Abstract
Single nucleotide polymorphisms in CYP3A5 (A6986G) and MDR-1 (C3435T) genes have been shown to be associated with the pharmacokinetics of tacrolimus in case of renal transplant recipients. Knowing these genotypes of the recipients before undergoing transplantation, is therefore essential for physicians to adjust the starting dose of tacrolimus in order to avoid drug induced nephrotoxicity. We have designed an allele specific PCR method for easier and rapid detection of these polymorphisms. 20 Indian renal transplant recipients on tacrolimus who developed nephrotoxicity within 1 month of transplantation and 58 Indian non-transplant subjects having the risk factors for kidney disease i.e. hypertension or diabetes or the family history of these, have been studied for these SNPs by allele specific PCR method. The data suggest that the heterozygosity of CYP3A5 and mutant allele frequency of MDR-1 SNP is higher in transplant patients as well as in general population.
Keywords: CYP3A5, MDR-1, Tacrolimus, Allele specific PCR
Introduction
Calcineurine inhibitor, tacrolimus has significantly improved the outcome of renal transplantation. However, it has a narrow therapeutic index and individual variations in metabolism. It also has it’s inherent nephrotoxic potential due to which, the success of transplant is limited. A6986G (CYP3A5*3) polymorphism of cytochrome P450 polypeptide 3A5 (CYP3A5) gene and C3435T polymorphism of Multidrug resistance-1 (MDR-1) gene have been shown to influence the metabolism and efflux of tacrolimus respectively. Due to the impaired metabolism and efflux, there is a risk of drug induced nephrotoxicity which may eventually lead to graft rejection [1]. Knowing the genotype of recipients before undertaking transplantation could guide clinicians upon the adjustment of the starting dose of tacrolimus, if necessary, so as to avoid nephrotoxicity. There are several methods for detection of these polymorphisms, out of which the restriction enzyme digestion based method is commonly used. However, it is time consuming and generally requires 2 days to complete entire procedure. Sequencing is the alternative however, it is costly. We, therefore, designed allele specific PCR method to study these two polymorphisms, which is rapid and low cost compared to the other methods.
Materials and Methods
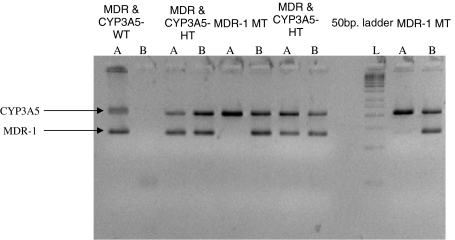
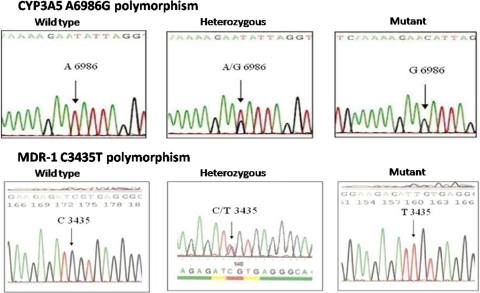
Allele specific primers were designed to amplify the desired region of CYP3A5 and MDR-1 genes (Table 1). 5cc EDTA blood sample was collected each from 20 Indian renal transplant recipients on tacrolimus (starting dose 0.15 mg/kg/day) who developed nephrotoxicity within 1 month of transplantation and 58 Indian non-transplant subjects having risk factors for kidney disease i.e. hypertension and/or diabetes or the family history of these. The blood sample was used for DNA extraction, which was performed as per Miller et al. [2] procedure. The extracted DNA was subjected to allele specific PCR and the amplified products were detected on 3% agarose gels for genotyping. Each sample’s reaction was in duplicate; one to amplify wild type allele and the other to amplify mutant. Forward primer was kept common for both the genes and in the reverse primer the 3′ end nucleotide was changed (Table 1) thereby, two reverse primers were designed to allow selective amplification of wild type or mutant alleles. Each 50 μl PCR reaction was carried out using 1× PCR buffer (Fermentas), 3 mM MgCl, 200 μM dNTP, 5 pmol of both forward and reverse primers and 1.5 units of the enzyme Taq DNA polymerase (Fermentas). 500 ng of DNA sample (Template) was used in each PCR reaction. For amplification, the initial denaturation at 95°C for 5 min was followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 60°C for both CYP3A5 and MDR-1 reactions for 1 min and extension at 72°C for 1 min followed by final extension at 72°C for 10 min. If the band was observed in both the reactions then the sample was considered to be heterozygous (Fig. 1). The specificity of primers was checked by the amplification of previously genotyped and sequenced representative samples (Fig. 2).
Table 1.
Details of the allele specific primer design for CYP3A5 and MDR-1 genes
| CYP3A5 | Forward primer: 5′ cac ttg atg att tac ctg cct tc 3′ | Amplified product length 218 bp |
| Reverse primer wild-type allele specific: 5′ ggt cca aac agg gaa gag ata t 3′ | ||
| Reverse primer mutant allele specific: 5′ ggt cca aac agg gaa gag ata c 3′ | ||
| MDR-1 | Forward primer: 5′ act ata ggc ca gaga ggc tgc 3′ | Amplified product length 134 bp |
| Reverse primer wild-type allele specific: 5′ gtg gtg tca cag gaa gag ctt 3′ | ||
| Reverse primer mutant allele specific: 5′ gtg gtg tca cag gaa gag ctc 3′ |
Bold emphasized nucleotide shows the change in allele specific primers
Fig. 1.
Genotyping of CYP3A5 (A6986G) and MDR-1 (C3435T) SNPs by allele specific PCR. Amplification of allele specific reactions for the detection of CYP3A5*3 and MDR-1 C3435T polymorphisms. The reactions are named as ‘A’ and ‘B’. ‘A’ reaction contains primers to amplify wild type alleles of both the genes and ‘B’ reaction contains primers to amplify mutant alleles of both the genes. L is the 50 bp DNA ladder
Fig. 2.
Sequencing of representative samples of CYP3A5 A6986G and MDR-1 C3435T SNPs. Sequencing results of the representative wild type, heterozygous and homozygous mutants of CYP3A5 (A6986G) and MDR-1 (C3435T) polymorphisms
Results and Discussion
Total 78 subjects comprised of 20 Indian renal transplant recipients between age range 35–62 years (17 males and 3 females) and 58 Indian non-transplant recipients between age range 25–81 years (33 males and 25 females) were genotyped by allele specific PCR method. Out of the 20 renal transplant recipients, 17 (85%) were found to be heterozygous for CYP3A5 A6986G polymorphism (mean tacrolimus trough level 12.2 ng/ml) and 50% were heterozygous and 30% were homozygous mutants for MDR-1 C3435T polymorphism. Their mean trough levels of tacrolimus were 13.3 and 9.1 ng/ml respectively (Table 2).
Table 2.
Demographic and clinical details of renal transplant recipients on tacrolimus
| Patient id | Age (years) | Sex | Tacrolimus level (ng/ml) | CYP3A5 genotype | MDR-1 genotype |
|---|---|---|---|---|---|
| 1 | 45 | M | 9 | w | m |
| 2 | 40 | M | 9.2 | h | h |
| 3 | 38 | M | 8.4 | h | w |
| 4 | 41 | F | 31.7 | h | w |
| 5 | 49 | M | 9 | h | h |
| 6 | 51 | M | 9.6 | h | w |
| 7 | 56 | M | 8.5 | h | h |
| 8 | 35 | M | 11.4 | m | h |
| 9 | 55 | M | 9 | h | m |
| 10 | 47 | M | 9.6 | h | m |
| 11 | 49 | F | 8.4 | h | m |
| 12 | 53 | M | 9.8 | h | h |
| 13 | 62 | F | 23.2 | h | h |
| 14 | 44 | M | 8.4 | h | h |
| 15 | 57 | M | 25.7 | h | h |
| 16 | 52 | M | 9 | h | w |
| 17 | 60 | M | 9.3 | h | h |
| 18 | 53 | M | 9.6 | h | m |
| 19 | 49 | M | 9 | h | m |
| 20 | 45 | M | 18.5 | w | h |
M male, F female, w wild type, h heterozygous, m mutant
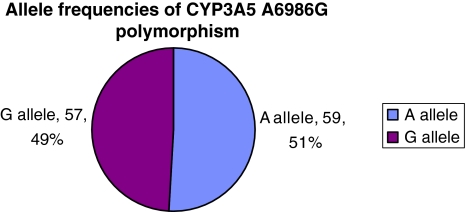
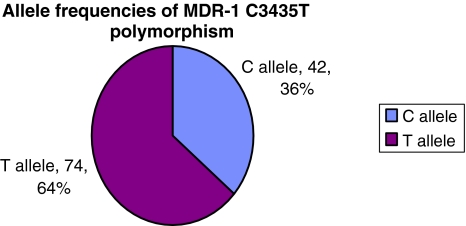
Studies on several populations have determined CYP3A5 G6986 allele frequency in healthy subjects. It is reported as 5–15% for Caucasians, 27% in Chinese and 15% in Japanese [3]. However, the frequency of T3435 allele of MDR-1 has been found to vary significantly in different ethnic groups. In case of African-Americans it has been found to be 17–32% in contrast to 75–79% in Caucasians and 62–71% in Asians [4]. The findings from our study suggest that out of the 58 Indian non-transplant subjects, 47 (81%) were found to be heterozygous for CYP3A5 A6986G SNP and 26 (44.83%), 24 (41.38%) were found to be heterozygous and/or homozygous mutants for MDR-1 C3435T SNP respectively. The distribution of allele frequencies of these SNPs in non-transplant subjects is given in Figs. 3 and 4. Out of these 58 subjects 23 had past history of Diabetes. On comparing the mutant allele distribution in the diabetic and non-diabetic subjects we found that it is almost similar in case of G6986 allele of CYP3A5 (0.4857 for non-diabetic and 0.500 for diabetic). However, it is higher in the diabetic group in case of T3435 allele of MDR-1 gene (0.7391 against 0.5714).
Fig. 3.
Allele frequencies of CYP3A5 A6986G polymorphism
Fig. 4.
Allele frequencies of MDR-1 C3435T polymorphism
Several studies have shown that the dose of tacrolimus is associated with CYP3A5 A6986G SNP [5–7]. Dose adjusted trough concentrations were 3 fold and 1.6 fold higher in mutants (G/G) than in heterozygous (A/G) patients for tacrolimus and Cyclosporine, respectively [8, 9]. Numerous studies have associated SNPs in the MDR-1 gene with dosage of tacrolimus in organ transplant recipients. Macphee et al. [10] reported only a weak association of MDR-1 C3435T with tacrolimus concentrations and hence the dose. However, Akbas et al. [11] in their study on 92 Turkish renal transplant recipients state that tacrolimus daily doses were significantly lower in the patients with 3435TT genotype at 1 and 6 month after transplant. The results from our study on 44 Indian renal transplant recipients on tacrolimus (findings accepted for publication) also suggest a significant association of the mutant alleles of CYP3A5 and MDR-1 SNPs with higher tacrolimus trough levels (P = 0.010 for CYP3A5 and P = 0.015 for MDR-1) Therefore, genotyping the patients for these SNPs before undergoing transplant by allele specific PCR method would guide physicians whether the patient is likely to develop higher trough tacrolimus levels which may lead to nephrotoxicity. The results also show that the heterozygosity of CYP3A5 and mutant allele homozygosity of MDR-1 SNPs is high in Indian population. It has also been the clinical observation that Indians require lower tacrolimus dose to achieve the same target drug levels compared to that in other populations. The general abundance of G6986 allele of CYP3A5 and T3435 allele of MDR-1 in Indians could be the factor contributing to this observation.
The findings from this study suggest that genotyping the renal transplant recipients for CYP3A5 and MDR-1 gene SNPs by allele specific PCR method could be helpful for physicians in adjusting tacrolimus dose. Therefore when the patients with homozygous mutant genotypes of both or heterozygous of CYP3A5 and mutant of MDR-1 go for renal transplant, they could be managed with lower tacrolimus dose, after the transplantation.
Acknowledgments
We acknowledge P. D. Hinduja National Hospital and Medical Research Center and the National Health Educations Society for funding this project. We also acknowledge staff of Muljibhai Patel Urological Hospital, Nadiad, Gujarat, India, for their kind help in providing the samples for this project.
References
- 1.Iwasaki K. Metabolism of tacrolimus (FK506) and recent topics in clinical pharmacokinetics. Drug Metab Pharmacokinet. 2007;22(5):328–355. doi: 10.2133/dmpk.22.328. [DOI] [PubMed] [Google Scholar]
- 2.Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acid Res. 1998;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamba J, Lin Y, Schuetz E, Thummel KE. Genetic contribution to variable human CYP3A mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/S0169-409X(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 4.Schwab M, Eichelbaum M, Fromm M. Genetic polymorphisms of the human MDR-1 drug transporter. Annu Rev Pharmacol Toxicol. 2003;43:285–307. doi: 10.1146/annurev.pharmtox.43.100901.140233. [DOI] [PubMed] [Google Scholar]
- 5.Zheng H, Webber S, Zeevi A, Schuetz E, Zhang J, Bowman P, et al. Tacrolimus dosing in pediatric heart transplant patients is related to CYP3A5 and MDR1 gene polymorphisms. Am J Transplant. 2003;3:477–483. doi: 10.1034/j.1600-6143.2003.00077.x. [DOI] [PubMed] [Google Scholar]
- 6.Zheng H, Zeevi A, Schuetz E, Lamba J, McCurry K, Griffith BP, et al. Tacrolimus dosing in adult lung transplant patients is related to cytochrome P4503A5 gene polymorphism. J Clin Pharmacol. 2004;44(2):135–140. doi: 10.1177/0091270003262108. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Liu ZH, Zheng JM, Chen ZH, Tang Z, Chen JS, Li LS. Influence of CYP3A5 and MDR1 polymorphisms on tacrolimus concentration in the early stage after renal transplantation. Clin Transplant. 2005;19(5):638–643. doi: 10.1111/j.1399-0012.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 8.Evans W, Relling M. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 9.Kamdem LK, Streit F, Zanger UM, Brockmöller J, Oellerich M, Armstrong VW, Wojnowski L. Contribution of CYP3A5 to the in vitro hepatic clearance of tacrolimus. Clin Chem. 2005;51(8):1374–1381. doi: 10.1373/clinchem.2005.050047. [DOI] [PubMed] [Google Scholar]
- 10.Macphee IA, Fredricks S, Tai T, Syrris P, Carter ND, Johnston A, et al. Tacrolimus pharmacogenetics: polymorphisms associated with expression of cytochrome p4503A5 and p-glycoprotein correlate with dose requirement. Transpl Proc. 2002;74(11):1486–1489. doi: 10.1097/00007890-200212150-00002. [DOI] [PubMed] [Google Scholar]
- 11.Akbas S, Bilgen T, Keser I, Tuncer M, Yucetin L, Tosun O, et al. The effect of MDR1 (ABCB1) polymorphisms on the pharmacokinetic of tacrolimus in Turkish renal transplant recipients. Transplant Proc. 2006;38(5):1290–1292. doi: 10.1016/j.transproceed.2006.02.079. [DOI] [PubMed] [Google Scholar]