Abstract
Background
Colobine monkeys constitute a diverse group of primates with major radiations in Africa and Asia. However, phylogenetic relationships among genera are under debate, and recent molecular studies with incomplete taxon-sampling revealed discordant gene trees. To solve the evolutionary history of colobine genera and to determine causes for possible gene tree incongruences, we combined presence/absence analysis of mobile elements with autosomal, X chromosomal, Y chromosomal and mitochondrial sequence data from all recognized colobine genera.
Results
Gene tree topologies and divergence age estimates derived from different markers were similar, but differed in placing Piliocolobus/Procolobus and langur genera among colobines. Although insufficient data, homoplasy and incomplete lineage sorting might all have contributed to the discordance among gene trees, hybridization is favored as the main cause of the observed discordance. We propose that African colobines are paraphyletic, but might later have experienced female introgression from Piliocolobus/Procolobus into Colobus. In the late Miocene, colobines invaded Eurasia and diversified into several lineages. Among Asian colobines, Semnopithecus diverged first, indicating langur paraphyly. However, unidirectional gene flow from Semnopithecus into Trachypithecus via male introgression followed by nuclear swamping might have occurred until the earliest Pleistocene.
Conclusions
Overall, our study provides the most comprehensive view on colobine evolution to date and emphasizes that analyses of various molecular markers, such as mobile elements and sequence data from multiple loci, are crucial to better understand evolutionary relationships and to trace hybridization events. Our results also suggest that sex-specific dispersal patterns, promoted by a respective social organization of the species involved, can result in different hybridization scenarios.
Background
With more than 50 species and due to some ecological adaptations, such as a ruminant-like chambered stomach to digest food rich in fiber, the Old World monkey subfamily Colobinae represents a diverse and enigmatic group of primates [1,2]. Colobines are predominantly arboreal and occur in forest and woodland habitats. They have experienced two major radiations, one in Africa with the genera Procolobus, Piliocolobus and Colobus, and a second in South and Southeast Asia comprising the langur genera Semnopithecus, Trachypithecus and Presbytis, and the odd-nosed monkey genera Rhinopithecus, Pygathrix, Nasalis and Simias [2]. However, their phylogenetic relationships are disputed [3-7], and recent molecular studies detected substantial gene tree discordance [8-10].
Traditionally, African and Asian genera are believed to form reciprocally monophyletic groups [1,2,11,12], though paraphyly has also been proposed [3-5]. Molecular investigations clearly confirm a common origin of Asian colobines and the odd-nosed monkey group [8-10], but evidence for monophyly of the langur group as well as for African colobines is still lacking. Moreover, nuclear and mitochondrial data indicate conflicting relationships among langur genera, and between langurs and the odd-nosed monkeys [8-10]. While nuclear data consistently link Semnopithecus and Trachypithecus to the exclusion of all other Asian colobines [9,10], mitochondrial data either do not resolve these relationships [9] or suggest a clade consisting of Presbytis and Trachypithecus [8].
Incongruent phylogenetic relationships among genes, like those detected among colobines are common in phylogenetic studies and could be explained by homoplasy, insufficient data, nucleotide composition, differential lineage sorting, or hybridization [13-21]. To ascertain which of these possibilities are responsible for the incongruence, information from various independent molecular loci can be helpful [22]. To date, only mitochondrial and X chromosomal data as well as presence/absence information of mobile elements, all based on an incomplete taxon sampling, are available for comparative phylogenetic studies in colobines [8-10,23]. Among all marker systems, mobile element insertions are a promising tool to uncover phylogenetic relationships among colobine genera. Compared to sole sequence data, mobile elements such as Short Interspersed Elements (SINEs) and Long Interspersed Elements (LINEs) exhibit advantages which make them ideal markers for phylogenetic reconstructions (for review see [24-30]). Accordingly, mobile elements are successfully applied in numerous primate phylogenetic studies [9,28,31-39].
In our study, we examined the presence/absence pattern of mobile elements and compared the inferred phylogeny with those derived from mitochondrial and nuclear sequence data (in total ~30,000 bp per genus). We extended available X chromosomal and mitochondrial genome data, and sequenced de novo five autosomal loci that map to different human chromosomes, and six Y chromosomal loci from all ten colobine genera. By combining results from different marker systems, we provide detailed insights into the evolutionary and biogeographic history of colobine monkeys, and show that different hybridization mechanisms might have been involved during the colobine radiation.
Results
Nuclear phylogeny
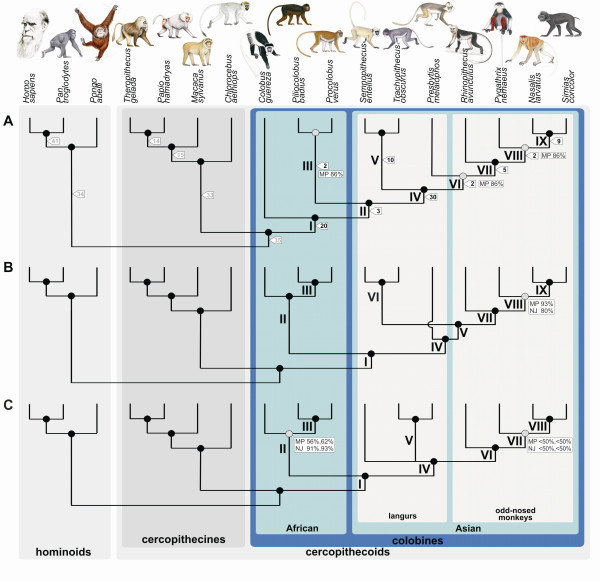
Eighty-three mobile elements are phylogenetically informative for colobines (Figure 1A, Additional file 1). Each of the following clades is strongly supported by at least five integrations: all colobines (clade I [A-I]), Asian colobines (A-IV), odd-nosed monkeys (A-VII), Trachypithecus and Semnopithecus (A-V), and Nasalis and Simias (A-IX). Three integrations were found in Piliocolobus and Procolobus and all Asian colobines (A-II), but not in Colobus. Two insertions suggested a sister grouping of Procolobus and Piliocolobus (A-III), Presbytis and the odd-nosed monkeys (A-VI), and a basal position of Rhinopithecus among the latter (A-VIII). Based on maximum-parsimony (MP) bootstrap analysis, most relationships were strongly supported (≥95%). Only the Piliocolobus/Procolobus (A-III), Presbytis/odd-nosed monkey (A-VI), and Pygathrix/Nasalis/Simias (A-VIII) clades gained relatively weak bootstrap values (86%). Based on alternative tree topology tests, different positions of the Piliocolobus/Procolobus clade and Presbytis among colobines were not rejected (P > 0.05), while relationships other than the most likely one were significantly rejected for all other taxa (P < 0.001, P < 0.05) (Additional file 2).
Figure 1.
Phylogenetic relationships among colobine and outgroup genera as inferred from different datasets. Panels refer to insertions of mobile elements (A), combined nuclear sequence data (B), and mitochondrial genome data (C). Roman numbers are used as branch identifiers and are discussed in the text. In A, numbers in flags represent the number of available mobile elements (black: colobine markers, grey: non-colobine markers). In B and C, all nodes are significantly supported by ML and Bayesian reconstructions (≥95%, 1.0). Black and grey dots on nodes indicate high (≥95%) and lower (<95%) branch support as obtained from MP (in A-C) and NJ (in B and C) reconstructions, respectively. Bootstrap values <95% are presented at respective nodes. In C, first and second values refer to those obtained from reconstructions using datasets mtDNA1 and mtDNA2, respectively.
Next, we performed phylogenetic analyses based on the concatenated nuclear sequence dataset, including five autosomal loci, six Y chromosomal loci and a fragment of the X chromosomal Xq13.3 region (see Methods for detailed locus description). We combined all nuclear sequence data, because heuristic search methods for individual loci produced no conflicting relationships (Additional file 3), and partition homogeneity tests revealed no significant difference in their evolutionary history (Y chromosomal loci combined: P = 0.2939; autosomal loci combined: P = 0.1543; all nuclear loci combined: P = 0.3559). Nucleotide composition of studied species was similar (Additional file 4). Phylogenetic reconstructions yielded identical and significantly supported branching patterns irrespectively of the applied algorithm (MP, neighbor-joining [NJ], maximum-likelihood [ML], Bayesian) (Figure 1B, for a phylogram see Additional file 5). Only the Pygathrix/Nasalis/Simias (B-VIII) clade had lower support values (MP: 93%, NJ: 80%, but ML: 98%, Bayesian posterior probabilities [PP]: 1.0). The resultant tree topology was mainly congruent with the mobile element-based phylogeny, but two cases of incongruence were obvious. First, in the nuclear sequence-based phylogeny, African (B-II) and Asian (B-IV) colobine genera formed reciprocally monophyletic clades and second, Presbytis represented a sister lineage to the other Asian genera (B-V). According to alternative tree topology tests (Additional file 2), paraphyly of African colobines with Piliocolobus/Procolobus being closer related to Asian colobines than to Colobus as well as various alternative positions of Presbytis among Asian colobines were not rejected (P > 0.05). However, affiliations of Presbytis to either Semnopithecus or Trachypithecus were rejected (P < 0.001).
Estimated divergence ages from the combined nuclear dataset (Table 1) and single loci (Additional file 6), both based on an a-priori fixed tree topology as obtained from mobile elements, differed slightly, most likely due to the general low variability in the studied loci (Additional file 7). However, estimates were in the same range suggesting that loci evolve at similar evolutionary rates (Additional file 8). According to our nuclear estimates, Colobus and Piliocolobus/Procolobus successively split off from Asian genera 10.93 million years ago (mya) and 10.73 mya, respectively (for 95% highest posterior densities see Table 1). The latter two separated 6.92 mya. In Asia, an initial split occurred 8.12 mya and led to a clade consisting of Trachypithecus and Semnopithecus, and a group containing Presbytis and the odd-nosed monkeys. Among the latter, Presbytis diverged 7.96 mya and the odd-nosed monkeys began differentiating 6.43 mya. The most recent splits among Asian genera occurred between Trachypithecus and Semnopithecus (2.56 mya) and between Nasalis and Simias (1.06 mya).
Table 1.
Estimation of divergence ages in mya (95% highest posterior density)
| node | nuclear DNA | mitochondrial DNA |
|---|---|---|
| cercopithecoids - hominoids | 24.39 (22.44-26.47) | 23.73 (21.88-25.94) |
| Pongo - Homo/Pan | 13.89 (12.80-14.95) | 13.58 (12.51-14.64) |
| Homo - Pan | 6.39 (5.85-7.01) | 6.18 (5.62-6.70) |
| cercopithecines - colobines | 15.50 (14.45-16.56) | 15.92 (14.11-17.79) |
| Cholorocebus - other cercopithecines | 9.47 (7.52-11.57) | 10.56 (8.78-12.29) |
| Macaca - Papio/Theropithecus | 6.59 (5.12-8.27) | 8.55 (6.82-10.03) |
| Papio - Theropithecus | 3.80 (3.20-4.38) | 3.97 (3.39-4.46) |
| Colobus - other colobines (A-I) | 10.93 (9.60-12.31) | - |
| Piliocolobus/Procolobus - Asian colobines (A-II) | 10.73 (9.38-12.04) | - |
| African - Asian colobines (C-I) | - | 10.90 (9.34-12.44) |
| Colobus - Piliocolobus/Procolobus (C-II) | - | 8.47 (6.83-9.88) |
| Piliocolobus - Procolobus (A-III, C-III) | 6.92 (4.38-9.35) | 6.58 (4.99-8.04) |
| Asian colobines (A-IV, C-IV) | 8.12 (7.14-9.16) | 8.91 (7.43-10.23) |
| Trachypithecus - Semnopithecus (A-V) | 2.56 (1.25-4.22) | - |
| Presbytis - odd-nosed monkeys (A-VI) | 7.96 (6.93-8.95) | - |
| Presbytis - Trachypithecus (C-V) | - | 7.45 (5.88-8.86) |
| odd-nosed monkeys (A-VII, C-VI) | 6.43 (5.03-7.75) | 6.91 (5.60-8.20) |
| Pygathrix - Nasalis/Simias (A-VIII, C-VII) | 5.66 (4.22-7.01) | 6.23 (5.11-7.38) |
| Nasalis - Simias (A-IX, C-VIII) | 1.06 (0.44-1.81) | 1.88 (1.21-2.45) |
Mitochondrial phylogeny
Mitochondrial and nuclear datasets were not combined, because the partition homogeneity test suggested that both track different evolutionary histories (P = 0.0002). Thus, mitochondrial sequence data were analyzed separately. For both alignments (mtDNA1, mtDNA2; for details about alignments see Methods), we observed a major shift in nucleotide composition between colobine and non-colobine representatives (Additional file 4). Both alignments produced identical and significantly supported branching patterns among genera (Figure 1C, for a phylogram see Additional file 5). Only the Pygathrix/Nasalis/Simias (C-VII) and African colobine (C-II) clades gained low MP (<50%, <50%, 56%, 62%) and NJ (<50%, <50%, 91%, 93%) bootstrap values, but ML and Bayesian reconstructions provided strong support for both nodes (96%, 100%; 1.0, 1.0). In principal, the tree topology was identical to those obtained from mobile elements and nuclear sequence data. However, as in the nuclear sequence tree, mitochondrial data suggested African (C-II) and Asian (C-IV) colobines as reciprocal monophyletic clades. Moreover, Asian colobines further diverged into a lineage leading to the odd-nosed monkeys (C-VI), a lineage comprising Trachypithecus and Presbytis (C-V), and finally a lineage with solely Semnopithecus, while the relationships among these three lineages remained unresolved.
According to alternative tree topology tests, paraphyly of African colobines with Piliocolobus/Procolobus being closer related to Asian colobines than to Colobus was rejected (P < 0.001, Additional file 2). Among Asian colobines, relationships in which Trachypithecus and Presbytis do not form a monophyletic clade were also rejected (P < 0.001, P < 0.05), as well as a close relationship of Trachypithecus and Semnopithecus (P < 0.01). In contrast, different positions of Semnopithecus among Asian colobines were similarly likely (P > 0.05).
Divergence age estimates from mitochondrial data were similar to nuclear estimates in case where identical branching patterns were obtained (Table 1). According to mitochondrial data, African and Asian colobine lineages were separated 10.90 mya. In Africa, Colobus represents the first split (8.47 mya), followed by the divergence of Piliocolobus and Procolobus (6.58 mya). The major Asian split leading to the three lineages Semnopithecus, Trachypithecus/Presbytis and the odd-nosed monkeys occurred 8.91 mya. Trachypithecus diverged from Presbytis 7.45 mya. The diversification of odd-nosed monkeys into genera started 6.91 mya and ended with the split between Nasalis and Simias 1.88 mya.
Inferring hybridization in the presence of incomplete lineage sorting
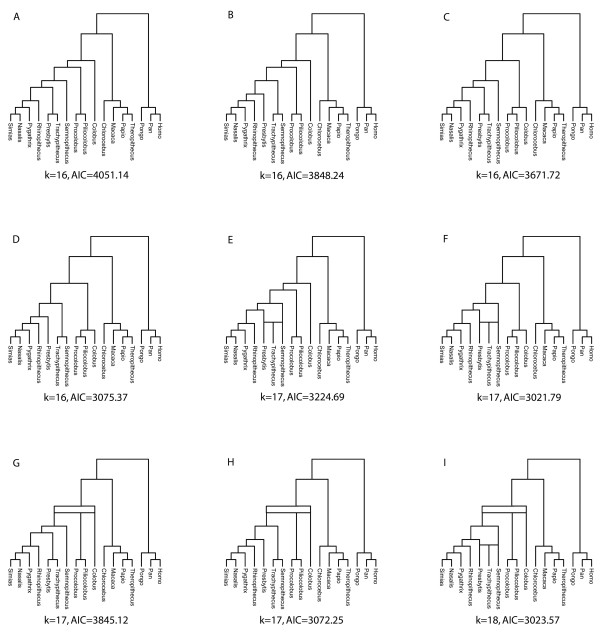
To assess the possible reasons for the incongruence between the nuclear and mitochondrial trees, we applied the method proposed by Kubatko [40]. The method assumes that incomplete lineage sorting (ILS) explains observed gene tree incongruence to some extent, and seeks to determine whether all variation in observed gene trees can be explained by ILS alone, as modeled by the coalescent process, or whether hybridization helps to explain significantly more the observed variation. Then, the Akaike information criterions (AIC) in each model (may or may not include hybridization scenarios) were compared to determine the best-fit model. For our data, two possible hybridization events were hypothesized. The first involved Trachypithecus, with parental taxa Semnopithecus and Presbytis, while the second involved the clade containing Piliocolobus and Procolobus, Colobus and the ancestor of Asian colobines.
By comparing the results from models with or without the hybridization events, the best-fit model (AIC = 3021.79, Figure 2F) was a tree in which Trachypithecus is the result of hybridization between Presbytis and Semnopithecus. The second best-fit model (AIC = 3023.57, Figure 2I) comprised the tree that includes both tested hybridization events. AIC values for all seven other models were considerably higher (3072.25 - 4051.14). Since AIC values for the scenarios presented in Figures 2F and 2I were the lowest and were within 2 of one another, both were considered plausible explanations for the observed gene tree discordances [41]. It is worth pointing out that the model used here to compute the AIC assumes that ILS is a possible source of gene trees incongruence. Since the two best-fit models include at least one hybridization event, it is clear that ILS alone does not adequately describe the extent of incongruence in the observed gene trees.
Figure 2.
The nine alternative hybridization scenarios compared in the coalescent framework. Beneath each tree, the number of parameters in the model (k) is given as well as the AIC. The lowest AIC values are observed for trees F and I, which indicate a similar fit for these scenarios.
Discussion
By combining presence/absence analysis of mobile elements with autosomal, X chromosomal, Y chromosomal and mitochondrial sequence data, the present study provides comprehensive insights into the evolutionary history of colobines. Most relationships are resolved and strongly supported by mobile elements and sequence data. Moreover, relationships and estimated divergence ages as obtained from different datasets are mainly congruent and in agreement with earlier studies [8-10,23,42-45]. Our study, however, also reveals significant discrepancies among gene trees. First, mitochondrial and nuclear sequence data suggest a monophyletic African colobine clade, while mobile elements provide evidence for a closer connection of the Piliocolobus/Procolobus clade to Asian genera than to Colobus. Second, mobile elements indicate close relationships between Semnopithecus and Trachypithecus, and between Presbytis and the odd-nosed monkeys. Nuclear sequence data support the former clade, but suggest Presbytis as basal among Asian colobines. In contrast, in the mitochondrial phylogeny, Presbytis and Trachypithecus are displayed as sister lineages, while the position of Semnopithecus remains ambiguous.
Possible explanations for gene tree discordance
Inadequate data, homoplasy, nucleotide composition, ILS or hybridization could be potential explanations for the observed differences [13-21]. For the mitochondrial dataset, at least for the African and Presbytis/Trachypithecus clades, incorrect branching patterns due to inadequate data or homoplasy are unlikely, since sufficient phylogenetic resolution with long internal branches is obtained. Likewise, a shift in nucleotide composition and differential sorting of ancestral mitochondrial lineages is implausible. Since the major shift in nucleotide composition was detected between colobines and non-colobines, it cannot be responsible for gene tree discordances among colobines. If the African and Presbytis/Trachypithecus clades are indeed the result of incomplete sorting of mitochondrial lineages, the mitochondrial divergence between respective genera should predate the nuclear splitting times, which is not the case (African colobines: 10.93 mya nuclear vs. 8.47 mya mitochondrial; Presbytis - Trachypithecus: 8.12 mya nuclear vs. 7.45 mya mitochondrial). However, the unresolved position of Semnopithecus among Asian colobines might have been affected by one or several of the above mentioned factors, or alternatively, might be the result of a true radiation-like divergence of lineages. For nuclear data, these factors are unlikely explanations as well for the branching of Trachypithecus and Semnopithecus, because ten independent insertions and sequence data from 12 nuclear loci clearly confirm their close relationship. More challenging are explanations for the discordant positions of Presbytis and the African genera among colobines in phylogenies revealed by mobile elements and nuclear sequence data. Mixed genomes due to differentially selected genes cannot be excluded, but interestingly, both mobile elements and nuclear sequence data (as revealed from single locus analysis) show no conflicting phylogenies themselves. Most prominent, however, the mobile element-based phylogeny is not rejected by nuclear sequence data, indicating that insufficient informative sites, as also suggested by the low resolution of phylogenetic relationships in single-locus analysis, in the latter dataset might display incorrect relationships. For the integration of mobile elements, homoplasy is typically regarded as minimal [25,28,30], but ILS has been reported [36,39]. Only two and three integrations support the branching of Presbytis with odd-nosed monkeys and the paraphyly of African colobines, and alternative relationships cannot be rejected statistically. However, no inconsistent elements were detected and subtractive hybridizations specifically set up to screen for African colobine and Trachypithecus/Presbytis monophyly markers revealed no equivalent insertions. Accordingly, ILS seems to be an unreasonable explanation for our findings. Since the mobile element-based phylogeny is not rejected by nuclear sequence data and due to their reliability as molecular-cladistic markers, the phylogeny suggested by mobile elements is assumed to reflect the true nuclear phylogeny of colobines, although we explicitly note that mosaic genomes cannot be excluded.
Because all above-mentioned factors provide no sufficient explanation for the herein detected discordances between mitochondrial and nuclear phylogenies, we favor ancestral hybridization as the main reason for the discordant pattern. Furthermore, comparisons of models with and without hybridization in a model selection framework strongly support hybridization in the presence of ILS over models of ILS alone. In other words, even after ILS was taken into account as a factor in the observed incongruence among gene trees, we still found support for hybridization in the evolutionary history of these taxa. This refers at least to Asian colobines, but hybridization among African colobines cannot be excluded either by the method we applied here.
Hybridization hypothesis
Although bidirectional hybridization, which would be indicated by mixed genomes, cannot be excluded with our data, a female introgression event is hypothesized for African colobines. The direction of gene flow remains obscure due to the rapid diversification of the colobine ancestor in Africa, but female introgression from Piliocolobus/Procolobus into Colobus is indicated and gains further support by some biological data [1,2]. In contrast to Colobus, females in Piliocolobus and Procolobus tend to leave their natal groups, which was most likely also the case in their ancestor [1], and Colobus males are on average larger than Piliocolobus and Procolobus males [1], thus increasing the chance of hybridization between Colobus males and Piliocolobus/Procolobus females. Moreover, hybridization between both ancestral lineages is in principal possible, because (at least nowadays) they occur in sympatry over wide ranges of their distribution [1,2]. Accordingly, after the successive separation of Colobus and Piliocolobus/Procolobus from the Asian colobine ancestor, Piliocolobus/Procolobus females might have entered Colobus populations and hybridized with their males. Backcrossing of hybrid females with resident Colobus males might has led to the fixation of the Piliocolobus/Procolobus mitochondrial lineage in the hybrid population, while the original nuclear genome of Colobus increased again in every generation.
For Asian langurs, we propose male introgression from Semnopithecus into Trachypithecus followed by nuclear swamping. Both genera are similar in their morphology and general appearance [2,46,47], but males in Semnopithecus are larger than in Trachypithecus [1]. Moreover, hybridization events due to (at least nowadays) partially overlapping ranges are generally possible [1,2]. Accordingly, after an initial separation, Semnopithecus males, which leave their natal group like most other primate males [1,48], might have invaded Trachypithecus populations and hybridized successfully with the resident females. By backcrossing with further invading Semnopithecus males over a longer period, the Trachypithecus population might have accumulated nuclear material of Semnopithecus (nuclear swamping), while the mitochondrial genome remained Trachypithecus-like.
Biogeographic implications
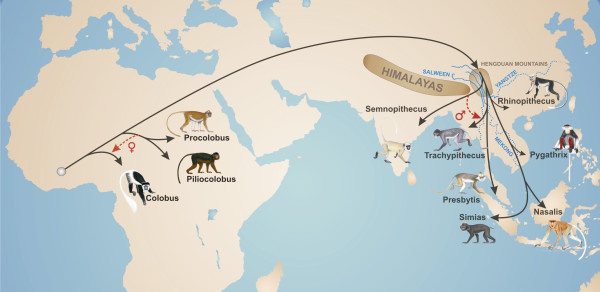
By combining the available information, we develop the following extended dispersal scenario for colobines (Figure 3). The origin of the subfamily is most likely in Africa, which is in agreement with earlier suggestions [1,49]. On the African continent, Colobus split off first from the main stem ~10.93 mya, followed shortly afterwards by the progenitor of Piliocolobus and Procolobus. After this initial separation, hybridization between both lineages might have lasted until finally both mitochondrial lineages diverged (~8.47 mya). Presumably, respective splitting and hybridization events took place in western Africa, because all three genera occur there in sympatry [1,2], and the most ancient splits among Piliocolobus and Colobus species are also found there [45]. The Asian colobine ancestor most likely invaded Eurasia via an emerging land bridge connecting Africa and the Arabian Peninsula in the late Miocene [49,50]. Whether a route into eastern Asia north or south of the Himalayas was chosen is a matter of speculation, but north of the Himalayas, on the Tibetan plateau, colobine fossils from the late Miocene were found, which is not the case south of the Himalayas [1]. Although not confirmed, the Hengduan Mountains in the border region of today's Burma, India and China might have been a possible diversification hotspot [4,51,52]. In the region, all the larger Southeast Asian rivers (Mekong, Salween, Yangtze) rise, which are all well-known as barriers for arboreal primates [53] and are all known to exist since at least the early Miocene [54]. Semnopithecus might have diverged as first lineage and invaded the Indian subcontinent. Subsequently, the progenitor of Presbytis and Trachypithecus separated from the odd-nosed monkey ancestor and migrated into southern mainland Asia. Afterwards, Presbytis diverged from Trachypithecus and entered first the Malaysian peninsular and later on Sundaland during periods of lowered sea levels [55]. Trachypithecus and Semnopithecus came into secondary contact and might have hybridized until the earliest Pleistocene. A potential contact zone could be the region of today's Bangladesh, Burma and the northeast of India, which is suggested as hybridization area for several primate species [9,44,56]. On the Asian mainland, odd-nosed monkeys successively migrated from China to the south and expanded their range into Indochina and Sundaland in the latest Miocene. The migration into Sundaland was probably via land bridges connecting the mainland with Sundaland islands during periods of lowered sea levels [55]. Finally, Nasalis on Borneo and Simias on the Mentawai islands west of Sumatra diverged in the Pleistocene. Due to the dating discrepancy (mitochondrial data: 1.88 mya, nuclear data: 1.06 mya), further gene flow between both genera after the initial separation cannot be excluded, especially considering that migration was repeatedly possible via land bridge connections during the Pleistocene [55].
Figure 3.
Dispersal scenario for colobine monkeys. Colobines most likely originated in western Africa. After the successive split of Colobus (~10.9 mya) and a progenitor of Piliocolobus/Procolobus (~10.7 mya) from the ancestor of Asian colobines, gene flow between both African lineages via female introgression from the Piliocolobus/Procolobus progenitor into Colobus occurred until ~8.5 mya (displayed by red-dashed arrow). During the late Miocene, colobines invaded eastern Asia most likely via a route north of the Himalayas. After their arrival at the Hengduan Mountains, Asian colobines diversified into a lineage comprising a progenitor of the odd-nosed monkeys and Trachypithecus/Presbytis, and of Semnopithecus, which later colonized the Indian subcontinent. Shortly afterwards, Trachypithecus/Presbytis split off from odd-nosed monkeys, and migrated to southern mainland Asia, before finally both genera diverged from each other. In the region of today's Burma, Bangladesh and India, Semnopithecus and Trachypithecus came into secondary contact and hybridized until ~2.6 mya (displayed by red-dashed arrow). In the latest Miocene, odd-nosed monkeys migrated from China to the south and expanded their range into Indochina and Sundaland. Nasalis and Simias finally separated from each other 1.1-1.9 mya.
Conclusion
Our study gives new and most comprehensive insights into the evolutionary history of colobine monkeys, and suggests hybridization among ancestral lineages as the most likely cause for the observed phylogenetic incongruences. Only the combination of maternally, paternally and bi-parentally inherited markers as well as the combination of sequence data with presence/absence patterns of mobile elements proved to be an adequate and reliable phylogenetic approach, particularly in revealing hybridization events. However, data from additional nuclear loci and a broader taxonomic sampling is required to fully understand hybridization mechanisms in colobines.
Hybridization among taxa is traditionally recognized as a factor leading to limited diversification, reproductive isolation and lowered fitness [57,58], whereas our and earlier studies clearly indicate that hybridization played a prominent role in diversification and speciation of primates (for review see [59,60]). Hybridization events are genetically confirmed within all major primate lineages, mainly among species (e.g., [56,61-65]) but also between genera (e.g., [9,44,66]). Even for the human lineage, hybridization has been suggested as an important evolutionary mechanism [67-69].
Since male dispersal and female philopatry predominates in primates [48], male introgression, and if intensive backcrossing of hybrids with more invading males occurs, followed by nuclear swamping would be the most likely hybridization scenario. In fact, the hybridization among Asian langur genera is most likely the result of such an event. However, as proposed for African colobines, alternative mechanisms (e.g. female introgression) could also occur, promoted by a respective social organization, where female migration predominates.
Methods
Sample collection and DNA extraction
Blood, tissue or fecal samples from representatives of all ten colobine genera (Colobus, Piliocolobus, Procolobus, Presbytis, Trachypithecus, Semnopithecus, Rhinopithecus, Pygathrix, Nasalis, Simias) and several non-colobine taxa (Macaca, Papio, Theropithecus, Chlorocebus, Pongo, Pan) were obtained from specimens kept in zoos or breeding facilities, or collected in the field (Table 2). Sample collection was conducted according to relevant German and international guidelines, including countries where we collected samples. Fecal samples were collected in a non-invasive way without disturbing, threatening or harming the animals. Blood samples were taken by veterinarians for diagnostic reasons to check the health status of the respective individuals, and tissue samples were obtained only from deceased specimens. Total genomic DNA was extracted with the DNeasy Blood & Tissue or QIAamp DNA Stool Mini kits from Qiagen following standard procedures.
Table 2.
Origin and sample type of studied species
| species | origin | sample type |
|---|---|---|
| Colobus guereza | Cologne zoo, Germany | tissue |
| Piliocolobus badius | Taï National Park, Ivory Coast | tissue |
| Procolobus verus | Taï National Park, Ivory Coast | tissue |
| Semnopithecus entellus | Dresden zoo, Germany | blood |
| Trachypithecus obscurus | Wuppertal zoo, Germany | blood |
| Presbytis melalophos | Howletts Wild Animal Park, Great Britain | tissue |
| Pygathrix nemaeus | Cologne zoo, Germany | tissue |
| Rhinopithecus avunculus | Endangered Primate Rescue Center, Vietnam | tissue |
| Nasalis larvatus | Wilhelma Stuttgart, Germany | blood |
| Simias concolor | Siberut Conservation Programme, Indonesia | feces |
| Macaca sylvanus | Nuremberg zoo, Germany | blood |
| Papio hamadryas | Munich zoo, Germany | blood |
| Theropithecus gelada | Duisburg zoo, Germany | blood |
| Chlorocebus aethiops | Paul-Ehrlich-Institute, Germany | blood |
| Pongo abelii | Nuremberg zoo, Germany | blood |
| Pan troglodytes | Munich zoo, Germany | blood |
Analysis of mobile elements
Due to their high copy number (~one million) and relatively small size (~300 bp), the primate specific Alu elements were selected as molecular-cladistic markers. The presence or absence of mobile elements in different colobines at specific loci was tested via PCR using primers occupying the flanking region of the insertion site. Details on analyzed loci, primers and presence/absence pattern of mobile elements in studied species are listed in Additional file 1. For most loci, sequencing was neglected, but in relevant cases the insertion orthology was confirmed by sequencing, and direct repeats flanking the insertion as well as the original target site prior to transposition were traced.
In our study, we included published markers [9,23,35], which were further examined in previously untested genera, and newly detected integration loci (Additional file 1). Therefore, we performed subtractive hybridizations following described methods [9]. To avoid biased hybridization results, various species combinations were used as tracer and driver (hybridization 1: tracer Nasalis/Pygathrix, driver Presbytis; hybridization 2: tracer Nasalis/Pygathrix, driver Semnopithecus; hybridization 3: tracer Trachypithecus/Presbytis, driver Pygathrix; hybridization 4: tracer Presbytis, driver Semnopithecus; hybridization 5: tracer Piliocolobus/Colobus, driver Pygathrix). Besides Alu insertions, a LINE present in Piliocolobus and Procolobus in the studied Xq13.3 fragment was additionally applied as marker (Additional file 1).
Phylogenetic reconstructions using the MP algorithm were conducted in PAUP v4.0b10 [70]. Presence of an integration was coded as 1, its absence as 0, and missing data as '?'. Internal node support was obtained via a heuristic search with 10,000 bootstrap replications. To evaluate the reliability of the depicted relationships among colobines, various alternative tree topologies (Additional file 2) were assessed with the Kishino-Hasegawa test [71] with full optimization and 1,000 bootstrap replications in PAUP.
Amplification and sequencing of nuclear loci
Inter-exonic intron and exonic sequences were generated for six single-copy genes of the Y chromosome, five autosomal loci, and a fragment of the X chromosomal Xq13.3 region. With exception of the SRY gene (sex-reversal, Y chromosome), all other Y chromosomal loci (DBY5: Dead Box, intron 5; SMCY7: SMC mouse homologue, intron 7; SMCY11: SMC mouse homologue, intron 11; UTY18: ubiquitous TPR motif, intron 18; ZFYLI: Zinc finger, last intron) have homologues on the X chromosome (X degenerate). As autosomal loci, we selected intron 11 of the von Willebrand Factor (vWF11), located on human chromosome 12, intron 3 of the serum albumin gene (ALB3, human chromosome 4), intron 3 of the interstitial retinol-binding protein (IRBP3, human chromosome 10), intron 1 of the transition protein 2 (TNP2, human chromosome 16) and intron 1 of the transthyretin gene (TTR1, human chromosome 18). SRY, DBY5, SMCY7, SMCY11, UTY18, vWF11 and a ~4,300 bp fragment of the Xq13.3 region were amplified using primers and PCR conditions as described [10,72-75] (Additional file 9). For the amplification of ZFYLI, ALB3, IRBP3, TNP2 and TTR1, new primers (Additional file 9) were designed on the basis of available primate sequences in GenBank. PCR conditions for the latter comprised a pre-denaturation step at 94°C for 2 min, followed by 40 cycles each with denaturation at 94°C for 1 min, annealing at varying temperatures (Additional file 9) for 1 min, and extension at 72°C for 2 min. At the end, a final extension step at 72°C for 5 min was added. The results of all PCR amplifications were checked on 1% agarose gels. PCR products were cleaned with the Qiagen PCR Purification kit and subsequently sequenced on an ABI 3130 × l sequencer using the BigDye Terminator Cycle Sequencing kit. Alignments and sequences are available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S11179) and GenBank, respectively (for GenBank accession numbers see Additional file 10).
Amplification and sequencing of mitochondrial genomes
To reduce the likelihood of amplifying nuclear pseudogenes (numts), complete mitochondrial genomes from four colobine genera (Rhinopithecus, Pygathrix, Nasalis, Procolobus) were generated following an approach in which two overlapping ~10,000 bp long fragments were amplified via long-range PCR [8,43]. Due to degradation of DNA extracted from faeces, the mitochondrial genome of Simias was amplified via five overlapping fragments, each with a size of ~5,000 bp. All long-range PCRs were performed with the SuperTaq Plus polymerase from Ambion following protocols of the supplier and primers as described [8,43]. Long-range PCR amplicons were separated on 1% agarose gels, excised from the gel, purified with the Qiagen Gel Extraction kit and used as template for nested PCRs. PCR conditions for all nested PCR amplifications were identical and comprised a pre-denaturation step at 94°C for 2 min, followed by 30 cycles each with denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1.5 min. At the end, a final extension step at 72°C for 5 min was added. Nested PCR products (900-1,200 bp in length) were cleaned with the Qiagen PCR Purification kit and sequenced on an ABI 3130 × l sequencer. Sequences were assembled with Geneious v4.6.1 [76]. No inconsistent positions in overlapping regions were detected and all protein-coding genes were correctly translated. Annotation of mitochondrial genomes was conducted with the online program DOGMA [77] and manually inspected. Alignment and sequences are available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S11179) and GenBank, respectively (for GenBank accession numbers see Additional file 10).
Statistical analysis of sequence data
For phylogenetic reconstructions, all datasets comprised 17 sequences including each one representative of the ten colobine genera (Colobus, Piliocolobus, Procolobus, Trachypithecus, Semnopithecus, Presbytis, Rhinopithecus, Pygathrix, Nasalis, Simias), four cercopithecine genera (Papio, Theropithecus, Macaca, Chlorocebus), and three hominoid genera (Homo, Pan, Pongo), which were used as outgroup taxa. To complete datasets, we partly implemented sequences from GenBank (Additional file 10). Alignments for individual loci were generated with MAFFT v6 [78] and corrected by eye. In all alignments, poorly aligned positions and indels were removed with Gblocks v0.91b [79] using default settings (Additional file 8). For the mitochondrial dataset, also the D-loop region was excluded (dataset mtDNA1) and a second alignment, generated in Mesquite v2.6 [80], included solely protein-coding genes (dataset mtDNA2). For all datasets, uncorrected pairwise differences were estimated in PAUP (Additional file 7). Nucleotide composition for all and only parsimony-informative positions for the combined nuclear and both mitochondrial alignments was also estimated in PAUP (Additional file 4). To test whether datasets can be combined, we performed partition homogeneity tests in PAUP with 10,000 replications.
Phylogenetic trees were constructed with MP and NJ algorithms as implemented in PAUP as well as with ML and Bayesian algorithms, using the programs GARLI v0.951 [81] and MrBayes v3.1.2 [82,83]. For MP analyses, all characters were treated as unordered and equally weighted throughout. A heuristic search was performed with the maximum number of trees set to 100. For NJ, ML and Bayesian reconstructions, the optimal nucleotide substitution models for each locus and concatenated datasets were chosen using AIC as implemented in MODELTEST v3.7 [84] (Additional file 8). Relative support of internal nodes was assessed by bootstrap analyses with 10,000 (MP, NJ) or 500 replications (ML). In GARLI, only the model specification settings were adjusted according to the respective concatenated dataset, while all other settings were left at their default value. ML majority-rule consensus trees were calculated in PAUP. For Bayesian reconstructions, the datasets were partitioned treating each locus separately and each with its own substitution model. The solely protein-coding alignment of the mitochondrial genome (mtDNA2) was partitioned into codon positions. We used four independent Markov Chain Monte Carlo (MCMC) runs with the default temperature of 0.1. Four repetitions were run for 10,000,000 generations with tree and parameter sampling occurring every 100 generations. The first 25% of samples were discarded as burnin, leaving 75,001 trees per run. PPs for each split and a phylogram with mean branch lengths were calculated from the posterior density of trees.
To evaluate the reliability of obtained relationships among colobines, various alternative tree topologies (Additional file 2) were tested with the Shimodaira-Hasegawa test [85] with full optimization and 1,000 bootstrap replications in PAUP.
Divergence age estimation
A Bayesian MCMC method, which employs a relaxed molecular clock approach [86], as implemented in BEAST v1.4.8 [87], was used to estimate divergence times. Therefore, a relaxed lognormal model of lineage variation and a Yule prior for branching rates was assumed. Divergence times were calculated for each locus separately and for the combined nuclear dataset. The latter was partitioned treating each locus as distinct unit. The mitochondrial alignment comprising solely protein-coding genes (mtDNA2) was partitioned into codon positions and the substitution model, rate heterogeneity and base frequencies were unlinked across codon positions. Optimal nucleotide substitution models were chosen using AIC in MODELTEST.
As calibrations we used the fossil-based divergence between Homo and Pan, which has been dated at 6-7 mya [88-90], the separation of Pongo from the Homo/Pan lineage ~14 mya [91], the split between Theropithecus and Papio ~4 mya [92,93], and the divergence of hominoids and cercopithecoids ~24 mya [94-96]. Instead of hardbounded calibration points, we used the published dates as a normal distribution prior for the respective node. For the Homo - Pan divergence, this translates into a normal distribution with a mean of 6.5 mya and a standard deviation (SD) of 0.5 mya, for the separation of Pongo from the Homo/Pan clade into a mean of 14.0 mya and a SD of 1.0 mya, for the Theropithecus - Papio split into a mean of 4.0 mya and a SD of 0.5 mya, and for the hominoid - cercopithecoid divergence into a mean of 24 mya and a SD of 2 mya.
Since the estimation of phylogenetic relationships was not the main aim of this analysis, we used an a-priori fixed tree topology as obtained from mobile elements (Figure 1A) for the calculation from nuclear sequence data. Four replicates were run for 10,000,000 generations with tree and parameter sampling occurring every 100 generations. The adequacy of a 10% burnin and convergence of all parameters were assessed by visual inspection of the trace of the parameters across generations using TRACER v1.4.1 [97]. Subsequently, the sampling distributions were combined (25% burnin) using the software LogCombiner v1.4.8 and a consensus chronogram with node height distribution was generated and visualized with TreeAnnotator v1.4.8 and FigTree v1.2.2 [98].
Inferring hybridization in the presence of incomplete lineage sorting
Statistical support for putative hybridization scenarios was assessed with the method proposed by Kubatko [40], in which statistical model selection techniques (e.g., AIC) are used to compare species trees that may or may not include hybridization scenarios. For our data, we hypothesized two possible hybridization events (for details see Results). The estimated gene trees used as input were those derived from single locus tree reconstructions (Additional file 3) and branch lengths as estimated in BEAST. To estimate evolutionary rates for individual loci, we followed the suggestion of Yang [99] (see also [100]) and computed for each gene the average pairwise sequence divergence of each ingroup (colobine) sequence to the outgroup (non-colobine) taxa. We then assigned to each locus a rate that was calculated by dividing the mean pairwise divergence for that locus by the median of the entire set of pairwise divergences (Additional file 8). To convert gene tree branch lengths to coalescent units, we considered two effective population sizes, 50,000 and 100,000, and used a generation time of 5 years. Since the results were identical in terms of the trees preferred, we show here the results only for effective population size 50,000. For haploid loci (mitochondrial genome, Y chromosomal loci), we additionally divided the rate by 2 (see [100]). We compared a total of nine species trees (four corresponding to no hybridization, four corresponding to single hybridization events, and one that included both hybridization scenarios, Figure 2). The AIC was computed for each tree using the STEM software [100]. Models with AIC values within 2 of one another were regarded as providing similar fit to the data [41].
Authors' contributions
CR designed the study, collected samples, did laboratory work, analyzed data, and wrote the paper. DZ designed the study, analyzed data, and wrote the paper. LSK, JX, MAB and MB analyzed data and wrote the paper. CS, MY, DM did laboratory work. SDN and LW wrote the paper. FHL, TZ, DPF and TN provided valuable samples and wrote the paper. MO did laboratory work, analyzed data and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Additional Table 1. Presence/absence pattern, location, primers and PCR product sizes of mobile elements
Additional Table 2. Alternative tree topology tests
Additional Figure 1. Single-locus phylogenetic trees (80% majority rule)
Additional Figure 2. Nucleotide composition of both mitochondrial and the combined nuclear datasets
Additional Figure 3. Phylograms based on the mitochondrial and combined nuclear datasets
Additional Table 3. Divergence ages in mya estimated for each locus separately
Additional Table 4. Uncorrected pairwise nucleotide differences for each locus
Additional Table 5. Locus-specific information including alignment length, number of variable sites, selected substitution model and estimated evolutionary rates
Additional Table 6. Primers and PCR conditions for the amplification of nuclear loci
Additional Table 7. GenBank accession numbers
Contributor Information
Christian Roos, Email: croos@dpz.eu.
Dietmar Zinner, Email: dzinner@gwdg.de.
Laura S Kubatko, Email: lkubatko@stat.osu.edu.
Christiane Schwarz, Email: cschwarz@dpz.eu.
Mouyu Yang, Email: myang@dpz.eu.
Dirk Meyer, Email: dirk.meyer@redape.de.
Stephen D Nash, Email: snash@ms.cc.sunysb.edu.
Jinchuan Xing, Email: jxing@genetics.utah.edu.
Mark A Batzer, Email: mbatzer@lsu.edu.
Markus Brameier, Email: brameier@dpz.eu.
Fabian H Leendertz, Email: LeendertzF@rki.de.
Thomas Ziegler, Email: tziegl@dpz.eu.
Dyah Perwitasari-Farajallah, Email: witafar@yahoo.com.
Tilo Nadler, Email: t.nadler@mail.hut.edu.vn.
Lutz Walter, Email: lwalter@gwdg.de.
Martin Osterholz, Email: mosterh@gwdg.de.
Acknowledgements
We thank the zoos in Cologne, Dresden, Duisburg, Howletts, Munich, Nuremberg, Stuttgart and Wuppertal, and Roland Plesker and Werner Schempp for providing samples. We are also grateful to our colleagues at Bogor Agricultural University (IPB) and the Indonesian Institute for Science (LIPI) as well as the Ivorian authorities, the Ministry of the Environment and Forests, the Ministry of Research, the directorship of Taï National Park and the Swiss Research Centre for providing the necessary field research permits and documents. No international and national rules and regulations have been violated during sampling and shipping. We thank Vanessa Roden, Laura Zidek, Linn Groeneveld, Daniel Stahl and two anonymous reviewers for valuable comments on an earlier version of the manuscript and statistical help. This work was supported by the German Primate Center and the Biodiversitäts-Pakt of the Wissenschaftsgemeinschaft Gottfried-Wilhelm Leibniz to CR, DZ and LW, and by grants from the National Institutes of Health to MAB (RO1 GM59290) and JX (K99 HG005846).
References
- Davies AG, Oates JF. Colobine Monkeys - Their Ecology, Behaviour and Evolution. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- Groves CP. Primate Taxonomy. Washington DC: Smithsonian Institution Press; 2001. [Google Scholar]
- Groves CP. A Theory of Human and Primate Evolution. Oxford: Clarendon Press; 1989. [Google Scholar]
- Jablonski NG. Natural History of the Doucs and Snub-Nosed Monkeys. New Jersey: World Scientific Publishing Company; 1998. [Google Scholar]
- Jablonski NG. Primate evolution - in and out of Africa. Curr Biol. 1999;9:R119–R122. doi: 10.1016/S0960-9822(99)80077-3. [DOI] [PubMed] [Google Scholar]
- Stewart CB, Disotell TR. Primate evolution - in and out of Africa. Curr Biol. 1999;9:R120–R122. doi: 10.1016/s0960-9822(07)00367-3. [DOI] [PubMed] [Google Scholar]
- Zhang YP, Ryder OA. Primate evolution - in and out of Africa. Curr Biol. 1999;9:R119–R120. doi: 10.1016/S0960-9822(99)80077-3. [DOI] [PubMed] [Google Scholar]
- Sterner KN, Raaum RL, Zhang YP, Stewart CB, Disotell TR. Mitochondrial data support an odd-nosed colobine clade. Mol Phylogenet Evol. 2006;40:1–7. doi: 10.1016/j.ympev.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C. Phylogenetic position of the langur genera Semnopithecus and Trachypithecus among Asian colobines, and genus affiliations of their species groups. BMC Evol Biol. 2008;8:58. doi: 10.1186/1471-2148-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting N, Tosi AJ, Li Y, Zhang YP, Disotell TR. Phylogenetic incongruence between nuclear and mitochondrial markers in the Asian colobines and the evolution of the langurs and leaf monkeys. Mol Phylogenet Evol. 2008;46:466–474. doi: 10.1016/j.ympev.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Napier JR, Napier PH. A Handbook of Living Primates. London: Academic Press; 1967. [Google Scholar]
- Szalay FS, Delson E. Evolutionary History of the Primates. New York: Academic Press; 1979. [Google Scholar]
- Barton NH. The role of hybridization on evolution. Mol Ecol. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- Nichols R. Gene trees and species trees are not the same. Trends Ecol Evol. 2001;16:358–364. doi: 10.1016/S0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- Funk D, Omland K. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Ann Rev Ecol Evol Syst. 2003;34:397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421. [DOI] [Google Scholar]
- Avise JC. Molecular Markers, Natural History, and Evolution. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- McCracken K, Sorenson M. Is homoplasy or lineage sorting the source of incongruent mtDNA and nuclear gene trees in the stiff-tailed ducks (Nomonyx-Oxyyura)? Syst Biol. 2005;54:35–55. doi: 10.1080/10635150590910249. [DOI] [PubMed] [Google Scholar]
- Pollard DA, Iyer VN, Moses AM, Eisen MB. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2006;2:e173. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblmüller S, Duftner N, Sefc KM, Aibara M, Stipacek M, Blanc M, Egger B, Sturmbauer C. Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika - the result of repeated introgressive hybridization. BMC Evol Biol. 2007;7:7. doi: 10.1186/1471-2148-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Laurent J. How good are deep phylogenetic trees? Curr Opin Genetics Dev. 1998;8:616–623. doi: 10.1016/S0959-437X(98)80028-2. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Han K, Ray DA, Huang CH, Chemnick LG, Stewart CB, Disotell TR, Ryder OA, Batzer MA. A mobile element based phylogeny of Old World monkeys. Mol Phylogenet Evol. 2005;37:872–880. doi: 10.1016/j.ympev.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9:481–487. doi: 10.1016/0888-7543(91)90414-A. [DOI] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991;1:498–504. doi: 10.1016/S0959-437X(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Batzer MA. Identity by descent and DNA sequence variation of human SINE and LINE elements. Cytogenet Genome Res. 2005;108:63–72. doi: 10.1159/000080803. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Roos C, Zischler H. Primate phylogeny: molecular evidence from retroposons. Cytogenet Genome Res. 2005;108:26–37. doi: 10.1159/000080799. [DOI] [PubMed] [Google Scholar]
- Van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Xing J, Callinan PA, Myers JS, Hedges DJ, Garber RK, Witherspoon DJ, Jorde LB, Batzer MA. Alu elements and hominid phylogenetics. Proc Natl Acad Sci USA. 2003;22:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci USA. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Xing J, Hedges DJ, Hall MA, Laborde ME, Anders BA, White BR, Stoilova N, Fowlkes JD, Landry KE, Chemnick LG, Ryder OA, Batzer MA. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Herke SW, Xing J, Ray DA, Zimmermann JW, Cordaux R, Batzer MA. A SINE-based dichotomous key for primate identification. Gene. 2007;390:39–51. doi: 10.1016/j.gene.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Zhang Y, Ray DA, Tosi AJ, Disotell TR, Batzer MA. A mobile element based evolutionary history of guenons (Tribe Cercopithecini) BMC Biol. 2007;5:5. doi: 10.1186/1741-7007-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007;45(Suppl):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C. Retropositional events consolidate the branching order among New World monkey genera. Mol Phylogenet Evol. 2009;50:507–513. doi: 10.1016/j.ympev.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Li J, Han K, Xing J, Kim HS, Rogers J, Ryder OA, Disotell T, Yue B, Batzer MA. Phylogeny of macaques (Cercopithecidae: Macaca) based on Alu elements. Gene. 2009;448:242–249. doi: 10.1016/j.gene.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS. Identifying hybridization events in the presence of coalescence via model selection. Syst Biol. 2009;58:478–488. doi: 10.1093/sysbio/syp055. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. New York: Springer; 2002. [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J Hum Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Karanth KP, Singh L, Collura RV, Stewart CB. Molecular phylogeny and biogeography of langurs and leaf monkeys of South Asia (Primates: Colobinae) Mol Phylogenet Evol. 2008;46:683–694. doi: 10.1016/j.ympev.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Ting N. Mitochondrial relationships and divergence dates of the African colobines: evidence of Miocene origins for the living colobus monkeys. J Hum Evol. 2008;55:312–325. doi: 10.1016/j.jhevol.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Brandon-Jones D. In: Encylopedia of Mammals. MacDonald ID, editor. London: George Allen and Unwin; 1984. Colobus and leaf monkeys; pp. 398–408. [Google Scholar]
- Strasser E, Delson E. Cladistic analysis of cercopithecid relationships. J Hum Evol. 1987;16:81–99. doi: 10.1016/0047-2484(87)90061-3. [DOI] [Google Scholar]
- Pusey AE, Packer C. In: Primate Societies. Smuts BB, Cheney DL, Seyfarth RM, Wrangham RW, Struhsaker TT, editor. Chicago: University of Chicago Press; 1987. Dispersal and philopatry; pp. 250–266. [Google Scholar]
- Stewart CB, Disotell TR. Primate evolution - in and out of Africa. Curr Biol. 1998;8:R582–R588. doi: 10.1016/S0960-9822(07)00367-3. [DOI] [PubMed] [Google Scholar]
- Whybrow PJ. In: The Cambridge Encyclopedia of Human Evolution. Jones S, Martin RE, Pilbeam D, editor. Cambridge: Cambridge University Press; 1992. Land movements and species dispersal; pp. 169–173. [Google Scholar]
- Peng YZ, Pan RL, Jablonski N. Classification and evolution of Asian colobines. Folia Primatol. 1993;60:106–117. doi: 10.1159/000156680. [DOI] [PubMed] [Google Scholar]
- Thinh VN, Mootnick AR, Geissmann T, Li M, Ziegler T, Agil M, Moisson P, Nadler T, Walter L, Roos C. Mitochondrial evidence for multiple radiations in the evolutionary history of small apes. BMC Evol Biol. 2010;10:74. doi: 10.1186/1471-2148-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijaard E, Groves CP. In: Primate Biogeography. Lehman SM, Fleagle JG, editor. New York: Springer; 2006. The geography of mammals and rivers in mainland Southeast Asia; pp. 305–329. full_text. [Google Scholar]
- Hallet B, Molnar P. Distorted drainage basins as markers of crustal strain east of the Himalayas. J Geophys Res. 2001;106:13697–13709. doi: 10.1029/2000JB900335. [DOI] [Google Scholar]
- Miller KG, Kominz MA, Browning JV, Wright JD, Mountain GS, Katz ME, Sugarman PJ, Cramer BS, Christie-Blick N, Pekar SF. The phanerozoic record of global sea-level change. Science. 2005;310:1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- Chakraborty D, Ramakrishnan U, Panor J, Mishra C, Sinha A. Phylogenetic relationships and morphometric affinities of the Arunachal macaque Macaca munzala, a newly described primate from Arunachal Pradesh, northeastern India. Mol Phylogenet Evol. 2007;44:838–849. doi: 10.1016/j.ympev.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle of Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Cambridge, MA: Belknap Press; 1963. [Google Scholar]
- Arnold ML, Meyer A. Natural hybridization in primates: one evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Zinner D, Arnold ML, Roos C. The strange blood: natural hybridization in primates. Evol Anthropol. in press . [DOI] [PubMed]
- Cortés-Ortiz L, Duda TF Jr, Canales-Espinosa D, Garcia-Orduna F, Rodriguez-Luna E, Bermingham E. Hybridization in large-bodied New World primates. Genetics. 2007;176:2421–2425. doi: 10.1534/genetics.107.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann O, Fischer A, Lankester F, Pääbo S, Vigilant L. The complex evolutionary history of gorillas: insights from genomic data. Mol Biol Evol. 2007;24:146–158. doi: 10.1093/molbev/msl160. [DOI] [PubMed] [Google Scholar]
- Rumpler Y, Warter S, Hauwy M, Fausser JL, Roos C, Zinner D. Comparing chromosomal and mitochondrial phylogenies of sportive lemurs (genus Lepilemur, Primates) Chromosome Res. 2008;16:1143–1158. doi: 10.1007/s10577-008-1265-z. [DOI] [PubMed] [Google Scholar]
- Merker S, Driller C, Perwitasari-Farajallah D, Pamungkas J, Zischler H. Elucidating geological and biological processes underlying the diversification of Sulawesi tarsiers. Proc Natl Acad Sci USA. 2009;106:8459–8464. doi: 10.1073/pnas.0900319106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner D, Groeneveld LF, Keller C, Roos C. Mitochondrial phylogeography of baboons (Papio spp.) - indication for introgressive hybridization? BMC Evol Biol. 2009;9:83. doi: 10.1186/1471-2148-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinner D, Arnold ML, Roos C. Is the new primate genus Rungwecebus a baboon? PLoS ONE. 2009;4:e4859. doi: 10.1371/journal.pone.0004859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo S. The mosaic that is our genome. Nature. 2003;421:409–412. doi: 10.1038/nature01400. [DOI] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, Hansen NF, Durand EY, Malaspinas AS, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burbano HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Höber B, Höffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PL, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Pääbo S. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis using Parsimony (*and other Methods), Version 4. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Whitfield LS, Lovell-Badge R, Goodfellow PN. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature. 1993;364:713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- Chaves R, Sampaio I, Schneider MP, Schneider H, Page SL, Goodman M. The place of Callimico goeldii in the callithrichine phylogenetic tree: evidence from von Willebrand factor gene intron 11 sequences. Mol Phylogenet Evol. 1999;13:392–404. doi: 10.1006/mpev.1999.0658. [DOI] [PubMed] [Google Scholar]
- Hellborg L, Ellegren H. Y chromsome conserved anchored tagged sequences (YCATS) for the analysis of mammalian male-specific DNA. Mol Ecol. 2003;12:283–291. doi: 10.1046/j.1365-294X.2003.01702.x. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Detwiler KM, Disotell TR. X-chromosomal window into the evolutionary history of the guenons (Primates: Cercopithecini) Mol Phylogenet Evol. 2005;36:58–66. doi: 10.1016/j.ympev.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Kearse M, Heled J, Moir R, Thierer T, Ashton B, Wilson A, Stones-Havas S. Geneious, version 4.6.1. 2009. http://www.geneious.com
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis, version 2.6. 2009. http://mesquite.org
- Zwickl DJ. PhD thesis. Texas University, Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud P, Duringer P, Mackaye HT, Likius A, Blondel C, Boisserie JR, De Bonis L, Eisenmann V, Etienne ME, Geraads D, Guy F, Lehmann T, Lihoreau F, Lopez-Martinez N, Mourer-Chauviré C, Otero O, Rage JC, Schuster M, Viriot L, Zazzo A, Brunet M. Geology and palaeontology of the Upper Miocene Toros-Menalla hominid locality, Chad. Nature. 2002;418:152–155. doi: 10.1038/nature00880. [DOI] [PubMed] [Google Scholar]
- Brunet M, Guy F, Pilbeam D, Lieberman DE, Likius A, Mackaye HT, Ponce de León MS, Zollokofer CP, Vignaud P. New material of the earliest hominid from the Upper Miocene of Chad. Nature. 2005;434:752–755. doi: 10.1038/nature03392. [DOI] [PubMed] [Google Scholar]
- Lebatard AE, Bourlès DL, Duringer P, Jolivet M, Braucher R, Carcaillet J, Schuster M, Arnaud N, Monié P, Lihoreau F, Likius A, Mackaye HT, Vignaud P, Brunet M. Cosmogenic nuclide dating of Sahelanthropus tchadensis and Australopithecus bahrelghazali: Mio-Pliocene hominids from Chad. Proc Natl Acad Sci USA. 2008;105:3226–3231. doi: 10.1073/pnas.0708015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J. In: The Primate Fossil Record. Hartwig WC, editor. Cambridge: Cambridge University Press; 2002. The hominoid radiation in Asia; pp. 369–384. [Google Scholar]
- Leakey MG. In: Theropithecus, the Rise and Fall of a Primate Genus. Jablonski NG, editor. Cambridge: Cambridge University Press; 1993. Evolution of Theropithecus in the Turkana Basin; pp. 85–124. full_text. [Google Scholar]
- Delson E. In: Encyclopedia of Human Evolution and Prehistory. Delson E, Tattersall I, Van Couvering JA, Brooks AS, editor. New York: Garland Publishing Inc; 2000. Cercopithecinae; pp. 166–171. 2000. [Google Scholar]
- Benefit BR, McCrossin ML. In: The Primate Fossil Record. Hartwig WC, editor. Cambridge: Cambridge University Press; 2002. The Victoriapithecidae, Cercopithecoidea; pp. 241–253. [Google Scholar]
- Young NM, MacLatchy L. The phylogenetic position of Moropithecus. J Hum Evol. 2004;46:163–184. doi: 10.1016/j.jhevol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Zalmout IS, Sanders WJ, MacLatchy LM, Gunnell GF, Al-Mufarreh YA, Ali MA, Nasser AAH, Al-Masari AM, Al-Sobhi SA, Nadhra AO, Matari AH, Wilson JA, Gingerich PD. New Oligocene primate from Saudi Arabia and the divergence of apes and Old World monkeys. Nature. 2010;466:360–365. doi: 10.1038/nature09094. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer: MCMC trace analysis tool, version 1.4.1. Institute of Evolutionary Biology, University of Edinburgh; 2007. http://tree.bio.ed.ac.uk/software/tracer/ [Google Scholar]
- Rambaut A. FigTree: Tree figure drawing tool, version 1.2.2. Institute of Evolutionary Biology, University of Edinburgh; 2008. http://tree.bio.ed.ac.uk/software/figtree/ [Google Scholar]
- Yang Z. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics. 2002;162:1811–1823. doi: 10.1093/genetics/162.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Carstens BC, Knowles LL. STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics. 2009;25:971–973. doi: 10.1093/bioinformatics/btp079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Table 1. Presence/absence pattern, location, primers and PCR product sizes of mobile elements
Additional Table 2. Alternative tree topology tests
Additional Figure 1. Single-locus phylogenetic trees (80% majority rule)
Additional Figure 2. Nucleotide composition of both mitochondrial and the combined nuclear datasets
Additional Figure 3. Phylograms based on the mitochondrial and combined nuclear datasets
Additional Table 3. Divergence ages in mya estimated for each locus separately
Additional Table 4. Uncorrected pairwise nucleotide differences for each locus
Additional Table 5. Locus-specific information including alignment length, number of variable sites, selected substitution model and estimated evolutionary rates
Additional Table 6. Primers and PCR conditions for the amplification of nuclear loci
Additional Table 7. GenBank accession numbers