, these observations suggest that ALS1 regulation in vivo is different from that in vitro. A study of C. albicans growth in the murine intestinal tract showed Efg1-independent expression of genes that are Efg1-regulated in vitro (White et al., 2007). Since Efg1 regulates ALS1 in vitro (Fu et al., 2002), perhaps this relationship accounts for the differences observed in vivo. Because anti-Als1 immunolabelling results are obviously different between in vitro- and in vivo-grown C. albicans cells, in vitro immunolabelling data will be of limited use to explain Als1 function in host–C. albicans interactions. Immunolabelling of cells harvested from additional disease models and perhaps from human clinical specimens is clearly the next step required to better understand the production and function of Als1 in host environments.
Acknowledgments
We thank Liping Wang and Rachel Breitenfeld of the University of Illinois Immunological Resource Center for producing anti-Als mAbs. Lou Ann Miller of the Center for Microscopic Imaging, and Barbara Pilas and Ben Montez of the University of Illinois Flow Cytometry Facility, also contributed to this work. This research was funded by the National Institute of Dental and Craniofacial Research, National Institutes of Health (grant R01 DE14158). The investigation was conducted in a facility constructed with support from Research Facilities Improvement Program grant number C06 RR16515-01 from the National Center for Research Resources, National Institutes of Health.
Abbreviations
BEC, buccal epithelial cell
References
- Banerjee, M., Thompson, D. S., Lazzell, A., Carlisle, P. L., Pierce, C., Monteagudo, C., López-Ribot, J. L. & Kadosh, D. (2008). UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas, R. J., Heitman, J. & Cardenas, M. E. (2009). The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog 5, e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher, B., Marot-Leblond, A., Billaud-Nail, S., Oh, S.-H., Hoyer, L. L. & Robert, R. (2009). Recognition of Candida albicans Als3 by the germ tube-specific monoclonal antibody 3D9.3. FEMS Immunol Med Microbiol 55, 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E. (2001). Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct Funct 26, 529–537. [DOI] [PubMed] [Google Scholar]
- Braun, B. R. & Johnson, A. D. (2000). TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle, P. L., Banerjee, M., Lazzell, A., Monteagudo, C., López-Ribot, J. L. & Kadosh, D. (2009). Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A 106, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin, W. L. (1984). Site selection for bud and germ tube emergence in Candida albicans. J Gen Microbiol 130, 431–440. [Google Scholar]
- Chant, J. & Pringle, J. R. (1995). Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol 129, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, G., Wozniak, K., Wallig, M. A., Fidel, P. L., Jr, Trupin, S. R. & Hoyer, L. L. (2005). Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect Immun 73, 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, D. A., Oh, S.-H., Zhao, X., Zhao, H., Hutchins, J. T., Vernachio, J. H., Patti, J. M. & Hoyer, L. L. (2009). Monoclonal antibodies specific for Candida albicans Als3 that immunolabel fungal cells in vitro and in vivo and block adhesion to host surfaces. J Microbiol Methods 78, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D. J., Adams, A. E. M., Fares, H., De Virgilio, C., Valle, G., Chuang, J. S. & Pringle, J. R. (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi, W. A. & Irwin, M. Y. (1993). Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Ibrahim, A. S., Sheppard, D. C., Chen, Y.-C., French, S. W., Cutler, J. E., Filler, S. G. & Edwards, J. E., Jr (2002). Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol 44, 61–72. [DOI] [PubMed] [Google Scholar]
- Gillum, A. M., Tsay, E. Y. & Kirsch, D. R. (1984). Isolation of the Candida albicans genes for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Goyard, S., Knechtle, P., Chauvel, M., Mallet, A., Prévost, M.-C., Proux, C., Coppée, J.-Y., Schwartz, P., Dromer, F. & other authors (2008). The Yak1 kinase is involved in the initiation and maintenance of hyphal growth in Candida albicans. Mol Biol Cell 19, 2251–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, C. B., Cheng, G., Chandra, J., Mukherjee, P., Ghannoum, M. A. & Hoyer, L. L. (2004). RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 150, 267–275. [DOI] [PubMed] [Google Scholar]
- Green, C. B., Zhao, X. & Hoyer, L. L. (2005a). Use of green fluorescent protein and reverse transcription-PCR to monitor Candida albicans agglutinin-like sequence gene expression in a murine model of disseminated candidiasis. Infect Immun 73, 1852–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, C. B., Zhao, X., Yeater, K. M. & Hoyer, L. L. (2005b). Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 151, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Green, C. B., Manfra Marretta, S., Cheng, G., Faddoul, F. F., Ehrhart, E. J. & Hoyer, L. L. (2006). RT-PCR analysis of Candida albicans ALS gene expression in a hyposalivatory rat model of oral candidiasis and in HIV-positive human patients. Med Mycol 44, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, A. B., López, M. C., Fernández-Lago, L. & Domínguez, A. (1999). Candida albicans and Yarrowia lipolytica as alternative models for analysing budding patterns and germ tube formation in dimorphic fungi. Microbiology 145, 2727–2737. [DOI] [PubMed] [Google Scholar]
- Hoyer, L. L. (2001). The ALS gene family of Candida albicans. Trends Microbiol 9, 176–180. [DOI] [PubMed] [Google Scholar]
- Hoyer, L. L., Scherer, S., Shatzman, A. R. & Livi, G. P. (1995). Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol Microbiol 15, 39–54. [DOI] [PubMed] [Google Scholar]
- Hoyer, L. L., Green, C. B., Oh, S.-H. & Zhao, X. (2008). Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family – a sticky pursuit. Med Mycol 46, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., Newport, G., Thorstenson, Y. R., Agabian, N. & other authors (2004). The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A 101, 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. L., Buckley, H. R. & Campbell, C. C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13, 148–153. [DOI] [PubMed] [Google Scholar]
- Lenardon, M. D., Whitton, R. K., Munro, C. A., Marshall, D. & Gow, N. A. R. (2007). Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol Microbiol 66, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, A. I., Castillo, L., Garcerá, A., Elorza, M. V., Valentín, E. & Sentandreu, R. (2004). Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology 150, 3151–3161. [DOI] [PubMed] [Google Scholar]
- Mitchell, L. H. & Soll, D. R. (1979). Commitment to germ tube or bud formation during release from stationary phase in Candida albicans. Exp Cell Res 120, 167–179. [DOI] [PubMed] [Google Scholar]
- Murad, A. M. A., Lee, P. R., Broadbent, I. D., Barelle, C. J. & Brown, A. J. P. (2000). CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16, 325–327. [DOI] [PubMed] [Google Scholar]
- Nobile, C. J. & Mitchell, A. P. (2005). Regulation of cell-surface genes and biofilm formation by the Candida albicans transcription factor Bcr1p. Curr Biol 15, 1150–1155. [DOI] [PubMed] [Google Scholar]
- Porta, A., Ramon, A. M. & Fonzi, W. A. (1999). PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol 181, 7516–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico, H., Herrero, E., Miragall, F. & Sentandreu, R. (1991). An electron microscopy study of wall expansion during Candida albicans yeast and mycelial growth using concanavalin A-ferritin labelling of mannoproteins. Arch Microbiol 156, 111–114. [DOI] [PubMed] [Google Scholar]
- Shannon, J. L. & Rothman, A. H. (1971). Transverse septum formation in budding cells of the yeastlike fungus Candida albicans. J Bacteriol 106, 1026–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery, P., Gow, N. & Berman, J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol 12, 317–324. [DOI] [PubMed] [Google Scholar]
- Sumita, T., Yoko-o, T., Shimma, Y. & Jigami, Y. (2005). Comparison of cell wall localization among Pir family proteins and functional dissection of the region required for cell wall binding and bud scar recruitment of Pir1p. Eukaryot Cell 4, 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, A. & Wendland, J. (2003). Septation and cytokinesis in fungi. Fungal Genet Biol 40, 187–196. [DOI] [PubMed] [Google Scholar]
- Warenda, A. J. & Konopka, J. B. (2002). Septin function in Candida albicans morphogenesis. Mol Biol Cell 13, 2732–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, S. J., Rosenbach, A., Lephart, P., Nguyen, D., Benjamin, A., Tzipori, S., Whiteway, M., Mescas, J. & Kumamoto, C. (2007). Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog 3, e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler, U., Lettner, T., Lassnig, C., Müller, M., Lajko, R., Hintner, H., Breitenbach, M. & Bito, A. (2009). UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans. FEMS Yeast Res 9, 126–142. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Oh, S.-H., Cheng, G., Green, C. B., Nuessen, J. A., Yeater, K., Leng, R. P., Brown, A. J. P. & Hoyer, L. L. (2004). ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150, 2415–2428. [DOI] [PubMed] [Google Scholar]