Abstract
Background
Purkinje cells (PCs) comprise the most distal component of the cardiac conduction system and their unique electrophysiological properties and the anatomic complexity of the Purkinje fiber network may account for the prominent role these cells play in the genesis of various arrhythmic syndromes.
Methods and Results
– Differential transcriptional profiling of murine Purkinje fibers and working ventricular myocytes was performed to identify novel genes expressed in PCs. The most highly enriched transcript in Purkinje fibers encoded Contactin-2 (Cntn2), a cell adhesion molecule critical for neuronal patterning and ion channel clustering. Endogenous expression of Cntn2 in the murine ventricle was restricted to a subendocardial network of myocytes that also express β-galactosidase in CCS-lacZ transgenic mice and the connexin40 gap junction protein. Both Cntn2-lacZ knockin mice and Cntn2-EGFP BAC transgenic reporter mice confirmed expression of Cntn2 in the Purkinje fiber network, as did immunohistochemical staining of single canine Purkinje fibers. Whole-cell patch-clamp recordings and measurements of Ca2+ transients in Cntn2-EGFP+ cells revealed electrophysiological properties indicative of PCs and distinctive from those of cardiac myocytes, including prolonged action potentials and frequent afterdepolarizations.
Conclusions
Cntn2 is a novel marker of the specialized cardiac conduction system. Endogenous expression of Cntn2 as well as Cntn2-dependent transcriptional reporters provides a new tool through which Purkinje cell biology and pathophysiology can now more readily be deciphered. Expression of a contactin family member within the CCS may provide a mechanistic basis for patterning of the conduction system network and the organization of ion channels within Purkinje cells.
Keywords: cell adhesion molecules, electrophysiology, genetics, Purkinje fiber
Introduction
Purkinje fibers (PFs) are the most distal component of the cardiac conduction system (CCS), first described by Purkinje in 1839 as gray, flat, gelatin-like ramifications, running under the endocardium1. Some seventy years later, Tawara more fully characterized the Purkinje system, identifying the left (anterior, septal and posterior) and right fascicular strands, which served to connect the distal PFs to the bundle branches proper2. Tawara was also the first to correctly suggest the functional role of the Purkinje system in rapidly transmitting the electrical wave of excitation to the ventricular muscle.
PFs appear to play a prominent role in the genesis of ventricular arrhythmias, (reviewed in 3). PF and anterior or posterior left fascicular triggers have been implicated in the initiation of monomorphic ventricular tachycardia in post myocardial infarction patients, as demonstrated by cure after focal ablation of Purkinje fiber or fascicular potentials4–6. PF-based triggers have also been described in patients with ventricular tachycardia associated with dilated forms of cardiomyopathy7, as well as idiopathic ventricular fibrillation (VF), in which ablation of premature beats arising from the PF-network resulted in significant reductions in the recurrence of VF8. PF-dependent triggering of arrhythmias has also been proposed in inherited syndromes including catecholaminergic polymorphic VT9, Brugada syndrome and long QT syndrome10. Despite growing evidence implicating PFs in ventricular arrhythmogenesis, our understanding of the cellular mechanisms underlying PF-dependent diseases is hampered by the lack of knowledge of the developmental biology of individual Purkinje cells (PCs), their patterning into a network of highly coupled cells, as well as their adaptive and maladaptive responses to pathologic stimuli. To some extent, this gap in knowledge reflects the anatomical complexity of the PF-network, which includes branching cells that couple not only with neighboring PCs, but also with working myocytes at Purkinje-muscle junctions. Furthermore, PFs are thought to include cells with functional and structural heterogeneity, such as the transitional J-cells11. In recent years a number of “conduction-system” markers have been reported12–14. These include transcription factors, gap junctional and ion channel associated proteins, with different specificities and different roles in the development and function of the CCS. However, the majority of these molecular markers, including HF-1b15 and Kcne116 preferentially identify cells of the proximal CCS and to date, only two, the CCS-lacZ gene (resulting from insertion of an Engrailed-2 reporter gene into the Slco3a1 locus17, 18) and gja5/connexin40 (Cx40), are robustly expressed in PFs.
In the developing mouse ventricle, CCS-lacZ expression is first observed in trabecular myocardium at embryonic day 10.5, a domain that appears to be the primary source of cells contributing to the developing PF network. By E13.5 the reporter gene’s expression pattern is similar to the neonate, with the appearance of a more discrete subendocardial fiber network19. Expression is also observed in other components of the CCS, including both the sinoatrial node (SAN) and atrioventricular node (AVN), as well as the His bundle and bundle branches20. Unfortunately, expression of β-galactosidase in the CCS-lacZ strain is the result of complex insertional mutation associated with substantial genomic rearrangement, complicating mechanistic studies of CCS-specific transcriptional regulation18.
Cx40 is a gap junction protein expressed in the developing and adult murine heart21 and Cx40-EGFP reporter mice have also been used to visualize the ventricular conduction system 22. Cx40 expression is observed in other components of the CCS (AVN, His, Bundle Branches), but is also abundantly expressed in atrial myocytes and coronary arteries. Examination of Cx40 deficient murine models has provided insight into its important role in development and function of the CCS. Loss of function of Cx40 results in an increased incidence of cardiac malformations (double-outlet right ventricle, dilated and hypertrophic cardiomyopathy)23 as well as conduction defects24. Interestingly, sequence variants within the human GJA5 locus have been described in association with Tetralogy of Fallot25.
There are also reports12, 14 describing PF-enriched expression of two cardiac homeobox transcription factors: Nkx2-5 and Homeodomain only protein (Hop). Nkx2-5+/− mice are characterized by ventricular conduction defects specifically attributable to a hypoplastic PF network, thought to be the result of impaired postnatal differentiation of myocytes12. However, Nkx2-5 is broadly expressed in many cell-types within the developing and adult heart, which diminishes its utility as a PF-specific marker. Hop is a cardiac homeobox transcription factor which acts downstream of Nkx2-514, 26. Hop-lacZ knockin mice show broad reporter gene expression in the developing heart, which becomes somewhat more restricted to the atria and proximal CCS in the adult.
Accordingly, with the goal of developing additional specific markers of the distal CCS that might be useful to facilitate studies elucidating the molecular circuitry regulating CCS specification, patterning and function, we performed transcriptional profiling of the murine PF network.
Methods
Methods are described in brief here (see the online-only DataSupplement for a full description).
Mutant Mice
All experiments were performed according to protocols approved by the NYU Institutional Animal Care and Use Committee and conformed to the National Institutes of Health (NIH) guidelines for the care and use of Laboratory Animals. CCS-lacZ+/− transgenic (backcrossed into the CD-1 background for at least nine generations)19 and Cntn2-lacZ knockin mice (on a C57BL/6x129SvEv mixed background)27 have been previously described. Cntn2-EGFP BAC transgenic mice [Tg(Cntn2-EGFP)344sat] were obtained from MMRRC and maintained on a CD-1 background.
Microarray Profiling
Purkinje fiber (PF) enriched samples were obtained by micro-dissecting the subendocardial layer of 6–8 month old CCS-lacZ murine hearts and PF depleted samples were micro-dissected from the sub-epicardial layer of the ventricular walls.
One-step Reverse Transcription and Polymerase Chain Reaction (RT-PCR) and Quantitative real time RT-PCR (QRT-PCR)
Total RNA was processed using QuantiTect SYBR Green One-Step RT-PCR kit (Qiagen) or One-Step SuperScript III (Invitrogen). In QRT-PCR experiments quantification of transcripts was performed using GAPDH as an internal reference and the 2−ΔΔCT method 28. For gene specific primers see Data Supplement.
Isolation of adult canine Purkinje Fibers (PFs) and individual Purkinje Cells (PCs)
PCs were enzymatically dispersed from PFs of the canine heart as previously described29, 30.
Immunohistochemistry and Immunocytochemistry, Antibodies
Mouse hearts or canine PFs were fixed in 4% PFA, then equilibrated in 10% sucrose and embedded in OCT. Cryosections, 6–10μM thick were processed for staining with individual antibodies as detailed in the Data Supplement.
Electrophysiological recordings in isolated ventricular myocytes
Ventricular myocytes were isolated using an established enzymatic digestion protocol31 yielding to 60–80% rod-shaped, calcium-tolerant myocytes. Action potential duration (APD) was measured at 90% and 50% of repolarization (APD90 and APD50). DADs were defined as phase 4 positive (depolarizing) deflections of the membrane potential. EADs were defined as positive (depolarizing) oscillations occurring during phase 2 or 3 of action potential.
Ca2+ imaging
Isolated myocytes from Cntn2-EGFP transgenic mice were loaded with the membrane-permeant Ca2+ indicator dye Rhod-2/AM and imaged by confocal microscopy (Leica SP5). In some experiments, the Ca2+ transients were measured by microfluorimetry (IonOptix, Milton, MA) using Indo-1/AM.
Statistics
Data are presented as mean ± SEM. Values were compared with a two-tailed t-test and P values less than 0.05 were considered statistically significant.
Results
Cntn2 Transcripts are Preferentially Expressed in Cardiac Purkinje Fibers
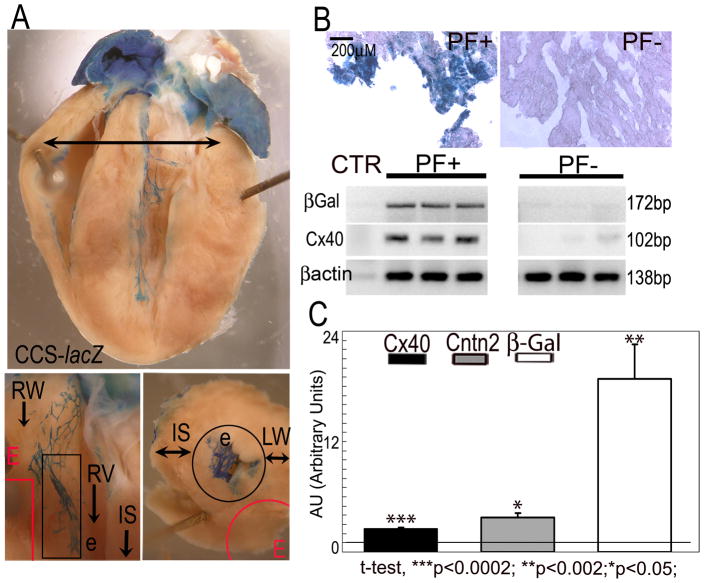
Using CCS-lacZ hearts, in which the distal conduction system is readily visualized, we performed comparative transcriptional profiling of mRNA prepared from Purkinje fiber (PF)-enriched subendocardial fractions harvested distal to the bundle branches proper and from PF-deficient subepicardial layers (Fig. 1A). Enrichment of PFs was confirmed by XGal staining of dissected tissue (Fig. 1B Top) and by RT-PCR, which showed preferential expression of transcripts encoding both β-galactosidase as well as connexin40 (Fig. 1B Bottom). Comparison of global gene expression patterns from three independent datasets of PF-enriched and PF-depleted samples by microarray analysis (Dataset Supplement) identified 163 probe sets that were significantly up-regulated (5.87-1.33 fold increase) and 251 that were down-regulated (0.77-0.26 fold decrease). The transcript encoding Contactin2 (Cntn2; UniGene Mm.34775), showed the highest level of enrichment (5.87). RT-PCR was used to confirm the microarray results identifying Cntn2 expression in cardiac PFs (Fig. 1C). Of note, two members of the Iroquois gene family, Irx5 (4.18) and Irx2 (2.89), were also enriched in PFs, as was Sema3b, a putative molecular partner of contactins. Interestingly, Nav1.8/SCN10A, which encodes the α-subunit of a TTX-resistant sodium channel isoform, was also enriched in the PF fraction (2.15). Sequence variation in the SCN10A gene has recently been associated with human conduction system disease32.
Fig. 1. Identification of novel Purkinje fiber-specific transcripts by comparative microarray analysis.
(A) PF+ and PF− samples were micro dissected distal to the bundle branches (Top, arrow) from the subendocardial and subepicardial regions (Bottom) of CCS-lacZ hearts. (B) Enrichment of PF-specific transcripts in PF+ samples was confirmed by increased expression of the PF markers CCS-lacZ and Cx40 compared to PF− samples, assessed by XGal staining (Top) and/or RT-PCR (Bottom). (C) QRT-PCR showing up-regulation of Cntn2 and the PF-specific markers CCS-lacZ (βGal) and Cx40 in micro dissected PFs. Values are expressed as Arbitrary Units (1AU= RV in PF− sample). PFs, Purkinje fibers; RW, right ventricular wall; LW, left ventricular wall; RV, right ventricle; LV, left ventricle; e, endocardium; E, epicardium. Black outline, subendocardial region; Red outline, subepicardial region; IS, interventricular septum. Size bar: 200μM.
Cntn2 is Expressed on the Sarcolemma of Murine Purkinje Cells
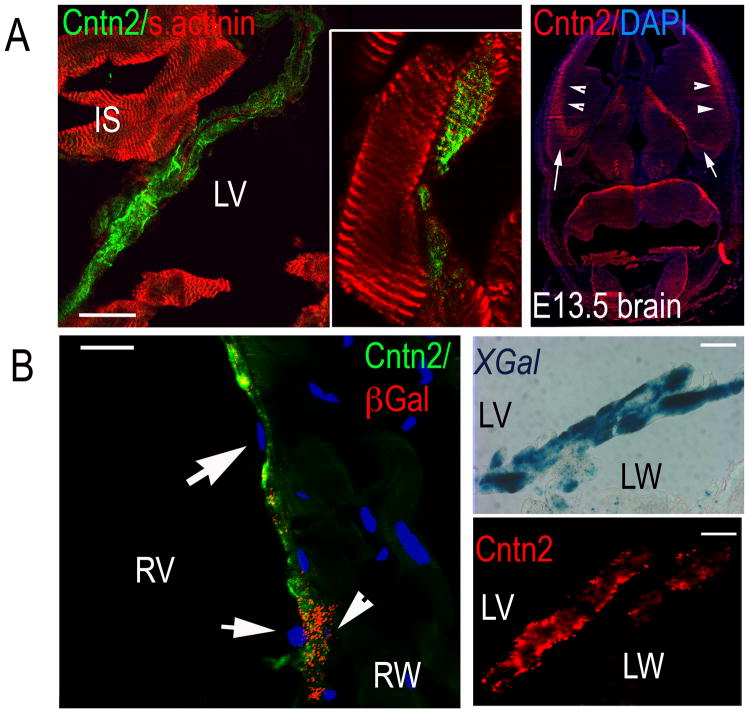
We performed immunohistochemical staining to specifically localize expression of Cntn2 in the heart. Cryosections of adult murine hearts were co-stained for Cntn2 as well as the cardiac-specific marker sarcomeric actinin, which revealed that Cntn2+ cells belong to the cardiac lineage (Fig. 2A Left). Co-staining of CCS-lacZ hearts for Cntn2 and β-galactosidase (βGal) or staining of serial sections for Cntn2 or XGal, confirmed that Cntn2+ cells were also βGal+ (Fig. 2B Left) or XGal+ (Fig. 2B Right). Furthermore, these images also demonstrated that the Cntn2 positive cells were localized just beneath the thin endocardial cell layer (Fig. 2A Right), similar to what we previously reported using the CCS-lacZ marker20. Taken together, these data indicate that Cntn2 is specifically expressed in adult murine PFs.
Fig. 2. Cntn2 is specifically expressed in Purkinje fibers.
(A Left) Confocal image of mouse heart sections confirming that Cntn2+ cells (green) express the cardiac marker sarcomeric actinin (red, Left). (Inset) High power image showing an optical slice of a Cntn2+/sarcomeric actinin+ myocyte. (B Left) Epifluorescent images of CCS-lacZ hearts showing a Cntn2+/βGal+ (green/red) cell (arrowhead) under a layer of Cntn2−/βGal− endocardial cells (arrows). (B Right) Images of CCS-lacZ heart serial sections stained for XGal (Upper) and Cntn2 (red, Lower) showing co-localization of Cntn2 and the PF-specific marker CCS-lacZ. (C) Epifluorescent image of E13.5 mouse brain used as positive control, showing Cntn2 expression (red) in the intermediate and marginal zones (arrowheads) and in the preplate region (arrows) of the ventral telenchephalon. LW, left ventricular wall; LV, left ventricle; RW, right ventricular wall; RV, right ventricle. Size bars: 20μM. A Left, Inset: 35μM.
Cntn2 is Expressed in the Proximal and Distal CCS
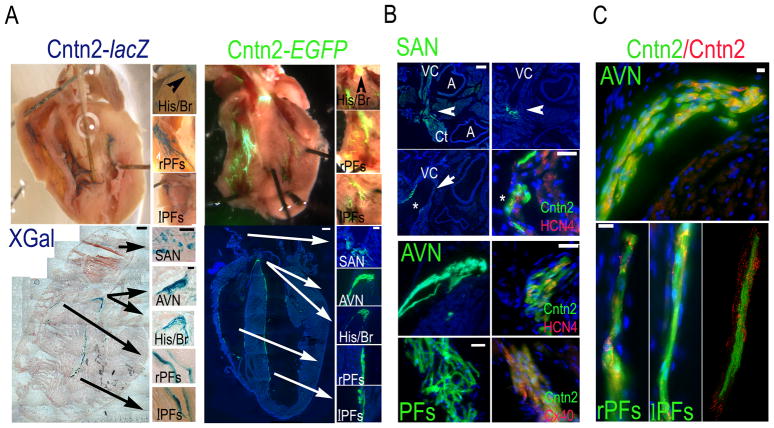
To more fully examine the spatial pattern of Cntn2 in the adult murine heart, we utilized two independent reporter gene mice: Cntn2-lacZ knockin mice and Cntn2-EGFP BAC transgenic mice. Examination of the endocardial surfaces of the ventricular cavities of whole mount Cntn2-lacZ hearts (Fig. 3A Top, Left) and Cntn2-EGFP (Fig. 3A Top, Right) hearts revealed dense networks of XGal+ and EGFP+ fluorescent PFs, which were also observed after sectioning (Fig. 3A Bottom). Detailed histologic examination of Cntn2-EGFP hearts also demonstrated expression in cells of the sinoatrial node (SAN; Fig. 3B Top)33, including cells close to the crista terminalis in the posterior part of the SAN; at the base of the vena cava in the central part of the SAN; and in the walls of the vena cava in the most anterior part of the SAN. Co-expression of Cntn2 and the nodal marker Hcn4 further confirmed that Cntn2 is indeed expressed in a sub-population of elongated Hcn4+ cells in the SAN. Detailed examination of the His bundle/atrioventricular (AVN) region revealed expression of Cntn2-EGFP in Hcn4+ cells of the AVN, localized posteriorly to the His Bundle (Fig. 3B Center). Co-staining for connexin40 (Cx40) also confirmed expression of Cntn2-EGFP in Cx40+ PCs of both ventricles (Fig. 3B Bottom). Staining with anti-Cntn2 antibodies confirmed that EGFP reporter gene expression in Cntn2-EGFP transgenic mice correctly reflected endogenous gene expression, as shown by co-localization of the two markers in components of the CCS (Fig. 3C).
Fig. 3. Cntn2 expression delineates the CCS in the adult heart.
(A) Whole mounts (Top) and cryosections (Bottom) of adult Cntn2-lacZ (Left; XGal staining) and Cntn2-EGFP hearts (Right) showing expression of Cntn2 throughout the CCS. (B) Epifluorescent images of Cntn2-EGFP hearts confirming expression of Cntn2 (green) in the SAN (Top), AVN (Center) and PFs (Bottom), identified by anatomical landmarks and expression of the nodal marker Hcn4 (red) or the PF marker Cx40 (red). (Top) Cntn2-EGFP+ cells (green) were found in the SAN, localized at the junction of the VC with the right atrium and extending from the CT (0 μM), posteriorly, to the base (+40 μM), and the walls (+190 μM) of the VC, anteriorly. Cntn2 identifies a sub-population of elongated Hcn4+ cells in the SAN. Cntn2-EGFP+/Hcn4+ cells were also found in the AVN, 20–40 μM posteriorly to the His Bundle (Center). (Bottom) Cntn2-EGFP+ (green)/Cx40+ (red) PFs were observed in both ventricles. (C) Cntn2-EGFP transgene expression recapitulates Cntn2 protein expression as shown by co-localization of anti-Cntn2-TR antibody (red) and EGFP (green) signals in the AVN (Top) and the PFs (Bottom). (Bottom Right) Confocal image detail of PFs. His/Br, His bundle and bundle branches; AVN, atrioventricular node; SAN, sinoatrial node; lPFs, left ventricle Purkinje fibers; rPFs, right ventricle Purkinje Fibers; Ct, crista terminalis; VC, vena cava; A, aorta and pulmonary artery; arrow, SAN (in B); CCS, cardiac conduction system. Blue fluorescence: DAPI nuclear counter stain, in A (Bottom, Right,), B and C. Size bars: (A) 50μM, all except for Bottom, Left, 500μM; (B) 20μM, all except for Top, Left, 200μM, and Bottom, Left, 50μM; (C), 100μM.
Cntn2 expression in canine Purkinje fibers
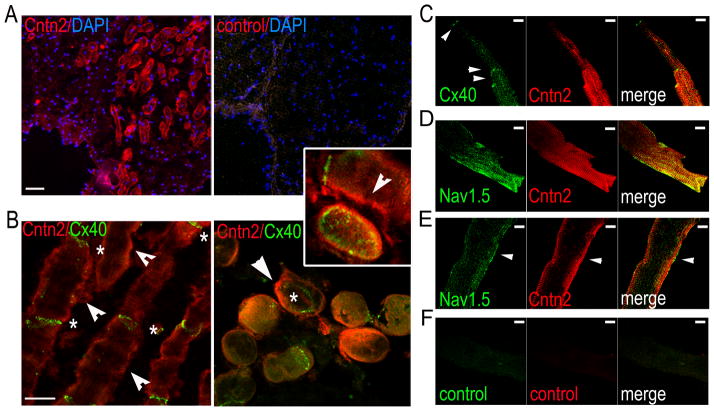
Immunohistochemistry of isolated canine PFs showed preferential expression of Cntn2 on the sarcolemma of individual Purkinje cells (PCs; Fig. 4A). Myocytes expressing Cntn2 on their cell membrane also expressed the PC-marker Cx40 at their intercalated disks, as observed in both longitudinal (Fig. 4B Left) and tangential (Fig. 4B Right) cell sections. Confocal images of dissociated canine PCs (Fig. 4C) confirmed the expression of Cntn2 on the sarcolemma of Cx40+ cells with a rod-shaped, Purkinje-like phenotype. The cardiac pore-forming sodium channel subunit, Nav1.5/Scn5a, was concentrated at the intercalated disks and along the cell membrane in PCs. Confocal images confirmed that Cntn2 is expressed in Nav1.5+ PCs; surface (Fig 4D) and sub-surface (Fig 4E) optical slices showed that, while co-expressed in the same cell, Cntn2 and Nav1.5 appeared to be targeted to distinct membrane domains with limited co-localization, most clearly seen in the sub-surface view.
Fig. 4. Cntn2 is expressed in canine Purkinje fibers.
(A) Epifluorescent images (Top) of micro dissected canine Purkinje fiber (PF) bundles showing expression of Cntn2 (red, Left). In the negative control (Right) the primary antibody was omitted. (B) Confocal images of the same canine Purkinje Fiber bundles, showing expression of Cntn2 (red) on the membrane (arrowheads) of Cx40+ (green) canine Purkinje cells (PCs), sectioned either longitudinally (Left) or transversally (Right). Cx40+ intercalated disks (stars) are clearly visible at the junction of Cntn2+ adjacent PCs. (Inset) High power en face image of an entire intercalated disk showing Cntn2 signal at the periphery of Cx40+ domains with typical gap junction organization with larger fluorescent patches at the periphery and smaller areas in the center. Cntn2 (arrowhead) is also visible on the sarcolemma of two longitudinal PCs, separated by a Cx40+ intercalated disk (star). (C) High power confocal image of cell surface optical slice of a single canine PC confirming expression of Cntn2 on the sarcolemma, identified by expression of Cx40 (green) at the intercalated disk junctions (arrowheads). Cntn2+ cells co-express the cardiac marker Nav1.5 as visualized at the cell surface (D), or in a subsurface slice (E). (F) Negative control: no primary antibody. Size bars: 20μM (A, B); 10μM (C–F).
Isolation and Functional Analysis of Cntn2-EGFP+ myocytes
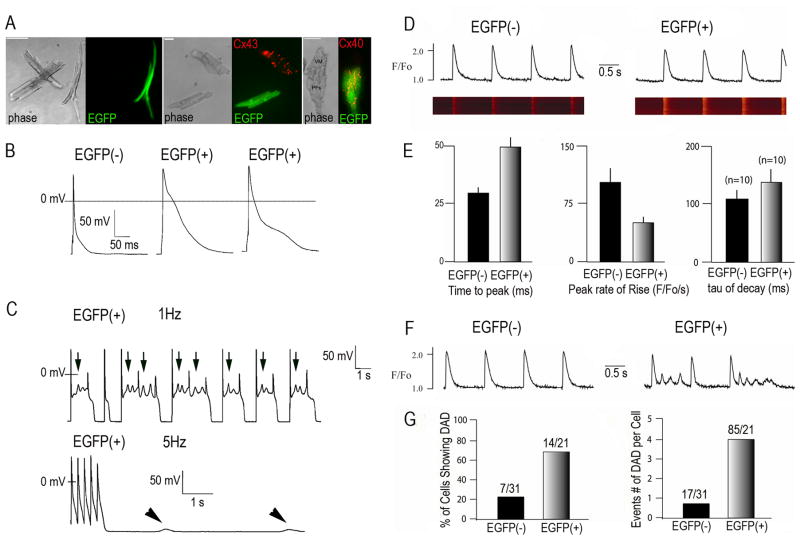
To further analyze the properties of the Cntn2-expressing myocytes, we dissociated myocytes from the ventricles of adult Cntn2-EGFP+ transgenic mice. Cntn2-EGFP+ myocytes were readily seen under epifluorescence microscopy, with an elongated, rod-shaped phenotype typical of PCs, easily distinguishable from the non-fluorescent working ventricular myocytes (Fig 5A Left). Cntn2-EGFP+ cells did not express Cx43 (Fig 5A Center), the most abundant connexin of working myocytes, but were strongly positive for Cx40. This specificity of Cx40 expression was most obvious in a cluster of myocytes that included both EGFP+ and EGFP− cells (Fig 5A Right). Action potentials (APs) from Cntn2-EGFP+ myocytes were characterized by a distinct plateau in phase 2 (Fig. 5B). APD50 and APD90 values were both significantly longer than those recorded from Cntn2-EGFP− myocytes (Table 1). Cntn2-EGFP+ myocytes also demonstrated, spontaneous electrical oscillations in phase 2, which resulted in early-after depolarizations (EADs) (Fig. 5C Top). In some instances (Fig. 5C Bottom), Cntn2-EGFP+ myocytes also developed delayed afterdepolarizations (DADs) at faster pacing rates (5 Hz); in contrast, none of Cntn2-EGFP− cells exhibited either EADs or DADs.
Fig. 5. Analysis of electrophysiological features of Cntn2-EGFP+ cells.
(A) Representative phase contrast and corresponding epifluorescence micrographs of dissociated myocytes from Cntn2-EGFP transgenic hearts. Cntn2-EGFP+ cells can be differentiated from surrounding cardiac myocytes by their rod-shaped morphology (Left) and their Cx43− (Center), Cx40+ (Right) phenotype. (B) Representative whole cell recordings of action potentials from Cntn2-EGFP− and Cntn2-EGFP+ cells. (C) APs from Cntn2-EGFP+ myocytes demonstrated spontaneous electrical oscillations in phase 2 which resulted in EADs (Top, arrows), at lower pacing rates (1Hz), and DADs (Bottom, arrowheads), at higher pacing rates (5Hz). (D) Line-scan fluorescence images and corresponding fluorescence profiles generated by Ca2+ transients in Cntn2-EGFP+ (Right) and Cntn2-EGFP− (Left) myocytes. (E) Comparison of kinetic parameters corresponding to Ca2+ transients in Cntn2-EGFP+ and Cntn2-EGFP− myocytes. (F) Line plots (G) and graphs showing that Cntn2-EGFP+ myocytes displayed a higher frequency of unstimulated Ca2+ events arising from DADs compared to Cntn2-EGFP− myocytes. EAD, early afterdepolarization; DAD, delayed afterdepolarization; AP, action potential. N=number of cells analyzed. VM, ventricular myocyte; PF, Purkinje Fiber; APs, action potentials; EADs, early afterdepolarizations; DADs, delayed afterdepolarizations.
Table 1.
Functional Properties of Cntn2-EGFP− and Cntn2-EGFP+ myocytes
| Action Potential Characteristics | Ca2+ Transient Kinetics | ||||||
|---|---|---|---|---|---|---|---|
| Cell Type | RMP (mV) | APA (mV) | APD50 (ms) | APD90 (ms) | Time to Max (ms) | Peak Rate of Rise (F/Fo/s) | Tau of decay (ms) |
| EGFP(−) (n=10) | −71.3±1.1 | 119.2±3.2 | 5.5±0.9 | 54.7±9.7 | 29.5±3.6 | 103.0±20 | 110.4±13.4 |
| EGFP(+) (n=10) | −71.5±1.3 | 128.3±3.7 | 24.7±4.3 | 139.3±17.8 | 49.4±4.8 | 49.4±5.9 | 136.7±23.2 |
| P value | .87 | .08 | .00035 | .00057 | < .000001 | < .00001 | 0.02 |
Data are expressed as mean values ± SEM. N, number of cells; RMP, resting membrane potential; APA, AP amplitude; APD50,90, AP duration at 50% and 90% of repolarization, respectively. APDs are significantly longer for Cntn2-EGFP+ myocytes compared to Cntn2-EGFP− myocytes. Ca2+ transient measurements demonstrated that Cntn2-EGFP+ myocytes have significantly slower Ca2+-handling kinetics compared to Cntn2-EGFP− myocytes.
Confocal microscopy and fluorescence photometry with calcium-sensitive dyes were also performed to study intracellular calcium dynamics. Compared to the Cntn2-EGFP− working ventricular myocytes, Cntn2-EGFP+ cells displayed significantly slower kinetics of activation and relaxation (Fig. 5D, E), with longer time to peak fluorescence, reduced peak rate of rise of fluorescence and slower decay (Table 1). EGFP+ myocytes were also significantly more likely to develop spontaneous Ca2+ events (Fig 5F, G).
Discussion
We identified Contactin2 (Cntn2) as a novel marker that is preferentially expressed in the cardiac conduction system, including myocytes of the murine and canine Purkinje fiber networks. Contactins are a subgroup of cell adhesion molecules of the immunoglobulin (Ig) superfamily. There are at least six contactins: Cntn1, Cntn2, BIG-1, BIG-2, NB-2 and NB-3; all include six Ig-like and four fibronectin III-like domains that are anchored to the plasma membrane by a glycosyl phosphatidylinositol (GPI) moiety. Cntn2, also known as transiently-expressed axonal glycoprotein 1 (TAG-1), in mouse and rat 34, transient axonal glycoprotein 1 (TAX1) in human and axonin-1 in chicken, has been most fully characterized in the central (CNS) and peripheral nervous systems (PNS) where it is expressed in the membrane of neurons and myelinating glial cells.
We used several complementary strategies to confirm the transcriptional profiling data demonstrating expression of Cntn2 within the Purkinje fiber (PF) network. These included immunohistochemical studies of murine and canine heart, as well as analyses of two genetically engineered reporter mice, i.e., Cntn2-lacZ knockin mice and Cntn2-EGFP BAC transgenic mice. Interestingly, while our initial transcriptional screen was focused on identifying transcripts enriched in Purkinje fibers, our studies demonstrated expression of endogenous Cntn2 (and reporter gene expression) throughout the entirety of the adult CCS, including nodal as well as fast-conducting cells of the His-Purkinje network. This observation, coupled with our previous description of widespread expression of the CCS-lacZ reporter gene throughout both the proximal and distal CCS, suggests that despite the functional diversity that exists within the heterogeneous elements of the CCS, these disparate components of the conductive network share some commonality with respect to transcriptional regulation.
In this study, we took advantage of Cntn2-EGFP reporter mice 35 since they provided us with a powerful approach to identify and functionally characterize individual Cntn2-expressing murine myocytes. These data, including patch clamp recordings and measurements of intracellular calcium dynamics, were consistent with a PC phenotype. Importantly, our functional studies indicated a significantly greater propensity of single murine PCs to develop spontaneous intracellular calcium release events (Fig. 5F) and afterdepolarizations (Fig. 5G). These observations are consistent with the purported role of PCs in the genesis and maintenance of various arrhythmic syndromes, including inherited diseases such as catecholaminergic polymorphic ventricular tachycardia 9 and idiopathic ventricular fibrillation36, as well as common acquired pathologies, such as post-myocardial infarction ectopy29, 37. Importantly, the Cntn2-EGFP mice will allow us to characterize the ventricular conduction system at varying levels of resolution, including studies of single cells (as in the current study), as well as analyses of the complex branching structure of the Purkinje fiber network. These data should be of benefit in the formulation of increasingly sophisticated computational models of cardiac excitation38, 39. These studies do not yet address the specific function of Cntn2 in the cardiac conduction system, although there are tantalizing hints from studies in the developing and adult nervous system. During development, Cntn2 participates in neural cell adhesion and migration, neurite outgrowth and fasciculation, axon pathfinding and myelination40. Even more relevant for the interpretation of our findings in PCs is the role of Cntn2 in adult neurons where it is involved in ion clustering with important implications for their electrophysiological properties.
Cntn2 is specifically involved in the clustering of delayed rectifier Shaker-type K+ channels, Kv1.1 and Kv1.2, and their Kvβ2 subunits at the juxtaparanodal region (JXP) of myelinated axons 27. Concentration of Kvs at the JXP and their segregation from domains enriched in voltage-gated Na+ channels (Navs), at the nodes of Ranvier plays an important role in limiting neuronal excitability by maintaining neuronal internodal resting potential or preventing rapid re-excitation41. Molecular mechanisms of Cntn2-mediated Kv clustering in neurons involve trans interaction with other Cntn2 molecules on the glial membrane and heterophilic cis interactions with Caspr2, a member of the neurexin superfamily in the neuronal membrane (axolemma). Caspr2 expression in the heart has not yet been examined, however in the nervous system, it influences the function of Cntn2 as demonstrated by studies showing that in Caspr2 deficient mice Cntn2 is unable to localize at the JXP and Kvs diffuse to the internodal region 27. Conversely, in Cntn2−/− mice, there is mislocalization of Caspr2 and Kvs at the internodal regions, demonstrating the interdependence of Cntn2 and Casp242. These observations suggest that the function of Cntn2 in PCs could also be to regulate Kv clustering through its interaction with Caspr2, thereby modulating PC excitability. Our data showing that Cntn2 and Nav1.5 appear to be targeted to distinct membrane domains support this functional parallel in PCs and neurons. Additional co-localization studies using Cntn-EGFP mice will be required to explore potential molecular interactions between Cntn2, Kvs, Navs, and Caspr2 in PCs.
Previous studies have also shown that Cntn2 is able to induce cis activation and binding of L1-CAM to the adapter protein ankyrin43, which, in neurons, is implicated in Nav clustering at the nodes (ankyrin-G) 44. In cardiomyocytes, mutations in ankyryn-G binding domains of Nav1.5 that result in impaired Nav1.5 localization to the intercalated disks were recently implicated in the pathogenesis of ventricular arrhythmias45–47. Furthermore, in the canine model of arrhythmia post-infarction, ankyrin-G has been implicated in sodium channel remodeling of epicardial border zone cells48. In light of its interaction with ankyrin it is conceivable that Cntn2 may play a mechanistic role in ankyrin-dependent regulation of ion channel targeting and arrhythmogenesis.
In conclusion, in the present study we have identified Cntn2 as a novel marker that is robustly expressed throughout the specialized conductive system of the heart, including the distal Purkinje fiber network. This discovery has enabled us to visualize the CCS, to isolate individual Purkinje cells, and to characterize their distinct physiological properties. One can now envision numerous strategies to decipher the mechanistic role of the Purkinje fiber network in the genesis and maintenance of clinically significant cardiac rhythm disturbances. Moreover, by analogy to neuronal systems, we suggest that Cntn2 may regulate CCS network formation and the excitability of its component cells.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Furley (University of Sheffield) for making the Cntn2-lacZ mice available to us and Dr. Takeshi Sakurai (Mount Sinai School of Medicine) for providing them.
Funding Sources: This work was supported by NIH R01HL64757, R01HL081336 and a New York State Stem Cell Science Award to G.I.F., a Glorney-Raisbeck Fellowship in Cardiovascular Disease to S.F.G, and NIH R01 HL58860 to P.A.B.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Jay V. The extraordinary career of Dr Purkinje. Arch Pathol Lab Med. 2000;124:662–663. doi: 10.5858/2000-124-0662-TECODP. [DOI] [PubMed] [Google Scholar]
- 2.Suma K. Sunao Tawara: a father of modern cardiology. Pacing Clin Electrophysiol. 2001;24:88–96. doi: 10.1046/j.1460-9592.2001.00088.x. [DOI] [PubMed] [Google Scholar]
- 3.Scheinman MM. Role of the His-Purkinje system in the genesis of cardiac arrhythmia. Heart Rhythm. 2009;6:1050–1058. doi: 10.1016/j.hrthm.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Bogun F, Good E, Reich S, Elmouchi D, Igic P, Tschopp D, Dey S, Wimmer A, Jongnarangsin K, Oral H, Chugh A, Pelosi F, Morady F. Role of Purkinje fibers in post-infarction ventricular tachycardia. J Am Coll Cardiol. 2006;48:2500–2507. doi: 10.1016/j.jacc.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi M, Kobayashi Y, Iwasaki YK, Morita N, Miyauchi Y, Kato T, Takano T. Novel mechanism of postinfarction ventricular tachycardia originating in surviving left posterior Purkinje fibers. Heart Rhythm. 2006;3:908–918. doi: 10.1016/j.hrthm.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Morishima I, Nogami A, Tsuboi H, Sone T. Verapamil-sensitive left anterior fascicular ventricular tachycardia associated with a healed myocardial infarction: changes in the delayed Purkinje potential during sinus rhythm. J Interv Card Electrophysiol. 2008;22:233–237. doi: 10.1007/s10840-008-9268-4. [DOI] [PubMed] [Google Scholar]
- 7.Sinha AM, Schmidt M, Marschang H, Gutleben K, Ritscher G, Brachmann J, Marrouche NF. Role of left ventricular scar and Purkinje-like potentials during mapping and ablation of ventricular fibrillation in dilated cardiomyopathy. Pacing Clin Electrophysiol. 2009;32:286–290. doi: 10.1111/j.1540-8159.2008.02233.x. [DOI] [PubMed] [Google Scholar]
- 8.Haissaguerre M, Shoda M, Jais P, Nogami A, Shah DC, Kautzner J, Arentz T, Kalushe D, Lamaison D, Griffith M, Cruz F, de Paola A, Gaita F, Hocini M, Garrigue S, Macle L, Weerasooriya R, Clementy J. Mapping and ablation of idiopathic ventricular fibrillation. Circulation. 2002;106:962–967. doi: 10.1161/01.cir.0000027564.55739.b1. [DOI] [PubMed] [Google Scholar]
- 9.Cerrone M, Noujaim SF, Tolkacheva EG, Talkachou A, O’Connell R, Berenfeld O, Anumonwo J, Pandit SV, Vikstrom K, Napolitano C, Priori SG, Jalife J. Arrhythmogenic mechanisms in a mouse model of catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2007;101:1039–1048. doi: 10.1161/CIRCRESAHA.107.148064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haissaguerre M, Extramiana F, Hocini M, Cauchemez B, Jais P, Cabrera JA, Farre J, Leenhardt A, Sanders P, Scavee C, Hsu LF, Weerasooriya R, Shah DC, Frank R, Maury P, Delay M, Garrigue S, Clementy J. Mapping and ablation of ventricular fibrillation associated with long-QT and Brugada syndromes. Circulation. 2003;108:925–928. doi: 10.1161/01.CIR.0000088781.99943.95. [DOI] [PubMed] [Google Scholar]
- 11.Li ZY, Wang YH, Maldonado C, Kupersmith J. Role of junctional zone cells between Purkinje fibres and ventricular muscle in arrhythmogenesis. Cardiovasc Res. 1994;28:1277–1284. doi: 10.1093/cvr/28.8.1277. [DOI] [PubMed] [Google Scholar]
- 12.Hatcher CJ, Basson CT. Specification of the cardiac conduction system by transcription factors. Circ Res. 2009;105:620–630. doi: 10.1161/CIRCRESAHA.109.204123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers DC, Fishman GI. Molecular and functional maturation of the murine cardiac conduction system. Trends Cardiovasc Med. 2003;13:289–295. doi: 10.1016/s1050-1738(03)00119-1. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S-S, Bruneau BG. Advances in developmental biology. Vol. 18. Elsevier B. V; 2008. Transcriptional control of the cardiac conduction system; pp. 219–258. [Google Scholar]
- 15.Nguyen-Tran VT, Kubalak SW, Minamisawa S, Fiset C, Wollert KC, Brown AB, Ruiz-Lozano P, Barrere-Lemaire S, Kondo R, Norman LW, Gourdie RG, Rahme MM, Feld GK, Clark RB, Giles WR, Chien KR. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell. 2000;102:671–682. doi: 10.1016/s0092-8674(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 16.Kupershmidt S, Yang T, Anderson ME, Wessels A, Niswender KD, Magnuson MA, Roden DM. Replacement by homologous recombination of the minK gene with lacZ reveals restriction of minK expression to the mouse cardiac conduction system. Circ Res. 1999;84:146–152. doi: 10.1161/01.res.84.2.146. [DOI] [PubMed] [Google Scholar]
- 17.Logan C, Khoo WK, Cado D, Joyner AL. Two enhancer regions in the mouse En-2 locus direct expression to the mid/hindbrain region and mandibular myoblasts. Development. 1993;117:905–916. doi: 10.1242/dev.117.3.905. [DOI] [PubMed] [Google Scholar]
- 18.Stroud DM, Darrow BJ, Kim SD, Zhang J, Jongbloed MR, Rentschler S, Moskowitz IP, Seidman J, Fishman GI. Complex genomic rearrangement in CCS-LacZ transgenic mice. Genesis. 2007;45:76–82. doi: 10.1002/dvg.20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE, Jalife J, Fishman GI. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128:1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers DC, Fishman GI. Toward an understanding of the genetics of murine cardiac pacemaking and conduction system development. Anat Rec A Discov Mol Cell Evol Biol. 2004;280:1018–1021. doi: 10.1002/ar.a.20077. [DOI] [PubMed] [Google Scholar]
- 21.Coppen SR, Kaba RA, Halliday D, Dupont E, Skepper JN, Elneil S, Severs NJ. Comparison of connexin expression patterns in the developing mouse heart and human foetal heart. Mol Cell Biochem. 2003;242:121–127. [PubMed] [Google Scholar]
- 22.Miquerol L, Meysen S, Mangoni M, Bois P, van Rijen HV, Abran P, Jongsma H, Nargeot J, Gros D. Architectural and functional asymmetry of the His-Purkinje system of the murine heart. Cardiovasc Res. 2004;63:77–86. doi: 10.1016/j.cardiores.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Gu H, Smith FC, Taffet SM, Delmar M. High incidence of cardiac malformations in connexin40-deficient mice. Circ Res. 2003;93:201–206. doi: 10.1161/01.RES.0000084852.65396.70. [DOI] [PubMed] [Google Scholar]
- 24.Kirchhoff S, Nelles E, Hagendorff A, Kruger O, Traub O, Willecke K. Reduced cardiac conduction velocity and predisposition to arrhythmias in connexin40-deficient mice. Curr Biol. 1998;8:299–302. doi: 10.1016/s0960-9822(98)70114-9. [DOI] [PubMed] [Google Scholar]
- 25.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ismat FA, Zhang M, Kook H, Huang B, Zhou R, Ferrari VA, Epstein JA, Patel VV. Homeobox protein Hop functions in the adult cardiac conduction system. Circ Res. 2005;96:898–903. doi: 10.1161/01.RES.0000163108.47258.f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Hirose M, Stuyvers BD, Dun W, Ter Keurs HE, Boyden PA. Function of Ca release Channels in Purkinje cells that survive in the Infarcted Canine Heart; a Mechanism for triggered Purkinje Ectopy. Circ Arrhythm Electrophysiol. 2008;1:387–395. doi: 10.1161/CIRCEP.107.758110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyden PA, Albala A, Dresdner KP., Jr Electrophysiology and ultrastructure of canine subendocardial Purkinje cells isolated from control and 24-hour infarcted hearts. Circ Res. 1989;65:955–970. doi: 10.1161/01.res.65.4.955. [DOI] [PubMed] [Google Scholar]
- 31.Liu N, Colombi B, Memmi M, Zissimopoulos S, Rizzi N, Negri S, Imbriani M, Napolitano C, Lai FA, Priori SG. Arrhythmogenesis in catecholaminergic polymorphic ventricular tachycardia: insights from a RyR2 R4496C knock-in mouse model. Circ Res. 2006;99:292–298. doi: 10.1161/01.RES.0000235869.50747.e1. [DOI] [PubMed] [Google Scholar]
- 32.Chambers J, Zhao J, Terracciano C, Bezzina C, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi J, Kooner M, Siedlecka U, Wass M, Dekker L, de Jong J, Sternberg M, McKenna W, Severs N, DeSilva R, Wilde A, Anand P, Yacoub M, Scott J, Elliot P, Wood J, Kooner J. Genetic Variation in SCN10a is Associated With Cardiac Conduction, Heart Block and Risk of Ventricular Fibrillation. Paper presented at: American Heart Association Annual Meeting; November 14–18 2009; Orlando, Florida. [Google Scholar]
- 33.Oosthoek PW, Viragh S, Mayen AE, van Kempen MJ, Lamers WH, Moorman AF. Immunohistochemical delineation of the conduction system. I: The sinoatrial node. Circ Res. 1993;73:473–481. doi: 10.1161/01.res.73.3.473. [DOI] [PubMed] [Google Scholar]
- 34.Walsh FS, Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- 35.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 36.Haissaguerre M, Shah DC, Jais P, Shoda M, Kautzner J, Arentz T, Kalushe D, Kadish A, Griffith M, Gaita F, Yamane T, Garrigue S, Hocini M, Clementy J. Role of Purkinje conducting system in triggering of idiopathic ventricular fibrillation. Lancet. 2002;359:677–678. doi: 10.1016/S0140-6736(02)07807-8. [DOI] [PubMed] [Google Scholar]
- 37.Wit AL, Bigger JT., Jr Possible electrophysiological mechanisms for lethal arrhythmias accompanying myocardial ischemia and infarction. Circulation. 1975;52:III96–115. [PubMed] [Google Scholar]
- 38.Deo M, Boyle P, Plank G, Vigmond E. Role of Purkinje system in cardiac arrhythmias. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:149–152. doi: 10.1109/IEMBS.2008.4649112. [DOI] [PubMed] [Google Scholar]
- 39.Vigmond E, Vadakkumpadan F, Gurev V, Arevalo H, Deo M, Plank G, Trayanova N. Towards predictive modelling of the electrophysiology of the heart. Exp Physiol. 2009;94:563–577. doi: 10.1113/expphysiol.2008.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen NR, Taylor JS, Scott LB, Guillery RW, Soriano P, Furley AJ. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr Biol. 1998;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- 41.Rasband MN, Shrager P. Ion channel sequestration in central nervous system axons. J Physiol. 2000;525(Pt 1):63–73. doi: 10.1111/j.1469-7793.2000.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traka M, Goutebroze L, Denisenko N, Bessa M, Nifli A, Havaki S, Iwakura Y, Fukamauchi F, Watanabe K, Soliven B, Girault JA, Karagogeos D. Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J Cell Biol. 2003;162:1161–1172. doi: 10.1083/jcb.200305078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malhotra JD, Tsiotra P, Karagogeos D, Hortsch M. Cis-activation of L1-mediated ankyrin recruitment by TAG-1 homophilic cell adhesion. J Biol Chem. 1998;273:33354–33359. doi: 10.1074/jbc.273.50.33354. [DOI] [PubMed] [Google Scholar]
- 44.Bhat MA. Molecular organization of axo-glial junctions. Curr Opin Neurobiol. 2003;13:552–559. doi: 10.1016/j.conb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Hashemi SM, Hund TJ, Mohler PJ. Cardiac ankyrins in health and disease. J Mol Cell Cardiol. 2009;47:203–209. doi: 10.1016/j.yjmcc.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohler PJ, Rivolta I, Napolitano C, LeMaillet G, Lambert S, Priori SG, Bennett V. Nav1.5 E1053K mutation causing Brugada syndrome blocks binding to ankyrin-G and expression of Nav1.5 on the surface of cardiomyocytes. Proc Natl Acad Sci U S A. 2004;10:17533–17538. doi: 10.1073/pnas.0403711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, Priori SG, Keating MT, Bennett V. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A. 2004;101:9137–9142. doi: 10.1073/pnas.0402546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dun W, Robinson R, Lowe J, Mohler P, Boyden P. Ankyrin-G participates in the remodeling of INa in myocytes of the EBZ of the infarcted heart. Biophys J. 2008;94:1. doi: 10.1371/journal.pone.0078087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.