Abstract
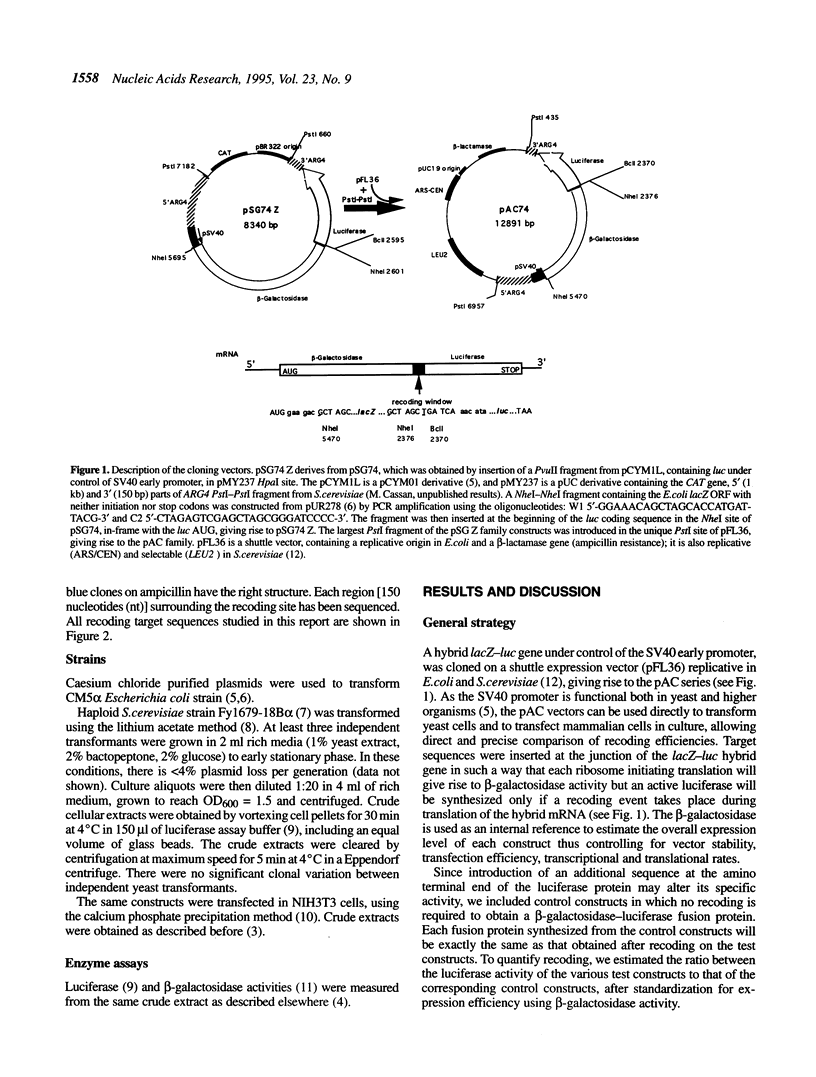
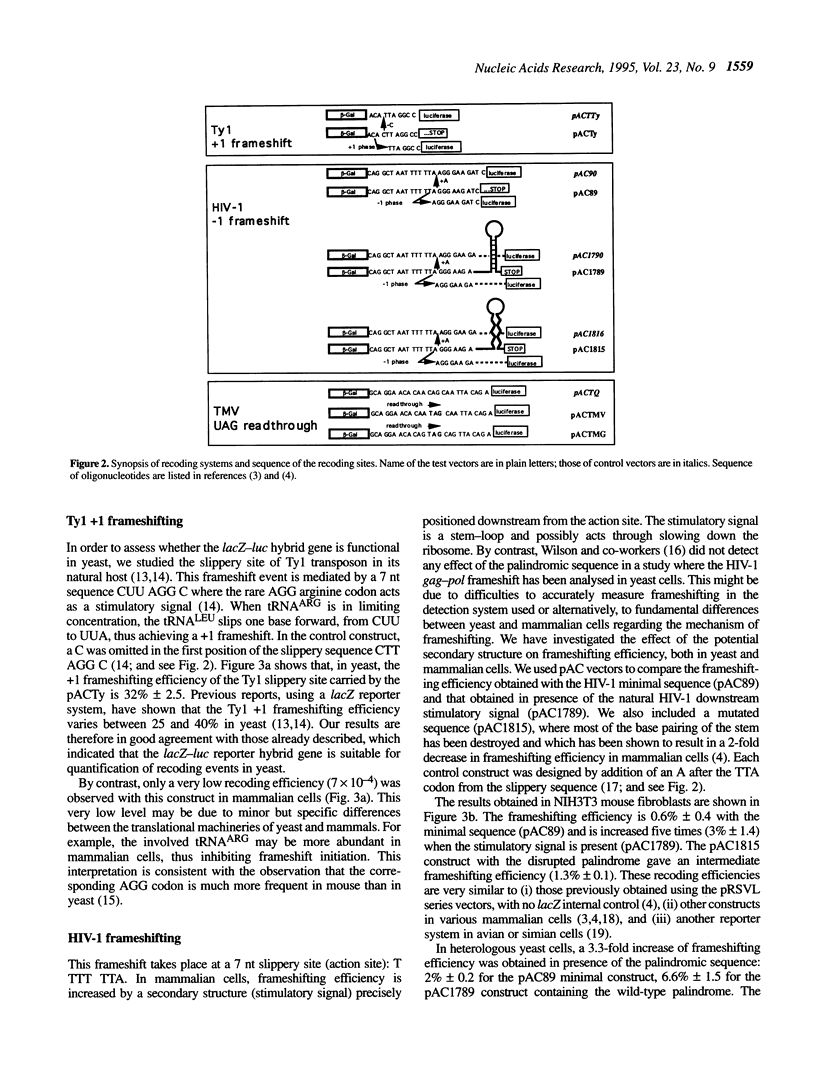
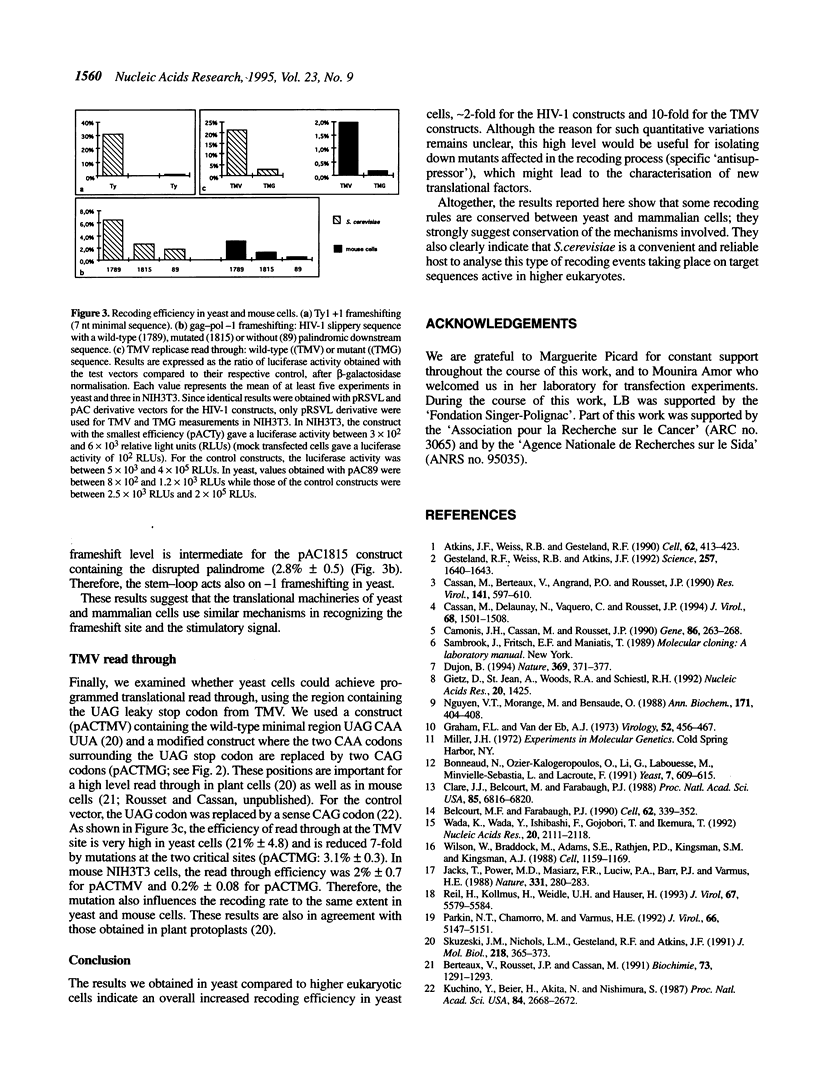
In many viruses and transposons, expression of some genes requires alternative reading of the genetic code, also called recoding. Such events depend on specific mRNA sequences and can lead to read through of an in-frame stop codon or to +1 or -1 frameshifting. Here, we addressed the issue of conservation of recoding rules between the yeast Saccharomyces cerevisiae and mammalian cells by establishing a versatile vector that can be used to study recoding in both species. We first assessed this vector by analysing the site of +1 frameshift of the Ty1 transposon. Two sequences from higher organisms were then tested in both yeast and mammalian cells: the gag-pol junction of human immunodeficiency virus type 1 (HIV-1) (a site of -1 frameshift), and the stop codon region of the replicase cistron from the tobacco mosaic virus (a site of UAG read through). We show that both sequences direct a high level of recoding in yeast. Furthermore, different mutations of the target sequences have similar effects on recoding in yeast and in mouse cells. Most notably, a strong decrease of frameshifting was observed in the absence of the HIV-1 stem-loop stimulatory signal. Taken together, these data suggest that mechanisms of some recoding events are conserved between lower and higher eukaryotes, thus allowing the use of S. cerevisiae as a model system to study recoding on target sequences from higher organisms.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berteaux V., Rousset J. P., Cassan M. UAG readthrough is not increased in vivo by Moloney murine leukemia virus infection. Biochimie. 1991 Oct;73(10):1291–1293. doi: 10.1016/0300-9084(91)90091-e. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos O., Li G. Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991 Aug-Sep;7(6):609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Cassan M., Rousset J. P. Of mice and yeast: versatile vectors which permit gene expression in both budding yeast and higher eukaryotic cells. Gene. 1990 Feb 14;86(2):263–268. doi: 10.1016/0378-1119(90)90288-3. [DOI] [PubMed] [Google Scholar]
- Cassan M., Berteaux V., Angrand P. O., Rousset J. P. Expression vectors for quantitating in vivo translational ambiguity: their potential use to analyse frameshifting at the HIV gag-pol junction. Res Virol. 1990 Nov-Dec;141(6):597–610. doi: 10.1016/0923-2516(90)90033-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan M., Delaunay N., Vaquero C., Rousset J. P. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J Virol. 1994 Mar;68(3):1501–1508. doi: 10.1128/jvi.68.3.1501-1508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J. J., Belcourt M., Farabaugh P. J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., Alexandraki D., André B., Ansorge W., Baladron V., Ballesta J. P., Banrevi A., Bolle P. A., Bolotin-Fukuhara M., Bossier P. Complete DNA sequence of yeast chromosome XI. Nature. 1994 Jun 2;369(6479):371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988 Jan 21;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Beier H., Akita N., Nishimura S. Natural UAG suppressor glutamine tRNA is elevated in mouse cells infected with Moloney murine leukemia virus. Proc Natl Acad Sci U S A. 1987 May;84(9):2668–2672. doi: 10.1073/pnas.84.9.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. T., Morange M., Bensaude O. Firefly luciferase luminescence assays using scintillation counters for quantitation in transfected mammalian cells. Anal Biochem. 1988 Jun;171(2):404–408. doi: 10.1016/0003-2697(88)90505-2. [DOI] [PubMed] [Google Scholar]
- Parkin N. T., Chamorro M., Varmus H. E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992 Aug;66(8):5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Kollmus H., Weidle U. H., Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J Virol. 1993 Sep;67(9):5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski J. M., Nichols L. M., Gesteland R. F., Atkins J. F. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991 Mar 20;218(2):365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- Wada K., Wada Y., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992 May 11;20 (Suppl):2111–2118. doi: 10.1093/nar/20.suppl.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]



