Abstract
Background/Aims: Bipolar disorder (BP) is a severe psychiatric illness, characterised by alternating episodes of depression and mania, which ranks among the top ten causes of morbidity and life-long disability world-wide. We have previously performed a whole-genome linkage scan on 6 pedigrees segregating severe BP from the well-characterised population isolate of Antioquia, Colombia. We recently collected genotypes for the same set of 382 autosomal microsatellite markers in 9 additional Antioquian BP pedigrees. Here, we report the analysis of the combined pedigree set.
Methods: Linkage analysis using both parametric and nonparametric approaches was conducted for 3 different diagnostic models: severe BP only (BPI); mood disorders (BPI, BPII and major depression); and psychosis (operationally defined by the occurrence of at least 1 episode of hallucinations and/or delusions). Results and Conclusion: For BPI only, the most interesting result was obtained for chromosome 7p21.1–p22.2 under a recessive model of inheritance (heterogeneity LOD score = 2.80), a region that had previously been linked to BP in a study on Portuguese Island families. For both BPI and mood disorders, nonparametric analyses identified a locus on chromosome 12ct–q14 (nonparametric linkage = 2.55 and 2.35, respectively). This locus has not previously been reported as a candidate region for BP. Additional candidate regions were found on chromosomes 1p22–31 (mood disorders) and 21q21–22 (BPI), 2 loci that have repeatedly been implicated in BP susceptibility. Linkage analysis of psychosis as a phenotype identified candidate regions on chromosomes 2q24–31 and 16p12–q12. The finding on chromosome 16p is noteworthy because the same locus has been implicated by genome-wide association analyses of BP.
Key Words: Bipolar disorder, Psychiatric genetics, Whole-genome linkage analysis
Introduction
Bipolar disorder (BP) is a severe and complex psychiatric condition characterised by alternating episodes of depression and mania [1]. Based on clinical severity, 2 variants of this disorder are commonly distinguished: BP type I (BPI), including at least 1 episode of full-blown mania, typically requiring hospitalization, and BP type II (BPII), characterised by hypomanic episodes, representing a milder form of mania [1]. With a life-time prevalence of 1–1.5% across populations and genders [2], BP is among the top ten causes of morbidity and life-long disability in both the developing and the developed world [3]. Family, twin and adoption studies provide strong evidence for a genetic contribution to the aetiology of this disorder [4,5]; however, progress in the identification of disease-causing variants has been slow. Family-based linkage studies have produced inconsistent results, and the first genome-wide association scans (GWAS) in BP have failed to live up to the expectations generated by the GWAS findings in other complex diseases [6,7]. These disappointing results are most likely related to the genetic heterogeneity and phenotypic variability resulting from the descriptive nature of psychiatric diagnoses. The study of a rigorously phenotyped pedigree collection from a population isolate should help reduce this phenotypic and genetic heterogeneity. We have previously implemented this approach by carrying out a whole-genome linkage scan in 6 extended pedigrees segregating severe BP from a well-characterised population isolate (Antioquia in Colombia) [8,9], and found genome-wide significant evidence of linkage on chromosome 5q31–34 [10]. This signal has subsequently been confirmed through fine-mapping of pedigrees and trios from both Antioquia and the related isolate of the Central Valley of Costa Rica [10,11].
Since reporting the results of this initial linkage scan, we have collected genome-wide linkage data for 9 additional Antioquian BP families. Here, we report the results of the joint analysis of our original linkage data [10] and the data for these additional pedigrees. In these extended analyses we also explore additional diagnostic and genetic models.
Subjects and Methods
Subjects
Fifteen extended pedigrees segregating severe BP were collected as part of an ongoing psychiatric genetics programme in the population isolate of Antioquia, Colombia (fig. 1). Six of these had been included in a previous linkage scan (CO3, CO4, CO7, CO14, CO18, CO27; fig. 1b) [10]. Index cases were recruited in the municipalities of Medellín and Envigado (Antioquia, Colombia) at the Hospital Mental de Antioquia, the Hospital San Vicente de Paúl, the Clínica Sameín, the Clínica Insam, and the Mental Health Centre of Envigado. All index cases had at least 6 great-grandparents born in Antioquia. Families with at least 3 individuals with a clinical diagnosis of BP were chosen for pedigree extension by a social worker or psychiatric nurse using the Family Interview for Genetic Studies [12]. Family CO24 includes only 2 cases of BP; it was studied because it also includes 1 case of major depression and 1 case of schizophrenia, which are phenotypes also considered in the analyses (see below).
Fig. 1.
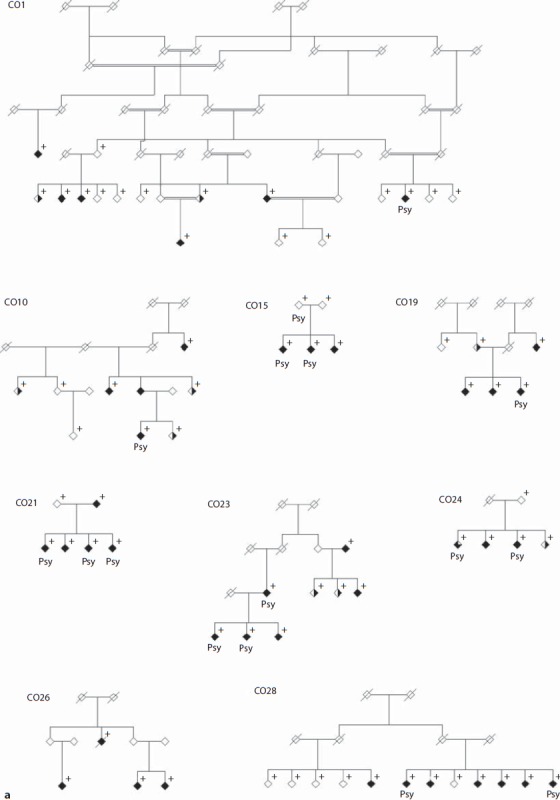
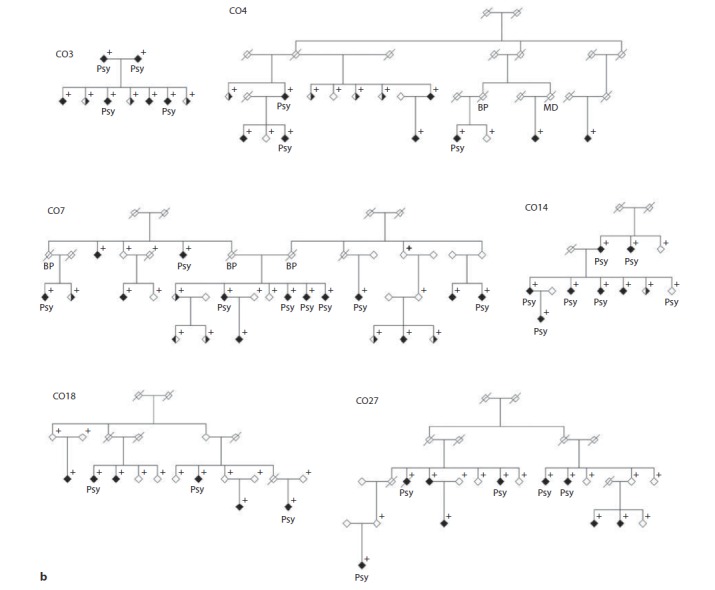
a Set of 9 Antioquian pedigrees segregating severe BP genotyped for this study. Individuals marked with a cross were available for genotyping. Both sets of pedigrees (shown here and in fig. 1b) were analysed together for this study. Filled symbols indicate BPI, half-filled symbols indicate major depression (right half filled) and BPII (left half filled). Filled symbols with a blank upper right quarter indicate schizophrenia. Individuals labelled ‘Psy’ have suffered episodes of psychosis. b Set of 6 Antioquian pedigrees genotyped in an initial linkage scan in the Antioquian population [10]. Both sets of pedigrees (shown here and in fig. 1a) were analysed together for this study. Individuals marked with a cross were available for genotyping. Filled symbols indicate BPI, half-filled symbols indicate major depression (right half filled) and BPII (left half filled). Filled symbols with a blank upper right quarter indicate schizophrenia. Individuals labelled ‘Psy’ have suffered episodes of psychosis.
Family members were assessed by a psychiatrist using the Spanish version of the Diagnostic Interview for Genetic Studies (DIGS version 3) that we validated in Colombia [12,13]. Final DSM-IV-TR diagnoses [14] were reached through consensus between 2 expert psychiatrists in a best estimate procedure, as described by Freimer and colleagues [15]. The occurrence of psychotic episodes was determined based on both the DIGS and medical records. Family members with a clinical diagnosis of BPI who were unavailable for interview (including deceased individuals) were considered affected if they had undergone at least 2 hospitalizations and clinical records allowed confirmation of symptoms through the best estimate process. No psychiatric diagnoses other than BPI were considered in family members that were unavailable for interview. In order to maintain a strict phenotypic definition, and to rule out cases where psychiatric symptoms might be secondary to other medical conditions, any psychiatric diagnoses in patients with mental retardation and/or neurological lesions, as well as cases related to substance abuse, were disregarded for the analyses. Altogether, the sample comprised 90 cases of BPI (37.8% males, 62.2% females; age at onset 22.2 ± 7.6 years), 22 cases of major depression (18.2% males, 82.8% females; age at onset 26.6 ± 13.8 years), and 1 case each of schizophrenia (female; age at onset 27 years) and BPII (male; age at onset could not be determined reliably). A total of 46 individuals (30.4% males, 69.6% females) had suffered from psychosis, operationally defined by the occurrence of at least 1 episode of hallucinations and/or delusions. These included 43 of the 90 BPI cases (47.8%), the schizophrenic individual and 2 individuals with a psychotic mood disorder that did not meet the full diagnostic criteria for BP (these 2 individuals are included in families CO14 and CO15; see fig. 1). Written informed consent was obtained from all subjects prior to enrolment in the study. This research was approved by the Ethics Committees of all participating institutions.
Genotyping
DNA was extracted from whole blood using standard laboratory procedures. Genotype data for 9 newly ascertained pedigrees from Antioquia were obtained for 382 evenly spaced autosomal microsatellite markers from the ABI Linkage Mapping Set v2.5 (Applied Biosystems, Warrington, UK; average distance ∼10 cM). Fragment length analysis was performed on a 96-capillary 3730xl DNA Analyzer (Applied Biosystems), and raw genotyping data were analysed using the GeneMapper® software v3.7 (Applied Biosystems). Genotypes were scored independently by 2 researchers blinded to affection status and relatedness between subjects (‘double scoring’). Any discordant genotypes were disregarded and, if necessary, repeated. Genotype data were checked for inconsistencies with mendelian inheritance using PedCheck [16], and the occurrence of non-mendelian errors was tested with SimWalk2 v2.9.1 [17,18]. The quality-checked genotype data for the 9 new families were combined with those from our previous study of 6 extended BPI families [10] by calibrating all genotypes using a Ceph sample of known genotype (Ceph 1347-02). All analyses were carried out on the combined data set. Because the patterns of disease transmission did not support an X-chromosomal mode of inheritance in the Antioquian pedigrees, the X-chromosome was not analysed. The present study therefore represents an autosome-wide linkage scan.
Data Analysis and Statistical Methods
We focused our ascertainment strategy on families segregating BPI. However, several other psychiatric diagnoses segregate in these pedigrees, most notably major depression. We therefore analysed our data under 3 different diagnostic models. In a conservative model, only cases of BPI were considered affected (‘narrow model’). In the second model, cases of BPI, BPII and major depression were considered affected (‘broad model’). In a third model, the affection status was defined by the occurrence of psychosis (‘psychosis model’). Because not all families included 2 or more individuals affected by psychosis, the data set for this last model comprised only 11 pedigrees. For analyses under all 3 models, all remaining family members were considered to be of unknown phenotype.
Multipoint parametric heterogeneity LOD (HLOD) score and nonparametric linkage (NPL) analyses were performed using SimWalk2 v2.9.1 [17]. Due to the uncertainties involved in specifying an inheritance model underlying the aetiologically complex phenotypes analysed here, NPL analysis was conducted for all 3 phenotypic models. For the narrow (BPI) model only, we also carried out parametric analyses in order to enable a direct comparison of the results from the present study to those from our previous study, which involved part of the families included here [10]. Parametric linkage analysis was performed under both a dominant and a recessive model. Analysis under the dominant model was carried out employing the same parameters used in our previous linkage scan; these parameters were estimated in previous studies of families from the Central Valley of Costa Rica [10,19]. The frequency of the disease allele was set at 0.003, the phenocopy rate at 0.01, and the penetrances for heterozygous and homozygous carriers of the disease allele were set to 0.81 and 0.9, respectively. Under the recessive model, the frequency of the disease allele was set to 0.1, the phenocopy rate to 0.01, and the penetrance for homozygous carriers of the disease allele was set to 0.80. These parameter values have been used in previous studies (e.g. [20]). In order to reduce the number of tests carried out and to facilitate comparison with our previous analysis of a subset of these families, the model-based linkage analysis was performed only for the narrow phenotypic model. Marker allele frequencies for all analyses were estimated from the complete pedigree data set using Mendel v8.0.1, correcting for relatedness between individuals [21]. Three-digit genotypes were recoded to a 2-digit format, and data files converted to different formats, as required, using Mega2 v4.0 [22]. All analyses used the Marshfield genetic map (http://research.marshfieldclinic.org/genetics/home/index.asp).
Results
Genotype data for 382 autosomal microsatellite markers were obtained for 75 samples from 9 pedigrees. Genotype data for a further 91 samples from 6 pedigrees from our previously published linkage study [10] were available for the same set of markers. For 10 markers, the genotypes from both data sets could not be calibrated unambiguously; these were excluded from further analyses (D1S498, D1S2836, D5S418, D6S262, D11S905, D13S159, D15S994, D17S798, D19S221, and D21S263). The overall data completeness was 94% across chromosomes and samples.
The genome-wide results of the parametric and nonparametric analyses are shown in figures 2 and 3, respectively. The most noticeable results have been extracted into table 1 (results for individual families are summarised in table 2).
Fig. 2.
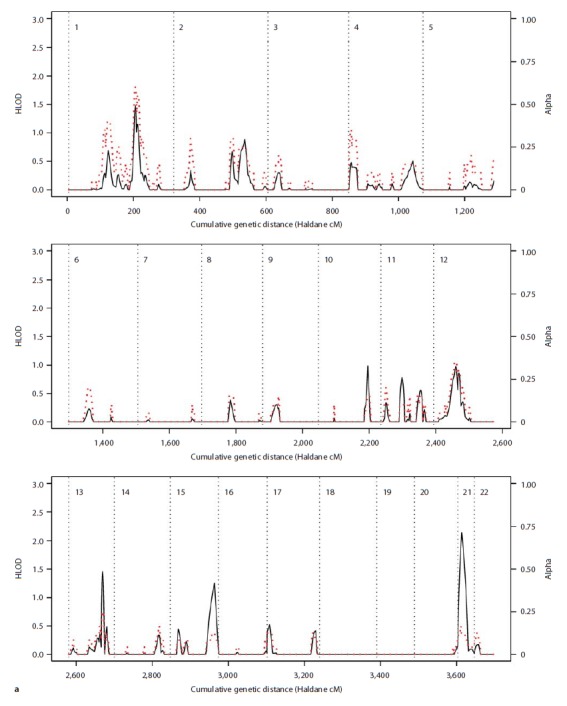
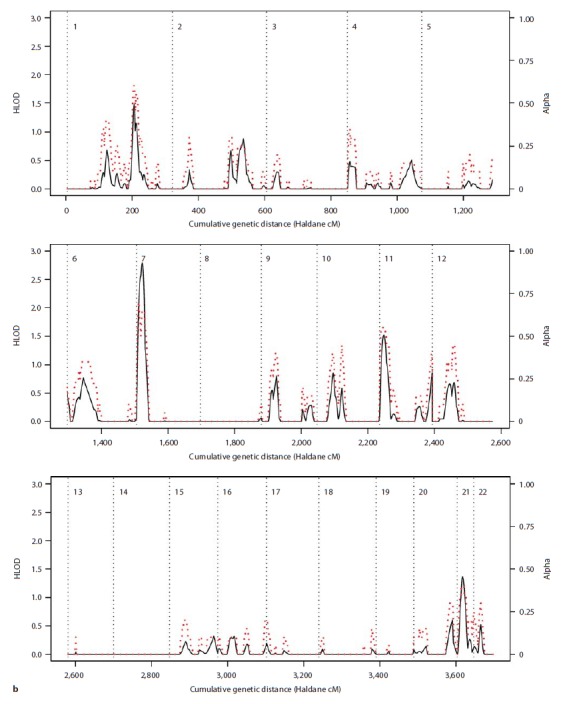
Results of the parametric linkage analysis under the narrow diagnostic model and near-dominant (a) and recessive (b) inheritance models. Multipoint HLOD scores (solid black line) and the estimated proportion of linked families (α; dotted red line) are plotted against the cumulative genetic distance in Haldane cM. Chromosomes are separated by dotted black lines.
Fig. 3.
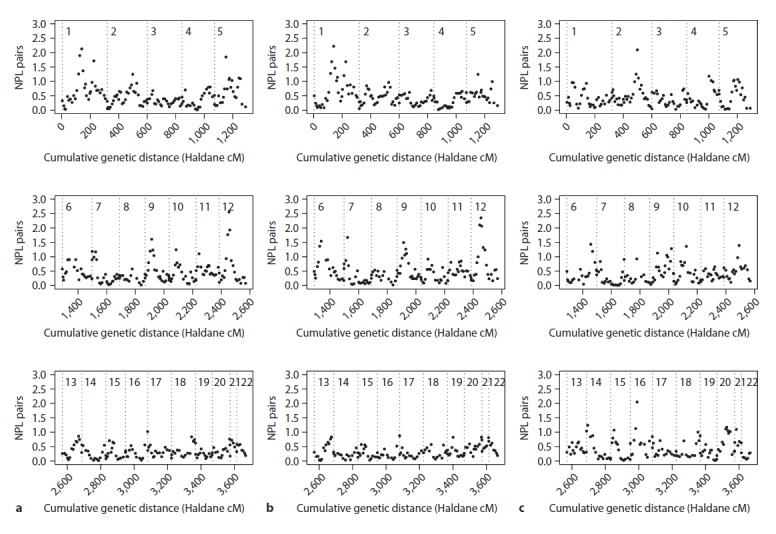
Results of the NPL analysis under narrow (a), broad (b) and psychosis models (c). NPL scores are plotted against the cumulative genetic distance in Haldane cM. Chromosomes are separated by dotted lines.
Table 1.
Chromosomal regions suggestive of linkage (LOD or NPL ≥1.3) in the collection of 15 Antioquian BP pedigrees
| Chromosome | Diagnostic model | Inheritance model | Maximum HLOD | Proportion of linked families, α | Marker interval | Chromosomal region | Position Mb |
|---|---|---|---|---|---|---|---|
| Parametric analysis | |||||||
| 1 | narrow | dominant | 1.32 | 0.35 | D1S2841–D1S2868 | 1p31.1–p22.1 | 79.49–93.35 |
| 1 | narrow | recessive | 1.47 | 0.6 | D1S196–D1S218 | Iq24.2–q25.1 | 167.60–174.50 |
| 7 | narrow | recessive | 2.80 | 0.65 | D7S517–D7S507 | 7p22.2–p21.1 | 4.50–17.60 |
| 13 | narrow | dominant | 1.46 | 0.3 | D13S158–D13S173 | 13q33.1–q33.3 | 103.98–107.81 |
| 21 | narrow | dominant | 2.14 | 0.15 | D21S1256–D21S1252 | 21q21.1–q22.13 | 19.32–37.83 |
| Chromosome | Diagnostic model | Inheritance model | Maximum NPL | p value | Marker interval | Chromosomal region | Position Mb |
|---|---|---|---|---|---|---|---|
| Nonparametric analysis | |||||||
| 1 | narrow | model-free | 2.13 | 0.0074 | D1S207-D1S206 | 1p31.1-p21.2 | 82.54–101.69 |
| 1 | broad | model-free | 2.22 | 0.0060 | D1S207-D1S206 | 1p31.1-p21.2 | 82.54–101.69 |
| 1 | narrow | model-free | 1.71 | 0.0194 | D1S238 | 1q31.1 | 188.15 |
| 1 | broad | model-free | 1.68 | 0.0207 | D1S238 | lq31.1 | 188.15 |
| 2 | psychosis | model-free | 2.09 | 0.0081 | D2S2330 | 2q24.3 | 166.70 |
| 5 | narrow | model-free | 1.85 | 0.0142 | D5S647 | 5q12.3 | 66.25 |
| 6 | broad | model-free | 1.54 | 0.0286 | D6S276-D6S1610 | 6p22.2-p21.2 | 24.19–39.26 |
| 6 | psychosis | model-free | 1.44 | 0.0367 | D6S441 | 6q25.2 | 153.81 |
| 7 | broad | model-free | 1.67 | 0.0212 | D7S507 | 7p21.1 | 17.60 |
| 9 | narrow | model-free | 1.61 | 0.0246 | D9S161 | 9p21.2 | 27.63 |
| 9 | broad | model-free | 1.50 | 0.0319 | D9S161 | 9p21.2 | 27.63 |
| 10 | psychosis | model-free | 1.36 | 0.0434 | D10S1652 | 10q21.2 | 64.41 |
| 12 | narrow | model-free | 2.55 | 0.0028 | D12S345–D12S368 | 12p11.21–q13.13 | 32.32–52.63 |
| 12 | broad | model-free | 2.35 | 0.0044 | D12S345–D12S83 | 12p11.21–q14.1 | 32.32–60.89 |
| 12 | psychosis | model-free | 1.39 | 0.0403 | D12S351 | 12q21.33 | 91.90 |
| 16 | psychosis | model-free | 2.05 | 0.0089 | D16S3068 | 16p12.1 | 25.56 |
Physical marker positions are from the February 2009 assembly of the UCSC genome browser (http://genome.ucsc.edu).
Table 2.
Chromosomal regions suggestive of linkage (LOD ≥2 or NPL >2) in individual families
| Chromosome | Diagnostic model | Inheritance model | Maximum LOD | Marker interval | Chromosomal region | Position Mb | Pedigree ID |
|---|---|---|---|---|---|---|---|
| Parametric analysis | |||||||
| 1 | narrow | dominant | 2.06 | D1S2890–D1S230 | 1p32.2–p31.3 | 57.87–62.60 | C027 |
| 3 | narrow | dominant | 2.57 | D3S1565–D3S1601 | 3q26.31–q28 | 173.48–191.68 | C07 |
| 11 | narrow | dominant | 2.19 | D11S905–D11S987 | 11p12–q13.2 | 40.97–67.89 | CO1 |
| 15 | narrow | dominant | 2.46 | D15S127–D15S120 | 15q26.1–15tel | 91.40–99.59 | C018 |
| 21 | narrow | dominant | 3.21 | D21S1256–D21S1252 | 21q21.1–q22.13 | 19.32–37.83 | C07 |
| Chromosome | Diagnostic Model | Inheritance model | Maximum NPL (p value) | Marker interval | Chromosomal region | Position Mb | Pedigree ID |
|---|---|---|---|---|---|---|---|
| Nonparametric analysis | |||||||
| 1 | broad | model-free | 2.00 (0.0101) | D1S207 | lp31.1 | 82.54 | C027 |
| 2 | broad | model-free | 2.23 (0.0059) | D2S117–D2S325 | 2q32.3–q33.3 | 195.62–208.27 | C023 |
| 3 | narrow | model-free | 2.32 (0.0048) | D3S1580 | 3q28 | 188.54 | C07 |
| 4 | psychosis | model-free | 2.30 (0.0051) | D4S424 | 4q31.21 | 142.20 | C014 |
| 16 | narrow | model-free | 2.49 (0.0032) | D16S3046–D16S3068 | 16pl2.2–12.1 | 20.89–25.56 | C07 |
| 16 | psychosis | model-free | 2.40 (0.0040) | D16S3046 | 16pl2.2 | 20.89 | C07 |
| 16 | broad | model-free | 2.12 (0.0075) | D16S3068 | 16pl2.1 | 25.56 | C07 |
| 21 | narrow | model-free | 2.32 (0.0048) | D21S1914 | 21q21.2 | 25.62 | C014 |
Physical marker positions are from the February 2009 assembly of the UCSC genome browser (http://genome.ucsc.edu).
The most interesting study-wide signals were found on chromosome 7p21.1–p22.2, with a HLOD score of 2.80 for the recessive analysis under the narrow diagnostic model, and at marker D12S85 on chromosome 12q13.11, where NPL scores of 2.55 and 2.35 were observed in nonparametric analysis under narrow and broad diagnostic models, respectively. A HLOD score of 2.14 was observed on chromosome 21q at marker D21S1914 in the dominant parametric analysis under the narrow diagnostic model. For this marker, a parametric LOD score of 3.21 was observed in family CO7, and a NPL score of 2.32 in family CO14 (seetable 2). For the dominant analysis, HLOD scores >1.3 were also observed on chromosomes 13q and 1p.
Nonparametric analysis under the narrow diagnostic model identified additional candidate regions on chromosomes 1p31.1–p21.2, 1q31.1, 5q12.3, and 9p21.2. Chromosome 1p was also highlighted by the parametric analysis (see above), and by the nonparametric analysis under the broad diagnostic model; results from all 3 analyses defined a common candidate region on chromosome 1p31.1–p21.2. Candidate regions on chromosomes 1q31.1 and 9p21.2 were also supported by the nonparametric analysis under the broad diagnostic model.
For the psychosis phenotype, the highest NPL scores were obtained for chromosomes 2q24.3 and 16p12.1 (2.09 and 2.05, respectively); NPL scores ≥1.3 were also identified on chromosomes 6q, 10q, and 12q. The signal on chromosome 12q was 34cM telomeric of the most significant signal obtained under narrow and broad models.
Discussion
We performed a whole-genome linkage scan in 15 extended pedigrees segregating severe BP from the population isolate of Antioquia, Colombia. While we identified no signals at a genome-wide level of significance, we found suggestive evidence for a range of candidate regions for BP, mood disorders in general, and psychosis. A lack of significant results at a genome-wide level has characterised linkage studies of BP for the past decades, reflecting the challenges implicated in the mapping of susceptibility variants for complex psychiatric disease, and making the replication of results all the more important. Interestingly, and encouragingly, many of our results replicate findings from previous studies, as discussed in detail below.
BP is a complex and heterogeneous condition, whose mode of inheritance does not easily conform to standard linkage analysis. The use of nonparametric methods provides a way of overcoming these difficulties, while the use of different diagnostic models can help explore the possible genetic overlap between BP, mood disorders and schizophrenia. Consequently, we performed NPL analysis under 3 different diagnostic models: BPI only; BPI, BPII and major unipolar depression; and psychosis (i.e. ‘narrow’, ‘broad’, and ‘psychosis models’, respectively). Additionally, parametric linkage analysis was used for the narrow diagnosis, using both a near-dominant and a recessive inheritance model. A similar combination of diagnostic definitions and genetic models has been previously used in a number of linkage studies of BP (e.g. [20]).
There was a substantial overlap in the results obtained with the narrow and broad diagnostic models, as both analyses highlighted loci on chromosomes 1p13–31, 1q25–31 and 12p11–q14. An overlap of linkage signals under these 2 diagnostic models has been previously reported [23,24,25,26,27,28]. These observations suggest the existence of loci predisposing to mood disorders in general rather than to BP in particular, consistent with the observation of an increased rate of major depression in relatives of BP patients [29].
The narrow diagnostic model (BPI) also produced some specific linkage signals on chromosomes 7p21–22, 5q12, 13q33 and 21q21–22, although results differed somewhat between parametric and nonparametric analyses (see table 1). The loci on chromosomes 7p21–22, 13q33 and 21q21–22 were highlighted by the parametric analysis, possibly implicating that segregation patterns at these loci were broadly consistent with the model used for analysis. Segregation on chromosome 7p21–22 may hence best fit the recessive model, while segregation of loci on chromosomes 13q33 and 21q21–22 may best fit the near-dominant mode of inheritance. It is possible that these regions were not consistently picked up by the nonparametric analysis because of its reduced power (although NPL analysis did produce a p value of 0.0212 for the chromosome 7 region under the broad phenotypic model).
Results under the psychosis phenotype implicated different regions from those suggested by the 2 other diagnostic models (chromosomes 2q24–31, 10q21 and 16p12). There are very few published analyses of the psychosis phenotype in bipolar pedigrees; available results are consistent with our findings in that they implicate regions different from those linked to BP as such [30,31], suggesting that there are genetic loci conferring susceptibility to psychosis independent of affective disorders per se. The concept of a genetic predisposition to psychosis is in line with evidence for a shared genetic susceptibility between BP and schizophrenia [32,33].
Several of the loci identified here have been reported in previous studies of BP. Both the 13q33 and 21q21–22 regions have repeatedly been implicated in susceptibility to BP [23,28,34,35,36,37]. The region on 21q21 was one of the best-supported loci in our previous linkage scan [10], and the present study provides further support for this region. Interestingly, most of the studies showing evidence for a locus on chromosome 21q also used a narrow diagnostic model similar to the one used here [23,28,35]. Lin and colleagues [38] also reported linkage of chromosome 21q22 for early-onset BP, a subtype of the disorder associated with increased severity of clinical symptoms [39]. Interestingly, the 13q33 region harbours the DAOA gene which has been found to be associated with both BP and schizophrenia [40,41,42,43,44].
The loci on 1p13–31 and 1q25–31 implicated here have also both been reported to be involved in BP susceptibility [23,45,46,47,48,49,50]. The chromosome 1p13–31 region had been highlighted by our initial genome scan [10], and the present study has further strengthened this signal. The locus on the long arm of chromosome 1 has frequently been linked to schizophrenia [33,51,52,53,54]. This region includes the genes encoding the regulator of G signalling protein, RGS4, and the nitric oxide synthase 1 (neuronal) adaptor protein, NOS1AP,which have both been associated with BP or schizophrenia [55,56,57,58,59]. Interestingly, NOS1AP expression has been found to be increased in the prefrontal cortex of bipolar subjects [60].
The strongest support for linkage in our study was observed for regions on chromosome 7p21.1–p22.2 (under the narrow affection model and assuming recessive inheritance), and 12q13 (under both narrow and broad affection models).
Evidence of linkage of BP to the chromosome 7p21 region has been reported in a set of 16 extended Portuguese Island pedigrees [61]. The replication of this finding in the Colombian families is of interest because both populations share ancestral contributions from the Iberian Peninsula.
The chromosome 12q13 region has been implicated in a study of French Canadian BP pedigrees. Like the present study, the signal from the Canadian pedigrees was obtained using a broad diagnostic model including BPI, BPII, schizoaffective disorder and recurrent unipolar depression [34]. However, the strongest support for linkage in the Canadian families appears to be in a region telomeric to the locus described here. An interesting candidate gene in the region identified here is Timeless. This gene plays a crucial role in the regulation of circadian rhythms [62], the dysfunction of which is a widespread phenomenon in BP [63].
Amongst the regions implicated under the psychosis model, both chromosomes 2q24 and 16p12 have previously been linked to psychiatric disease. The 2q24 region has been implicated in both BP and schizophrenia [53,64,65], and the chromosome 16p12 region has been implicated in severe psychotic BP [31]. Interestingly, the most significant signal in the WTCCC BP genome-wide association study was for a SNP in the 16p12 region [6]. This region harbours several interesting candidate genes, including DCTN5, whose product appears to interact with that of DISC1 (Disrupted in Schizophrenia 1), a gene repeatedly linked to schizophrenia [66,67].
In this expanded pedigree sample we failed to find further linkage support for the 5q31–34 region implicated by a subset of the families analysed previously [10]. A possible explanation is interfamily heterogeneity. In particular the two largest families examined here (CO1 and CO28) stem from an isolated village, which could represent a subisolate within Antioquia. Neither of these two families provides support for a locus on chromosome 5q31–34, and due to their size, these two pedigrees have a strong impact on the total LOD-scores calculated. For a complex disorder like BP, genetic heterogeneity is not unlikely even within a population isolate.
Such difficulties highlight the need to readjust how results from linkage studies should be interpreted. Most individual studies might not have sufficient power to detect susceptibility loci for psychiatric disease on their own, and replication studies and meta-analyses are essential to evaluate the true significance of linkage findings. In this respect, it is interesting that regions highlighted by meta-analyses of genome-wide linkage studies of BP often do not coincide with the most significant signals of the individual studies included [47,68,69]. Results from an individual study might therefore represent relatively larger genetic effects of importance only for a specific family or population. On the other hand, individually weaker signals could show greater consistency across studies, hence only reaching statistical significance through meta-analyses. Family studies have contributed, and continue to contribute, important information to the emerging picture of genetic predisposition to BP. The present study, which has provided further supporting evidence for a range of previously reported loci, but in which we have also identified a possible novel locus on chromosome 12q13, adds to this picture.
Acknowledgements
This work was partly funded by Wellcome Trust grant 086052 and a NARSAD young investigator award to A.R.-L, and by NIH grant 5R01MH075007 to N.B.F. B.K. would like to thank the Annals of Human Genetics under the editorial leadership of Professor Sue Povey for financial support. We would also like to thank Émilie Boucher and Julie Pivard for technical assistance, and two anonymous reviewers for their comments. We are grateful to all family members for their participation in this study.
References
- 1.Muller-Oerlinghausen B, Berghofer A, Bauer M. Bipolar disorder. Lancet. 2002;359:241–247. doi: 10.1016/S0140-6736(02)07450-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsuang MT, Faraone SV. The Genetics of Mood Disorders. Baltimore: Johns Hopkins University Press; 1990. [Google Scholar]
- 3.Murray CJL, Lopez AD. The Global Burden of Disease: a Comprehensive Assessment of Mortality and Disability from Diseases, Injuries, and Risk Factors in 1990 and Projected to 2020. Cambridge, MA: The Harvard School of Public Health; 1996. [Google Scholar]
- 4.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 5.Jones I, Kent L, Craddock N, McGuffin P, Owen MJ, Gottesman II. Genetics of affective disorders. In: McGuffin P, Owen MJ, Gottesman II, editors. Psychiatric Genetics and Genomics. Oxford, New York: Oxford University Press; 2004. pp. 211–245. [Google Scholar]
- 6.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, DeFelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, Macintyre DJ, McLean A, Vanbeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM. Whole-genome association study of bipolar disorder. Mol Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvajal-Carmona LG, Ophoff R, Service S, Hartiala J, Molina J, Leon P, Ospina J, Bedoya G, Freimer N, Ruiz-Linares A. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet. 2003;112:534–541. doi: 10.1007/s00439-002-0899-8. [DOI] [PubMed] [Google Scholar]
- 9.Bedoya G, Montoya P, Garcia J, Soto I, Bourgeois S, Carvajal L, Labuda D, Alvarez V, Ospina J, Hedrick PW, Ruiz-Linares A. Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci USA. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzberg I, Jasinska A, Garcia J, Jawaheer D, Service S, Kremeyer B, Duque C, Parra V, Vega J, Ortiz D, Carvajal L, Polanco G, Restrepo GJ, Lopez C, Palacio C, Levinson M, Aldana I, Mathews C, Davanzo P, Molina J, Fournier E, Bejarano J, Ramirez M, Araya Ortiz C, Araya X, Sabatti C, Reus V, Macaya G, Bedoya G, Ospina J, Freimer N, Ruiz-Linares A. Convergent linkage evidence from two Latin American population isolates supports the presence of a susceptibility locus for bipolar disorder in 5q31–34. Hum Mol Genet. 2006;15:3146–3153. doi: 10.1093/hmg/ddl254. [DOI] [PubMed] [Google Scholar]
- 11.Jasinska A, Service S, Jawahar S, Deyoung J, Levinson M, Kremeyer B, Muller H, Polanco G, Carvajal L, Aldana I, Garcia J, Restrepo G, Lopez C, Palacio C, Ospina J, Duque C, Parra M, Vega J, Ortiz D, Bedoya G, Mathews C, Davanzo P, Fournier E, Bejarano J, Ramirez M, Araya Ortiz C, Araya X, Sabatti C, Reus V, Macaya G, Ruiz-Linares A, Freimer N. A narrow and highly significant linkage signal for severe bipolar disorder in the chromosome 5q33 region in Latin American pedigrees. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:998–1006. doi: 10.1002/ajmg.b.30956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- 13.Palacio CA, García J, Arbeláez MP, Sánchez R, Aguirre B, Garcés I, Montoya GJ, Gómez J, Agudelo A, López CA, Calle JJ, Cardeño CA, Cano JF, López MC, Montoya P, Herrera CP, González N, González A, Bedoya G, Ruiz A, Ospina J. Validación de la entrevista diagnóstica para estudios genéticos (DIGS) en Colombia. Biomedica. 2004;24:56–62. [PubMed] [Google Scholar]
- 14.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 15.Freimer NB, Reus VI, Escamilla M, Spesny M, Smith L, Service S, Gallegos A, Meza L, Batki S, Vinogradov S, Leon P, Sandkuijl LA. An approach to investigating linkage for bipolar disorder using large Costa Rican pedigrees. Am J Med Genet. 1996;67:254–263. doi: 10.1002/(SICI)1096-8628(19960531)67:3<254::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 18.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S, Rojas E, Spesny M, Baharloo S, Blankenship K, Peterson A, Tyler D, Shimayoshi N, Tobey C, Batki S, Vinogradov S, Meza L, Gallegos A, Fournier E, Smith LB, Barondes SH, Sandkuijl LA, Freimer NB. A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci USA. 1996;93:13060–13065. doi: 10.1073/pnas.93.23.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Juo SH, Dewan A, Grunn A, Tong X, Brito M, Park N, Loth JE, Kanyas K, Lerer B, Endicott J, Penchaszadeh G, Knowles JA, Ott J, Gilliam TC, Baron M. Evidence for a putative bipolar disorder locus on 2p13–16 and other potential loci on 4q31, 7q34, 8q13, 9q31, 10q21–24, 13q32, 14q21 and 17q11–12. Mol Psychiatry. 2003;8:333–342. doi: 10.1038/sj.mp.4001254. [DOI] [PubMed] [Google Scholar]
- 21.Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69S:A1886. [Google Scholar]
- 22.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 23.Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI, Jr, Gershon ES. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA. 1999;96:5604–5609. doi: 10.1073/pnas.96.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekholm JM, Kieseppa T, Hiekkalinna T, Partonen T, Paunio T, Perola M, Ekelund J, Lonnqvist J, Pekkarinen-Ijas P, Peltonen L. Evidence of susceptibility loci on 4q32 and 16p12 for bipolar disorder. Hum Mol Genet. 2003;12:1907–1915. doi: 10.1093/hmg/ddg199. [DOI] [PubMed] [Google Scholar]
- 25.McInnis MG, Dick DM, Willour VL, Avramopoulos D, MacKinnon DF, Simpson SG, Potash JB, Edenberg HJ, Bowman ES, McMahon FJ, Smiley C, Chellis JL, Huo Y, Diggs T, Meyer ET, Miller M, Matteini AT, Rau NL, DePaulo JR, Gershon ES, Badner JA, Rice JP, Goate AM, Detera-Wadleigh SD, Nurnberger JI, Reich T, Zandi PP, Foroud TM. Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health genetics initiative bipolar pedigrees. Biol Psychiatry. 2003;54:1265–1273. doi: 10.1016/j.biopsych.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Schumacher J, Kaneva R, Jamra RA, Diaz GO, Ohlraun S, Milanova V, Lee YA, Rivas F, Mayoral F, Fuerst R, Flaquer A, Windemuth C, Gay E, Sanz S, Gonzalez MJ, Gil S, Cabaleiro F, Del Rio F, Perez F, Haro J, Kostov C, Chorbov V, Nikolova-Hill A, Stoyanova V, Onchev G, Kremensky I, Strauch K, Schulze TG, Nurnberg P, Gaebel W, Klimke A, Auburger G, Wienker TF, Kalaydjieva L, Propping P, Cichon S, Jablensky A, Rietschel M, Nothen MM. Genomewide scan and fine-mapping linkage studies in four European samples with bipolar affective disorder suggest a new susceptibility locus on chromosome 1p35-p36 and provides further evidence of loci on chromosome 4q31 and 6q24. Am J Hum Genet. 2005;77:1102–1111. doi: 10.1086/498619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcheco-Teruel B, Flint TJ, Wikman FP, Torralbas M, Gonzalez L, Blanco L, Tan Q, Ewald H, Orntoft T, Kruse TA, Borglum AD, Mors O. A genome-wide linkage search for bipolar disorder susceptibility loci in a large and complex pedigree from the eastern part of Cuba. Am J Med Genet B Neuropsychiatr Genet. 2006;141:833–843. doi: 10.1002/ajmg.b.30314. [DOI] [PubMed] [Google Scholar]
- 28.Cassidy F, Zhao C, Badger J, Claffey E, Dobrin S, Roche S, McKeon P. Genome-wide scan of bipolar disorder and investigation of population stratification effects on linkage: support for susceptibility loci at 4q21, 7q36, 9p21, 12q24, 14q24, and 16p13. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:791–801. doi: 10.1002/ajmg.b.30524. [DOI] [PubMed] [Google Scholar]
- 29.McGuffin P, Katz R. The genetics of depression and manic-depressive disorder. Br J Psychiatry. 1989;155:294–304. doi: 10.1192/bjp.155.3.294. [DOI] [PubMed] [Google Scholar]
- 30.Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M. Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry. 2004;9:1091–1099. doi: 10.1038/sj.mp.4001541. [DOI] [PubMed] [Google Scholar]
- 31.Cheng R, Juo SH, Loth JE, Nee J, Iossifov I, Blumenthal R, Sharpe L, Kanyas K, Lerer B, Lilliston B, Smith M, Trautman K, Gilliam TC, Endicott J, Baron M. Genome-wide linkage scan in a large bipolar disorder sample from the National Institute of Mental Health genetics initiative suggests putative loci for bipolar disorder, psychosis, suicide, and panic disorder. Mol Psychiatry. 2006;11:252–260. doi: 10.1038/sj.mp.4001778. [DOI] [PubMed] [Google Scholar]
- 32.Craddock N, O'Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craddock N, Owen MJ. Rethinking psychosis: the disadvantages of a dichotomous classification now outweigh the advantages. World Psychiatry. 2007;6:20–27. [PMC free article] [PubMed] [Google Scholar]
- 34.Morissette J, Villeneuve A, Bordeleau L, Rochette D, Laberge C, Gagne B, Laprise C, Bouchard G, Plante M, Gobeil L, Shink E, Weissenbach J, Barden N. Genome-wide search for linkage of bipolar affective disorders in a very large pedigree derived from a homogeneous population in quebec points to a locus of major effect on chromosome 12q23-q24. Am J Med Genet. 1999;88:567–587. doi: 10.1002/(sici)1096-8628(19991015)88:5<567::aid-ajmg24>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 35.Kaneva RP, Chorbov VM, Milanova VK, Kostov CS, Nickolov KI, Chakarova CF, Stoyanova VS, Nikolova-Hill AN, Krastev SK, Onchev GN, Kremensky IM, Kalaydjieva LV, Jablensky AV. Linkage analysis in bipolar pedigrees adds support for a susceptibility locus on 21q22. Psychiatr Genet. 2004;14:101–106. doi: 10.1097/01.ypg.0000128766.92096.ad. [DOI] [PubMed] [Google Scholar]
- 36.Kelsoe JR, Spence MA, Loetscher E, Foguet M, Sadovnick AD, Remick RA, Flodman P, Khristich J, Mroczkowski-Parker Z, Brown JL, Masser D, Ungerleider S, Rapaport MH, Wishart WL, Luebbert H. A genome survey indicates a possible susceptibility locus for bipolar disorder on chromosome 22. Proc Natl Acad Sci USA. 2001;98:585–590. doi: 10.1073/pnas.011358498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw SH, Mroczkowski-Parker Z, Shekhtman T, Alexander M, Remick RA, Sadovnick AD, McElroy SL, Keck PE, Jr, Kelsoe JR. Linkage of a bipolar disorder susceptibility locus to human chromosome 13q32 in a new pedigree series. Mol Psychiatry. 2003;8:558–564. doi: 10.1038/sj.mp.4001267. [DOI] [PubMed] [Google Scholar]
- 38.Lin PI, McInnis MG, Potash JB, Willour VL, MacKinnon DF, Miao K, DePaulo JR, Zandi PP. Assessment of the effect of age at onset on linkage to bipolar disorder: evidence on chromosomes 18p and 21q. Am J Hum Genet. 2005;77:545–555. doi: 10.1086/491602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suominen K, Mantere O, Valtonen H, Arvilommi P, Leppamaki S, Paunio T, Isometsa E. Early age at onset of bipolar disorder is associated with more severe clinical features but delayed treatment seeking. Bipolar Disord. 2007;9:698–705. doi: 10.1111/j.1399-5618.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 40.Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, Thomas MG, Gehrig C, Radhakrishna U, Snyder SE, Balk KG, Neufeld K, Swartz KL, DeMarchi N, Papadimitriou GN, Dikeos DG, Stefanis CN, Chakravarti A, Childs B, Housman DE, Kazazian HH, Antonarakis S, Pulver AE. Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 41.Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, Puech A, Tahri N, Cohen-Akenine A, Delabrosse S, Lissarrague S, Picard FP, Maurice K, Essioux L, Millasseau P, Grel P, Debailleul V, Simon AM, Caterina D, Dufaure I, Malekzadeh K, Belova M, Luan JJ, Bouillot M, Sambucy JL, Primas G, Saumier M, Boubkiri N, Martin-Saumier S, Nasroune M, Peixoto H, Delaye A, Pinchot V, Bastucci M, Guillou S, Chevillon M, Sainz-Fuertes R, Meguenni S, Aurich-Costa J, Cherif D, Gimalac A, van Duijn C, Gauvreau D, Ouellette G, Fortier I, Raelson J, Sherbatich T, Riazanskaia N, Rogaev E, Raeymaekers P, Aerssens J, Konings F, Luyten W, Macciardi F, Sham PC, Straub RE, Weinberger DR, Cohen N, Cohen D. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet. 2003;72:1131–1140. doi: 10.1086/374822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumacher J, Jamra RA, Freudenberg J, Becker T, Ohlraun S, Otte AC, Tullius M, Kovalenko S, Bogaert AV, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S. Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry. 2004;9:203–207. doi: 10.1038/sj.mp.4001421. [DOI] [PubMed] [Google Scholar]
- 44.Prata DP, Breen G, Osborne S, Munro J, St Clair D, Collier D. Association of DAO and G72(DAOA)/G30 genes with bipolar affective disorder. Am J Med Genet B Neuropsychiat Genet. 2007 doi: 10.1002/ajmg.b.30682. [DOI] [PubMed] [Google Scholar]
- 45.Ewald H, Flint T, Kruse TA, Mors O. A genome-wide scan shows significant linkage between bipolar disorder and chromosome 12q24.3 and suggestive linkage to chromosomes 1p22–21, 4p16, 6q14–22, 10q26 and 16p13.3. Mol Psychiatry. 2002;7:734–744. doi: 10.1038/sj.mp.4001074. [DOI] [PubMed] [Google Scholar]
- 46.Rice JP, Goate A, Williams JT, Bierut L, Dorr D, Wu W, Shears S, Gopalakrishnan G, Edenberg HJ, Foroud T, Nurnberger J, Jr, Gershon ES, Detera-Wadleigh SD, Goldin LR, Guroff JJ, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Reich T. Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 1, 6, 8, 10, and 12. Am J Med Genet. 1997;74:247–253. doi: 10.1002/(sici)1096-8628(19970531)74:3<247::aid-ajmg3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 47.Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI, Jr, Craddock N, DePaulo JR, Baron M, Gershon ES, Ekholm J, Cichon S, Turecki G, Claes S, Kelsoe JR, Schofield PR, Badenhop RF, Morissette J, Coon H, Blackwood D, McInnes LA, Foroud T, Edenberg HJ, Reich T, Rice JP, Goate A, McInnis MG, McMahon FJ, Badner JA, Goldin LR, Bennett P, Willour VL, Zandi PP, Liu J, Gilliam C, Juo SH, Berrettini WH, Yoshikawa T, Peltonen L, Lonnqvist J, Nothen MM, Schumacher J, Windemuth C, Rietschel M, Propping P, Maier W, Alda M, Grof P, Rouleau GA, Del-Favero J, Van Broeckhoven C, Mendlewicz J, Adolfsson R, Spence MA, Luebbert H, Adams LJ, Donald JA, Mitchell PB, Barden N, Shink E, Byerley W, Muir W, Visscher PM, Macgregor S, Gurling H, Kalsi G, McQuillin A, Escamilla MA, Reus VI, Leon P, Freimer NB, Ewald H, Kruse TA, Mors O, Radhakrishna U, Blouin JL, Antonarakis SE, Akarsu N. Genome scan meta-analysis of schizophrenia and bipolar disorder. 3. Bipolar disorder. Am J Hum Genet. 2003;73:49–62. doi: 10.1086/376547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curtis D, Kalsi G, Brynjolfsson J, McInnis M, O'Neill J, Smyth C, Moloney E, Murphy P, McQuillin A, Petursson H, Gurling H. Genome scan of pedigrees multiply affected with bipolar disorder provides further support for the presence of a susceptibility locus on chromosome 12q23-q24, and suggests the presence of additional loci on 1p and 1q. Psychiatr Genet. 2003;13:77–84. doi: 10.1097/01.ypg.0000056684.89558.d2. [DOI] [PubMed] [Google Scholar]
- 49.Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, Liang KY, Pulver AE. Genomewide linkage scan for bipolar-disorder susceptibility loci among Ashkenazi Jewish families. Am J Hum Genet. 2004;75:204–219. doi: 10.1086/422474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savitz J, Cupido CL, Ramesar RK. Preliminary evidence for linkage to chromosome 1q31–32, 10q23.3, and 16p13.3 in a South African cohort with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2007;144:383–387. doi: 10.1002/ajmg.b.30461. [DOI] [PubMed] [Google Scholar]
- 51.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D. Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21–22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3–24 and 20q12.1–11.23. AmJ Hum Genet. 2001;68:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O'Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O'Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder. 2. Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulayeva KB, Glatt SJ, Bulayev OA, Pavlova TA, Tsuang MT. Genome-wide linkage scan of schizophrenia: a cross-isolate study. Genomics. 2007;89:167–177. doi: 10.1016/j.ygeno.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Craddock N, O'Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Owen MJ, Craddock N, O'Donovan MC. Schizophrenia: genes at last? Trends Genet. 2005;21:518–525. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS. Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet. 2004;74:1057–1063. doi: 10.1086/420774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kremeyer B, Garcia J, Kymalainen H, Wratten N, Restrepo G, Palacio C, Miranda AL, Lopez C, Restrepo M, Bedoya G, Brzustowicz LM, Ospina-Duque J, Arbelaez MP, Ruiz-Linares A. Evidence for a role of the NOS1AP (CAPON) gene in schizophrenia and its clinical dimensions: an association study in a South American population isolate. Human Heredity. 2009;67:163–173. doi: 10.1159/000181154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordeiro Q, Talkowski ME, Chowdari KV, Wood J, Nimgaonkar V, Vallada H. Association and linkage analysis of RGS4 polymorphisms with schizophrenia and bipolar disorder in Brazil. Genes Brain Behav. 2005;4:45–50. doi: 10.1111/j.1601-183x.2004.00096.x. [DOI] [PubMed] [Google Scholar]
- 60.Xu B, Wratten N, Charych EI, Buyske S, Firestein BL, Brzustowicz LM. Increased expression in dorsolateral prefrontal cortex of CAPON in schizophrenia and bipolar disorder. PLoS Med. 2005;2:e263. doi: 10.1371/journal.pmed.0020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pato CN, Pato MT, Kirby A, Petryshen TL, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Verner A, Hudson TJ, Morley CP, Kennedy JL, Azevedo MH, Daly MJ, Sklar P. Genome-wide scan in Portuguese Island families implicates multiple loci in bipolar disorder: fine mapping adds support on chromosomes 6 and 11. Am J Med Genet B Neuropsychiatr Genet. 2004;127:30–34. doi: 10.1002/ajmg.b.30001. [DOI] [PubMed] [Google Scholar]
- 62.Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, Gillette MU. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–442. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 63.Mansour HA, Monk TH, Nimgaonkar VL. Circadian genes and bipolar disorder. Ann Med. 2005;37:196–205. doi: 10.1080/07853890510007377. [DOI] [PubMed] [Google Scholar]
- 64.Cichon S, Schumacher J, Muller DJ, Hurter M, Windemuth C, Strauch K, Hemmer S, Schulze TG, Schmidt-Wolf G, Albus M, Borrmann-Hassenbach M, Franzek E, Lanczik M, Fritze J, Kreiner R, Reuner U, Weigelt B, Minges J, Lichtermann D, Lerer B, Kanyas K, Baur MP, Wienker TF, Maier W, Rietschel M, Propping P, Nothen MM. A genome screen for genes predisposing to bipolar affective disorder detects a new susceptibility locus on 8q. Hum Mol Genet. 2001;10:2933–2944. doi: 10.1093/hmg/10.25.2933. [DOI] [PubMed] [Google Scholar]
- 65.Zandi PP, Badner JA, Steele J, Willour VL, Miao K, MacKinnon DF, Mondimore FM, Schweizer B, McInnis MG, Depaulo JR, Jr, Gershon E, McMahon FJ, Potash JB. Genome-wide linkage scan of 98 bipolar pedigrees and analysis of clinical covariates. Mol Psychiatry. 2007;12:630–639. doi: 10.1038/sj.mp.4002027. [DOI] [PubMed] [Google Scholar]
- 66.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, Okawa M, Yamada N, Hatten ME, Snyder SH, Ross CA, Sawa A. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gogos JA, Gerber DJ. Schizophrenia susceptibility genes: emergence of positional candidates and future directions. Trends Pharmacol Sci. 2006;27:226–233. doi: 10.1016/j.tips.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 69.McQueen MB, Devlin B, Faraone SV, Nimgaonkar VL, Sklar P, Smoller JW, Abou Jamra R, Albus M, Bacanu SA, Baron M, Barrett TB, Berrettini W, Blacker D, Byerley W, Cichon S, Coryell W, Craddock N, Daly MJ, DePaulo JR, Edenberg HJ, Foroud T, Gill M, Gilliam TC, Hamshere M, Jones I, Jones L, Juo SH, Kelsoe JR, Lambert D, Lange C, Lerer B, Liu J, Maier W, Mackinnon JD, McInnis MG, McMahon FJ, Murphy DL, Nothen MM, Nurnberger JI, Pato CN, Pato MT, Potash JB, Propping P, Pulver AE, Rice JP, Rietschel M, Scheftner W, Schumacher J, Segurado R, Van Steen K, Xie W, Zandi PP, Laird NM. Combined analysis from eleven linkage studies of bipolar disorder provides strong evidence of susceptibility loci on chromosomes 6q and 8q. Am J Hum Genet. 2005;77:582–595. doi: 10.1086/491603. [DOI] [PMC free article] [PubMed] [Google Scholar]


