This study investigated the role of Sas-4 in basal body duplication in Paramecium and found that Sas-4 proteins are required to assemble and stabilize the germinative disk and cartwheel, which suggests that Sas-4 plays an essential role in basal body duplication.
Abstract
Centrioles and basal bodies are structurally related organelles composed of nine microtubule (MT) triplets. Studies performed in Caenorhabditis elegans embryos have shown that centriole duplication takes place in sequential way, in which different proteins are recruited in a specific order to assemble a procentriole. ZYG-1 initiates centriole duplication by triggering the recruitment of a complex of SAS-5 and SAS-6, which then recruits the final player, SAS-4, to allow the incorporation of MT singlets. It is thought that a similar mechanism (that also involves additional proteins) is present in other animal cells, but it remains to be investigated whether the same players and their ascribed functions are conserved during basal body duplication in cells that exclusively contain basal bodies. To investigate this question, we have used the multiciliated protist Paramecium tetraurelia. Here we show that in the absence of PtSas4, two types of defects in basal body duplication can be identified. In the majority of cases, the germinative disk and cartwheel, the first structures assembled during duplication, are not detected. In addition, if daughter basal bodies were formed, they invariably had defects in MT recruitment. Our results suggest that PtSas4 has a broader function than its animal orthologues.
INTRODUCTION
Centrioles and basal bodies are related cell structures that contain a well-conserved ninefold symmetry. In animals, just before mitosis, centrioles recruit and organize the pericentriolar material to form the mitotic centrosome. In differentiated vertebrate cells or in protists such as Paramecium tetraurelia, basal bodies localize at the plasma membrane and recruit many proteins to assemble cilia.
Recent genome-wide studies performed in Caenorhabditis elegans have identified a core duplication module composed of five different proteins—SPD-2, ZYG-1, SAS-5, SAS-6, and SAS-4—that act in a sequential order to form a daughter centriole (O’Connell et al., 2001; Kirkham et al., 2003; Leidel and Gonczy, 2003; Leidel et al., 2005; Delattre and Gonczy, 2004; Delattre et al., 2006; Kemp et al., 2004; Pelletier et al., 2006). Interestingly, a conserved role for proteins involved in centriole duplication has been found only for Sas-6 and Sas-4 orthologues (Leidel et al., 2005; Basto et al., 2006; Cho et al., 2006; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007; Culver et al., 2009; Tang et al., 2009). Additional key proteins such as Sak/Plk4 (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), Bld-10/Cep135 (Matsuura et al., 2004; Hiraki et al., 2007), Asterless/Cep152 (Blachon et al., 2008), and γ-tubulin (Ruiz et al., 1999; Dammermann et al., 2004; Kleylein-Sohn et al., 2007) have also been implicated in this process in other model systems. Sas-4 is of particular interest because it is present in all organisms that contain centrioles, suggesting its appearance very early in evolution and its essential role as a key factor in centriole duplication (Carvalho-Santos et al., 2010; Hodges et al., 2010). Furthermore, mutations in the human gene CPAP/CenpJ are associated with the neural developmental disorder microcephaly (Bond et al., 2005).
Importantly, most of the recent studies have exclusively focused on the role of Sas-4 in centriole duplication and, hence, centrosome function in animals. The role of Sas-4 proteins in basal body duplication and ciliogenesis remains therefore to be investigated. We have analyzed the role of Sas-4 in the multiciliated protist P. tetraurelia, an excellent model organism to investigate both basal body duplication and motile cilia assembly.
RESULTS
PtSAS4 genes in Paramecium
Blast sequence search identified three SAS-4 genes in Paramecium: PtSAS4a, PtSAS4b, and PtSAS4c. These three genes are 63% identical and encode proteins of 1113, 1103, and 1153 residues, respectively. The most conserved of PtSas4 protein exhibits 30% identity and 50% similarity to the C terminus of Drosophila DSas-4 and 38% identity and 60% similarity to the C terminus of human CPAP (Supplemental Figure 1).
PtSas4 is required for basal body duplication but not ciliogenesis
To investigate the function of PtSas4 in basal body duplication, we performed RNA interference (RNAi) experiments to silence the PtSAS4 genes. In Paramecium this technique is widely applied with great success rates by using bacteria expressing double-stranded RNA homologous to the inactivated gene in feeding experiments (Galvani and Sperling, 2002). This potent technique offers an excellent opportunity to analyze the primary defects that result from target gene inactivation, very rapidly, just after the first division.
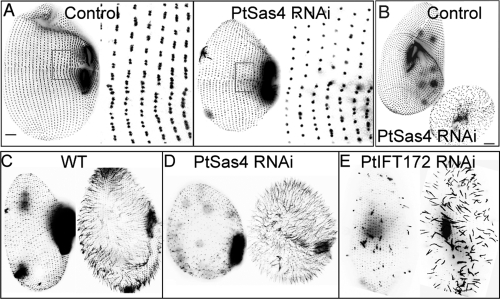
Silencing any of the PtSAS4 genes alone, or even in pair combination, resulted in mild basal body duplication defects that could be detected only after several cell divisions (Supplemental Figure 2). Because the three PtSAS4 genes share between 63% and 80% sequence identity, this mild phenotype might reflect just the presence of a robust redundant mechanism in which any of the PtSAS4 genes can compensate for the lack of the others. We decided, therefore, to silence the three PtSAS4 genes simultaneously and readily observed a strong basal body duplication defect immediately after the first division following silencing (16–20 h), with a clear reduction in the number of basal bodies (Figure 1, A and B). This defect could be easily identified in immunostaining experiments using antibodies that label basal bodies but also by analyzing cell morphology (Figure 1, A and B). PtSas4-depleted Paramecium cells, which we will refer to simply as PtSas4 cells, showed reduced size and round morphology, which invariably resulted in growth arrest after an attempted second division (Figure 1B). Defects in cell cycle progression have been observed after depletion of other basal body components in Paramecium and are normally associated with defects in establishment of cell symmetry and polarity (Ruiz et al., 1999; Ruiz et al., 2005; Jerka-Dziadosz et al., 2010). In addition, the oral apparatus, containing in control cells ∼900–1000 basal bodies (Iftode et al., 1997), appeared abnormal in size and shape. PtSas4 cells also presented an abnormal swimming behavior that could result from defects in basal body duplication or in cilia assembly.
FIGURE 1:
PtSAS4 depletion causes basal body duplication defects but does not affect ciliogenesis. (A, B) Cells labeled with the ID5 antibody, to visualize all basal bodies (old and new). (A) The control cell (left) displays groups of two and/or three dots (inset on the right), corresponding to duplicating basal bodies. In a PtSas4 cell (right), a defect in basal body duplication can be perceived by the presence of single dots (inset on the right). (B) Comparison between control (left) and PtSas4 (right) cell after three divisions. The PtSas4 cell is much smaller than the control, and the basal body cortical organization is lost. Bar = 10 μm. (C–E) Cells were deciliated and labeled with a polyclonal antitubulin antibody, which decorates basal bodies and cilia (Cohen et al., 1982) either immediately after deciliation (left of each panel) or 4 h after (right of each panel). (C, D) In control cells (C) and PtSas4 cells (D), complete reciliation can be observed after 4 h. Note the round abnormal shape of PtSas4 cells due to defects in basal body duplication. In the absence of PtSas4, cilia are able to regrow from the preexisting basal bodies. (E) In PtIFT172 cells only a small percentage of cilia can regrow. Shorter cilia can also be observed. Bar = 10 μm.
To investigate whether PtSas4 also has a role in cilia assembly, we performed deciliation experiments in control cells, PtSas4 cells, and PtIFT172-depleted cells, an essential intraflagellar transport component (Pedersen et al., 2005) fulfilling the same function in Paramecium (Laligne et al., 2010). In PtIFT172-depleted cells, reciliation was severely delayed, and after 4 h, less than half of the basal bodies were able to assemble cilia (Figure 1E). In both control cells and PtSas4 cells, cilia assembled (albeit only from old basal bodies in PtSas4 cells) at an identical rate (Figure 1, C and D), suggesting that PtSas4 is dispensable for ciliogenesis.
PtSas4 localization
To characterize PtSas4 localization, we fused the green fluorescent protein (GFP) coding region to the N terminal of two of the PtSAS4 genes (PtSAS4a and PtSAS4c) under the control of the Paramecium calmodulin promoter and regulatory sequences. Transformation by injection of any of these constructs into Paramecium nuclei allowed the identification of GFP-positive clones. Essentially, the two constructs behave in an identical way in the cell, and we will simply refer to these as GFP-PtSas4 cells. GFP-PtSas4 was found associated with all basal bodies in the cortex and oral apparatus but seems to be excluded from cilia (Figure 2A) just like in C. elegans (Dammermann et al., 2009).
FIGURE 2:
PtSas4 is associated with the basal body and its localization is cell cycle regulated. (A) GFP-PtSas4–expressing cell double labeled with the monoclonal anti-tubulin ID5 (shown in red and in the top panel on the right), which labels all basal bodies, and a polyclonal anti-GFP (shown in green and in the middle panel on the right). PtSas4 localizes to the basal bodies’ oral apparatus (asterisks, merged pictures) and the contractile vacuoles (white arrows). (B) Cells were isolated during fission and then fixed at different times. Pictures were taken with the same exposition conditions. Just after division (T = 0, first panel) and 1 h after (T = 1 h), GFP-PtSas4 is associated with all the basal bodies. During interphase (T = 2 and 3 h), GFP-PtSas4 cannot be detected at the basal bodies; a weak signal is seen at the fibers associated with the oral apparatus (white arrows). An increase in the intensity signal appears as cells prepare for mitosis (T = 4.5 h). During cell division, note the presence of two oral apparatuses (asterisks); GFP-PtSas4 is detected localizing to all basal bodies (old and newly assembled ones) (T = 5.5 h). Bars = 10 μm.
In C. elegans, Drosophila and vertebrate cell culture Sas-4 proteins are associated with the centrosome throughout the cell cycle (Kirkham et al., 2003; Leidel and Gonczy, 2003; Basto et al., 2006; Dammermann et al., 2008; Tang et al., 2009). In contrast, in Paramecium, the association of PtSas4 with basal bodies seems to be quite transient (Figure 2B). In interphase cells, PtSas4 could not be detected at the cell cortex (Figure 2B, T = 2 and 3 h). As cells start to divide and to duplicate their basal bodies, PtSas4 is recruited first to old basal bodies (Figure 2B, T = 4.5 h) and can then be seen rapidly associating with newly assembled basal bodies (Figure 2B, T = 5.5 h). PtSas4 remains associated with both the old and new basal bodies for 1 h after cell division is completed (Figure 2B, T = 0 and 1 h). The transient localization of GFP-PtSas4 reported here is significantly different from GFP-PtSas4 fusions described in animal cells. We decided therefore to test whether this fusion is functional and whether the results obtained are not simply an artifact due to the overexpression of GFP-PtSas4 in the presence of endogenous proteins. As the depletion of the three PtSas4 isoforms (genes a–c) in the GFP-PtSas4a cells resulted in a complete reduction of the GFP signal and basal body duplication defects (Supplemental Figure 2A), we decided to reduce the amount of endogenous Sas4 proteins without affecting our GFP-PtSas4 signal. For this, we used two different strategies. In the first approach, we depleted only two of the three PtSAS4 genes (PtSAS4b and PtSAS4c) in cells expressing GFP-PtSas4a. As a control, we also depleted PtSas4b and PtSas4c in cells that did not express GFP-PtSas4a. In control cells, basal body duplication defects appeared, but the phenotype was milder than the one observed with the depletion of the three PtSAS4 genes: Cells could progress through the cell cycle for 4 d (more than 10 cell divisions), but basal body duplication defects progressively accumulated that caused a reduction of cell size (Supplemental Figure 2D). In GFP-PtSas4a cells, defects in basal body duplication were not observed upon depletion of PtSas4b and PtSas4c, suggesting that an increase in the levels of PtSas4a (by the presence of GFP-PtSas4a) was sufficient to rescue basal body duplication defects and viability. Importantly, careful analysis of GFP-PtSas4a, upon PtSas4b and PtSas4c depletion, showed the same dynamics for GFP-Sas4a as described previously (Supplemental Figure 2B).
In the second approach, we replaced a 400–base pair fragment within the GFP-PtSAS4a gene to make it resistant to RNAi (see Materials and Methods). We then target, by RNAi, the three SAS4 genes, PtSAS4(a–c), in cells expressing this altered form of GFP-PtSAS4a, here referred to as GFP-PtSAS4a*. Once again, the GFP-PtSAS4a* construct was able to rescue the cell from the depletion of endogenous, and we could see GFP-PtSas4a* being recruited to basal bodies, transiently, during cell division, but it was absent during interphase (Supplemental Figure 2, C and E).
Our data suggest therefore that PtSas4 localization at the basal body is markedly different from its centrosome counterpart in all animal species described so far. Interestingly, Sas4-GFP seems to follow similar dynamics in the Drosophila male germ line (Blachon et al., 2009). Sas4-GFP could be seen localizing proximally, in the long centrioles from meiosis until the early spermatid stage. However, this localization disappeared at later stages of spermatogenesis during sperm tail assembly and elongation (Blachon et al., 2009).
PtSas4 is required to stabilize other basal body components at the site of assembly
We wanted to know whether the recruitment of basal body components implicated in centriole duplication was perturbed upon PtSas4 depletion. Recent work described PtSas6 and PtBld10 proteins in Paramecium and their role in basal body duplication (Jerka-Dziadosz et al., 2010). We coexpressed fusions to GFP-PtBld10 and red fluorescent protein (RFP)–PtSas6 in Paramecium and silenced all three PtSAS4(a–c) genes. In control cells, GFP-PtBld10 and RFP-PtSas6 could be seen associated with the basal body marker ID5 at both old and newly assembled basal bodies (Figure 3A). However, in PtSas4 cells, even if we detected GFP-PtBld10 and RFP-PtSas6 associated with old basal bodies, we failed to detect any of these proteins at new sites of assembly (Figure 3B). The fact that neither GFP-PtBld10 nor RFP-PtSas6 could be observed at the new sites of assembly does not necessarily imply that PtSas4 is required to recruit these proteins. Another explanation could be that these proteins are transiently recruited but not maintained at the new sites of basal body assembly because they lack PtSas4. In either case, our results suggest that the recruitment or maintenance of PtBld10 and PtSas6 at new sites of assembly requires PtSas4.
FIGURE 3:
Recruitment and maintenance of PtSas6 and PtBld10 are perturbed in the absence of PtSas4. (A, B) Cells labeled with ID5 antibody (left and shown in blue), RFP-PtSas6 (shown in red), and GFP-PtBld10 (shown in green). In the last panel, the red and green channels have been shifted to allow better visualization of the three colors. (A) In control cells all basal bodies, both old (O) and new (N), are decorated by RFP-PtSas6, GFP-PtBld10, and ID5. (B) In PtSas4 cells, defects in basal body duplication are detected. Next to the old basal body (O), empty spaces that do not contain RFP-PtSas6, GFP-PtBld10, or ID5 can be identified. Note that in the PtSas4 cell the organized arrangement of basal body rows is lost. Bars = 10 μm.
PtSas4 is essential for the assembly or stability of the cartwheel
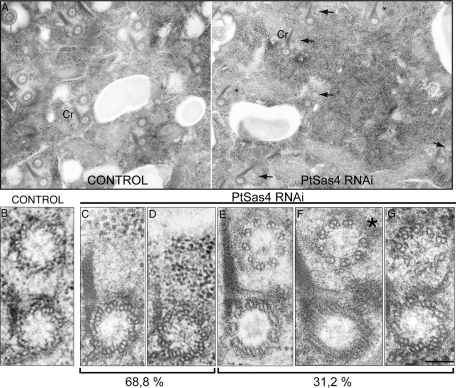
We next decided to characterize the defects in basal body duplication at the ultrastructural level. To avoid the observation of a phenotype resulting from an accumulation of defects, we examined cells during their first division following RNAi treatment. To do so, we carefully chose cells by their typical cell shape. During division, on both sides of the fission furrow, two or three successive rounds of duplication take place that result in the formation of highly ordered rows along which new basal bodies are inserted next to old basal bodies. In Paramecium, old basal bodies can be easily identified by the presence of an electron-dense ciliary rootlet (Cr), also known as the kinetodesmal fiber (Figure 4A) (Dippell, 1968). The new basal body forms at right angles to the old basal body, and toward the end of duplication the new basal body is tilted toward the membrane to assume a parallel position relative to the old basal body (Figure 5A) (Dippell, 1968). Newly assembled basal bodies localize near the Cr, to the anterior side of old basal bodies (Figure 4A). In control cells, 86.3% (n = 242) of old basal bodies were seen associated with new basal bodies. All newly assembled basal bodies were structurally similar to old basal bodies, containing nine microtubule (MT) triplets (Figure 4B). However, in PtSas4 cells, only 31.2% of old basal bodies could be seen associated with new basal bodies (Figure 4, A and E–G, n = 450). Invariably, these newly assembled basal bodies were structurally abnormal. Although the large majority of daughter basal bodies presented nine MT sets, occasionally one MT set was missing and a gap could be identified (Figure 4F, asterisk). This observation suggests that the ninefold symmetry is present in newly (but partially) assembled basal bodies. In agreement, we identified normal-sized cartwheels with ninefold symmetry, but in which only MT singlets or doublets were present (Figure 4G). It is worth noting that we have never observed newly assembled basal bodies with nine MT triplets (Figure 4, E and F).
FIGURE 4:
Ultrastructural defects of basal bodies upon PtSas4 depletion. Electron micrographs of control and PtSas4 cells. (A) Low magnification of transversal views of control (left) and PtSas4 (right) cells. Old basal bodies can be recognized by their electron-dense Cr. In the control cell, all old basal bodies are associated with one daughter. In the PtSas4 cell, fixed during the first division postsilencing, two major types of defects in basal body duplication can be identified: total absence of daughter basal bodies (asterisks) or partially assembled basal bodies (arrow). Bar = 200 nm. (B–G) High magnifications of transversal sections of duplicating basal bodies. In all cells Cr is to the left and the old basal bodies are on the bottom. In control cells (B) a complete daughter basal body (top) can be seen next to an old basal body. In PtSas4 cells no intermediate structure could be identified in 69% of cases (C, D). (D) The section shown passes through the old basal body cartwheel. (E–G) Partially assembled basal bodies can be identified in ∼31% of cases. These contain either nine MT doublets or singlets (E) or eight MT doublets or singlets (F); the asterisk marks the missing singlet or doublet. (G) A section passing through the cartwheel of the daughter basal body. The cartwheel structure appears to contain the normal ninefold symmetry. Bar = 100 nm.
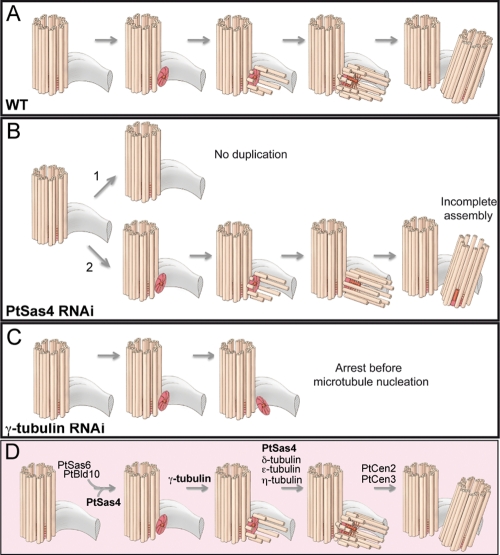
FIGURE 5:
Model for basal body assembly in Paramecium. (A) Model of basal body assembly in WT cells as first described by Dippell (1968). The old basal body (orange cylinder) is represented with its Cr (gray structure). The germinative disk and cartwheel of the new basal body (dark orange) start to be assembled perpendicularly to the old basal body. At the early steps of duplication, the germinative disk and cartwheel are stabilized. Afterward, individual MTs, the A-tubules, are inserted and continue to elongate as the B- and C-MTs are added. The final step of basal body duplication involves the tilting of the newly assembled basal body toward the cell cortex (top). (B) In the majority of PtSas4 cells (∼69%), an intermediate daughter basal body cannot be detected associated with the old basal body (1). In the remaining 31% of cases, partially assembled daughter basal bodies can be seen (2). (C) In γ-tubulin–depleted cells, cartwheels that lack MTs can be assembled. (D) Model for basal body duplication in Paramecium according to our study. PtSas4, together with PtSas6 and PtBld10, seems to be involved in the early steps of basal body duplication for the assembly and stabilization of the germinative disk and cartwheel. γ-Tubulin allows the nucleation of the first MTs singlets. The addition of the other MTs seems to require PtSas4, δ-tubulin (Garreau de Loubresse et al., 2001), η-tubulin (Ruiz et al., 2000), and ε-tubulin (Dupuis-Williams et al., 2002). PtCen2 and PtCen3 act at the last step of duplication, in the tilting and positioning of the newly assembled basal body (Ruiz et al., 2005).
If only around one-third of old basal bodies can be seen associated with newly abnormal basal bodies in PtSas4 dividing cells, what happens to the remaining duplicating basal bodies? We analyzed several hundred electron microscopy (EM) sections to attempt to distinguish any type of structure such as the germinative disk or cartwheel (discussed later in this article), but we were unable to identify any. In PtSas4 cells, new basal bodies could not be detected associated with the large majority of old basal bodies in 68.8% of cases (Figure 4A). Higher magnifications of EM sections revealed the absence of any recognizable structure, and simply empty spaces could be identified next to old basal bodies (Figure 4, C and D). Importantly, our results also suggest that PtSas4 does not seem to be required to maintain the structure of old basal bodies. In PtSas-4–depleted cells, we did not detect any structural defects in preexisting basal bodies (Figure 4).
The phenotype observed and described above could not have resulted from premature observations in the process of basal body duplication. In Paramecium, daughter basal bodies start to be assembled at right angles of preexisting basal bodies (Figure 5A), and MTs are added to the daughter basal body before and during the tilting toward the cell surface (Dippell, 1968). Therefore, in EM cross sections, the parallel position of old basal bodies and partially assembled basal bodies tells us that tilting has already occurred and that the assembly process is completed, even if not correctly.
Almost 70% of old basal bodies in PtSas4 cells were not seen associated with new basal bodies whereas in the remaining 30% incomplete new basal bodies could be detected. Importantly, within the same cell we could identify abnormally duplicating basal bodies and basal bodies that were not being duplicated at all (Figure 4A). We think that the capacity to duplicate is probably related to the level of depletion and the amount of Sas-4 protein available. It is worth mentioning that all our data were obtained at the first division postsilencing, in which the levels of depletion might not be as high as in subsequent divisions. However, after the first division, the abnormal shape of PtSas4 cells precludes the identification of cells undergoing division.
EM tomography analysis of C. elegans embryos depleted for SAS-4 has elegantly shown that centriole duplication is arrested after the formation of the central tube that lacks any MT singlets (Pelletier et al., 2006). These studies implied, then, a role for SAS-4 at the last step of centriole duplication in the assembly and incorporation of MT singlets but not in the formation of the central tube or any earlier structure. In light of these results, we would expect that PtSas4 depletion would affect only later stages of basal body duplication in Paramecium, such as the assembly and incorporation of MTs. The lack of germinative disk and cartwheel at sites where the new basal bodies would be expected suggests, however, that this might not be the case. One possible explanation relies on the fact that, in Paramecium, basal bodies do not contain a central tube. EM studies of basal body duplication in Paramecium have shown that duplication is initiated with the assembly of a ring of electron-dense material, the germinative disk (Dippell, 1968) (Figure 5, red disk). Within the germinative disk, nine spokes connect the central hub to the nine A-tubules of each MT triplet, forming the cartwheel (Dippell, 1968; Jerka-Dziadosz et al., 2010), a structure also present in mammalian procentrioles (Lange and Gull, 1996; Azimzadeh and Bornens, 2007). Interestingly, unlike mammalian mature centrioles, in Paramecium and other protists (Azimzadeh and Bornens, 2007) the cartwheel is maintained throughout the life of the basal body (Dippell, 1968). Our findings suggest, therefore, that Sas-4 proteins in Paramecium have a functional role at the very early steps of basal body duplication possibly contributing to the assembly or stabilization of the germinative disk and cartwheel and to the recruitment and/or stabilization of PtSas6 and PtBld10. Importantly, our results also suggest that PtSas4 proteins are required only to assemble or stabilize the cartwheel, as we have never detected basal bodies with abnormal symmetry. The structural differences found between centrioles and basal bodies could therefore justify a different requirement for Sas-4 proteins during basal body biogenesis.
PtSas4 overexpression generates supernumerary germinative disks
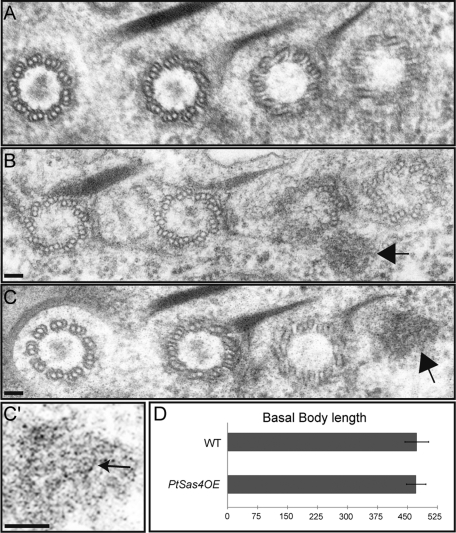
To further test the function of PtSas4 at the early stages of basal body assembly, we analyzed the consequences of PtSas4 overexpression. In animal cells, overexpression of certain centriole replication proteins often results in the formation of centrosomes with supernumerary centrioles or supernumerary centrosomes (Kleylein-Sohn et al., 2007; Peel et al., 2007; Basto et al., 2008; Kohlmaier et al., 2009). Surprisingly, even if in GFP-PtSas4–overexpressing cells (PtSas4OE cells) extra rows of basal bodies could not be detected, we were able to detect, by EM, the presence of extra germinative disks lying next to old basal bodies (Figure 6, A, B, C, and C′). Analysis of 41 cortical basal body units of PtSas4OE cells (n = 4 cells), in a total of 130 basal bodies, revealed the presence of 27 extra germinative disks, which means that 65% of cortical basal body units contain an extra germinative disk. We have never observed supernumerary germinative disks close to preexisting basal bodies in control cells. Sometimes these extra-germinative disks seem to contain a hub with only a few spokes, but unfortunately we could not get better resolution of such structures to be able to interpret them unequivocally (Figure 6C′, arrow). Nevertheless, the size of this structure is comparable to the size of the hub in control cartwheels (unpublished data).
FIGURE 6:
PtSas4 overexpression generates supernumerary germinative disks but not longer basal bodies. (A) Electron micrograph of a control cell displaying a duplicating cortical unit. Extra germinative disks are never seen in control cells. (B, C) Electron micrographs of cells overexpressing GFP-PtSas4 (PtSas4OE). In 65% of cortical basal body units from PtSas4OE cells, we identified extra germinative disks. Next to a row of duplicating basal bodies (note the Cr on the top of the two old basal bodies in B and a maturing daughter basal body in C), extra germinative disks can be seen (arrows). Bar = 50 nm. (C′) Enlargement of the germinative disk seen in (C). By modulating the contrast, it is possible to detect a structure that resembles a hub (arrow) inside the germinative disk. Bar = 50 nm. (D) A graph showing measurements of the length of basal bodies from control cells and PtSas4OE cells.
In Paramecium, PtBld10 or PtSas6 overexpression generates extra basal bodies (Jerka-Dziasdosk et al., 2010). Together, our results suggest that overexpression of PtSas4 is not sufficient to generate supernumerary basal bodies. Other proteins such as PtBld10 or PtSas6 might also have to be in excess for the formation or stabilization of extra basal bodies.
In vertebrate cell culture, centriole size is regulated by Sas-4 and CP110. Overexpression of Sas-4 or depletion of CP110 results in the formation of abnormally long centrioles that affect centrosome function (Tsang et al., 2008; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009). In Paramecium, we did not observe basal bodies of abnormal length upon Sas-4 overexpression (Figure 6C) (n = 36 for wild-type [WT] and n = 60 for PtSas4OE cells). We were not able to identify a CP110 orthologue in the Paramecium genome that supports our initial idea that Sas-4 proteins behave differently during basal body duplication.
Naked cartwheels can be stabilized in the absence of γ-tubulin
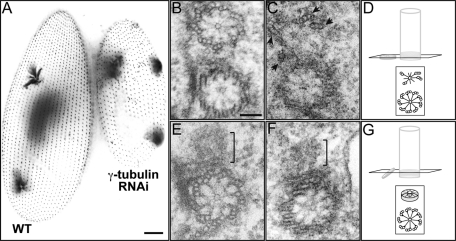
The results obtained in Paramecium upon PtSas4 depletion suggest that Sas-4 proteins might play an earlier role in basal body duplication than the one played by Sas-4 homologues in centriole assembly. We can therefore envisage at least two hypotheses to explain these observations. The lack of any earlier structure, such as the germinative disk or cartwheel, could result from a direct role of PtSas4 in the process of assembly itself where, in its absence, these structures cannot be assembled or stabilized. Alternatively, PtSas4 proteins could also have a more indirect role in which they would not be involved in the assembly per se but rather in basal body stabilization, for example, allowing the addition or incorporation of MT singlets. In this case, the lack of germinative disk or cartwheel would result from the destabilization of incomplete basal bodies. However, previous results reporting the characterization of δ-tubulin mutants in Chlamydomonas or η- and ε-tubulins in Paramecium (Dutcher and Trabuco, 1998; Garreau de Loubresse et al., 2001; Dupuis-Williams et al., 2002) have shown that basal bodies without B- and C-tubules can still be stabilized. Nevertheless, it is not known whether germinative disks and cartwheels can be stabilized in the complete absence of MTs, including the A-tubule. To answer to this question, we decided to deplete γ-tubulin, an essential basal body component and MT nucleator (Oakley, 1992; Ruiz et al., 1999). In Ptγ-tubulin RNAi cells, defects in basal body duplication could be detected between 24 and 48 h of silencing (three to four divisions at 18°C) (Figure 6A). EM analysis showed that next to preexisting old basal bodies, new and incomplete basal bodies could be detected in some cases (Figure 7, B–D). In 42% of Ptγ-tubulin–depleted cells (n = 24 in a total of 56 basal bodies analyzed), a smaller structure resembling a cartwheel of reduced diameter could be identified whereas in the remaining 58% we could not identify a cartwheel. In 2 out of the 24 basal bodies analyzed, a few MTs could be seen (Figure 7D, arrowhead). In some cases this small cartwheel was not tilted (Figure 7, D–F). This analysis suggests that a cartwheel can be stabilized in the absence of γ-tubulin and MTs but somehow it cannot be tilted or achieve its normal size. Because we have never observed such structures in PtSas4–depleted cells, we conclude that in the absence of MTs (albeit with Sas-4 still present) the cartwheel is a stable structure and suggest that the observed phenotype, upon PtSas4 depletion, likely does not result from destabilization of a preassembled basal body.
FIGURE 7:
γ-Tubulin depletion results in the formation of daughter basal bodies that contain only cartwheels. (A) Control (left) and γ-tubulin–depleted (right) cells labeled with ID5 antibody. γ-Tubulin depletion leads to an inhibition of basal body duplication, as previously described (Ruiz et al., 1999). Bar = 10 μm. (B) Electron micrograph of a duplicating basal body in a control cell showing the mother (bottom) and daughter (top) cartwheels. (C, E, and F) Electron micrographs of γ-tubulin cells, fixed 48 h postsilencing. (C) The cross section passes through the cartwheel of mother and daughter basal bodies. In 42% of γ-tubulin–depleted cells (n = 24 in a total of 56 basal bodies analyzed), a cartwheel of reduced diameter could be identified. The structure of the new basal body is severely altered. Only 4 MT sets (triplets/doublets) are present (arrows). We detected MTs in only 2 out of the 24 basal bodies that contained a daughter cartwheel. (D) Schematic representation of the plane of section and (B). (E, F) The transverse sections passing through the cartwheel of the old basal body allow the visualization of a smaller daughter cartwheel (brackets). These cartwheels lack MTs. In these sections, the cartwheel is not completely tilted, as shown in the scheme in (G). Bar = 100 nm.
DISCUSSION
In the past few years significant progress has been made in the identification of proteins required for centriole duplication in animals. Sas-4 is conserved in all the organisms bearing centrioles (Carvalho-Santos et al., 2010; Hodges et al., 2010). Sas-4 has been recognized as an essential protein in the centriole duplication because depletion or mutations in Sas-4 invariably lead to a failure in centriole duplication in all model systems studied so far (Kirkham et al., 2003; Leidel and Gonczy, 2003; Basto et al., 2006; Cho et al., 2006). Epistatic genetic studies and EM tomography in C. elegans have placed SAS-4 at the end of centriole duplication after SAS-6 to allow the incorporation of MT singlets (Delattre et al., 2006; Pelletier et al., 2006). In human cells, Sas-4 also seems to be required after Sas-6 and together with γ-tubulin and Cep135 participates in the incorporation of centriolar MTs and centriole elongation (Kleylein-Sohn et al., 2007). In addition, Sas-4 levels have to be highly controlled as overexpression of this protein results in the formation of abnormal centrosome structures in Drosophila unfertilized eggs and longer daughter centrioles in vertebrate cells (Peel et al., 2007; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009).
Our work shows that Sas-4 proteins are also essential during basal body duplication in the multiciliated protist Paramecium, but, unexpectedly, we have found that PtSas-4 is required not only to incorporate MTs to the nascent daughter basal body. In the majority of PtSas4–depleted cells, the initial stages of duplication and assembly of a germinative disk and cartwheel fail, and PtSas6 and PtBld10 proteins cannot be recruited or stabilized at new sites of assembly. Unfortunately, our analysis did not allow us to distinguish whether PtSas4 proteins are required for germinative disk and cartwheel assembly per se or whether PtSas4 plays a role in the stabilization of these structures once they are assembled. We propose, however, that the role of Sas-4 in maintaining and stabilizing these structures is not related to its function in the incorporation of stable MTs to the nascent basal body. In γ-tubulin–depleted cells, naked stabilized cartwheels could be found, which suggests that these structures can be maintained in the absence of MTs.
A role in cartwheel stabilization has been attributed to other proteins in multiciliated protists, Drosophila, and vertebrates (Blachon et al., 2009; Keller et al., 2009; Pearson et al., 2009). In Chlamydomonas, the cartwheel component Bld10 is essential for both extending the cartwheel diameter and attaching the spokes to the MT triplets (Hiraki et al., 2007). In Paramecium, cartwheel assembly is also affected, and abnormal hub and spoke organization could be identified in PtBld10-depleted cells. Moreover, PtBLD10 (or even PtSAS6) overexpressions induced the formation of extra basal bodies of abnormal morphology, which, in the case of PtBld10 overexpression, could be formed at wrong positions within the cell (Jerka-Dziadosz et al., 2010). In Tetrahymena, Poc1 also play an important role in basal body stability (Pearson et al., 2009). New basal bodies can still assemble in poc1 mutant cells, but they are shorter and contain structural abnormalities. Conversely, Poc1 overexpression results in the formation of longer centriole-like structures, consistent with a role for Poc1 in determining length control. In Paramecium, PtSas4 depletion or overexpression defects overlap only partially with the defects found in PtBld10-depleted cells or Poc1 Tetrahymena mutant cells. It is therefore possible that all these molecules play a major role in stabilizing the nascent basal body but at different levels. Considering this possibility, we think that Sas-4 is probably required to stabilize the germinative disk and cartwheel but does not play a role in determining its structure. The cartwheel in PtSas4-depleted cells (present in the 30% of cells where new basal bodies can be detected) seems to be correctly assembled, displaying the ninefold symmetry just like in control cells. Unlike Poc-1 or Sas-4 in vertebrate cells, the overexpression of PtSas4 does not generate longer basal body MTs; instead, extra germinative disks could be detected assembled next to duplicating basal bodies. We have never observed the presence of fully assembled supernumerary basal bodies in PtSAS4-overexpressing cells, suggesting that other factors, probably PtBld10 and PtSas6, also need to be in excess to produce extra basal bodies.
One of the most surprising observations of our results is related to the behavior of Sas-4 proteins. In all studies described so far, Sas-4 proteins have been associated with the centrioles throughout the cell cycle, whereas in Paramecium, Sas-4 is recruited only transiently during basal body duplication (Kirkham et al., 2003; Leidel and Gonczy, 2003; Basto et al., 2006; Dammermann et al., 2008; Tang et al., 2009). It has also been shown that DSas-4 disappears from the Drosophila sperm basal body during sperm maturation (Blachon et al., 2009). Together, these observations suggest that Sas-4 does not remain associated with basal bodies that nucleate motile cilia, and its permanent association with centrioles (at the centrosome) might reflect a second function for Sas-4 at the centrosome not related to its role in centriole duplication (Dammermann et al., 2008).
It is worth mentioning that while we have identified several differences involving Sas-4 behavior in Paramecium, we have also found a role for PtSas4 in the incorporation of MTs to the nascent basal body, just like in C. elegans (Delattre et al., 2006; Pelletier et al., 2006).
The significance of our findings in the context of the present literature suggests at least two hypotheses. Basal bodies are ancient structures that were already present in the flagellate common ancestor of eukaryotes, whereas centrioles appeared only later in evolution with the development of multicellularity (Cavalier-Smith, 2002; Beisson and Wright, 2003; Azimzadeh and Bornens, 2004; Carvalho-Santos et al., 2010; Hodges et al., 2010). It is therefore possible that Sas-4 function at early steps of duplication has been lost through evolution, with the appearance of novel proteins implicated in the duplication process. In light of this hypothesis, our findings then suggest that the same set of proteins can be used differently to duplicate basal bodies and centrioles.
Alternatively, a function of Sas-4 proteins in cartwheel assembly and stabilization might be required only if procentriole assembly is initiated with the formation of a cartwheel and if this structure is stabilized in the mature centriole. In this context, Sas-4 proteins would then be required to stabilize the germinative disk and the cartwheel in both basal bodies and centrioles. Because C. elegans centrioles contain a central tube and not a cartwheel (Pelletier et al., 2006), we hypothesize that this earlier Sas-4 function has not been maintained in this organism. In vertebrate cells, on the other hand, daughter centriole duplication is initiated with the formation of a cartwheel that does not seem to be maintained as the centriole matures (Lange and Gull, 1996; Azimzadeh and Bornens, 2007). It is worth noting, however, that no centriole intermediates were found in serial sections of DSas-4 mutant neuroblasts (Basto et al., 2006), which might therefore support, in this cell type, a role for Sas-4 at early steps of duplication in the stabilization of the cartwheel.
Overall, this study clearly sustains the importance of studying proteins implicated in basal body and centriole duplication in a variety of model systems.
MATERIALS AND METHODS
Strains and culture conditions
Stock d4-2 of P. tetraurelia, the wild-type reference strain, was used in all feeding experiments. The nd7-1 mutant, carrying a recessive monogenic mutation preventing trichocyst discharge (Skouri and Cohen, 1997), was used for the expression of GFP-fusions. Cells were grown at 27°C or at 18°C (feeding experiments) in a wheatgrass infusion (BHB, L’arbre de vie, Luçay Le Male, France), bacterized with Klebsiella pneumoniae, and supplemented with 0.8 μg/ml β-sitosterol according to standard procedures (Sonneborn, 1970).
Gene identification
By BLAST search in ParameciumDB (http://paramecium.cgm.cnrs-gif.fr), we identified three Paramecium genes encoding proteins homologous to SAS-4: PtSAS4a, GSPATP00004862001; PtSAS4b, GSPATP00003188001; and PtSAS4c, GSPATP00002681001. The genes PtSAS4a and PtSAS4b result from the last whole genome duplication (Aury et al., 2006).
Gene cloning
The coding regions of PtSas4a and PtSAS4c were amplified from genomic DNA by PCR using specific primers containing KpnI restriction sites. Because a KpnI restriction site was already present in PtSas4a and c, we made first a directed mutagenesis by PCR (change of the GGTACC to GGTTCC at position 2519 of PtSAS4a and change of the GGTACC to GGAACC at position 1086 of PtSAS4c). These fragments were then cloned into the KpnI restriction site at the 3′ end of the GFP synthetic (Cohen and Meyer, unpublished data) present in the pPXV vector under control of the calmodulin promoter and regulating sequences. A modified version of PtSAS4a, resistant to gene silencing, was generated by inserting silent mutations every 15–18 base pairs in the region 132–565 of PtSAS4a between the BstXI and BsrGI restriction sites (25 mutations in total; the details of the sequence can be obtained from the corresponding author). This modified version was assembled by generating a synthetic fragment of 400 base pairs (GenScript, Piscataway, NJ). The modified version of Ptsas4a (GFP-Ptsas4a*) was then fused to the GFP coding sequence in the pPXV vector. The RFP-PtSas6 and GFP-PtBld10 constructs are described in Jerka-Dziadosz et al. (2010).
Nd7-1 cells were transformed by microinjection into their macronucleus (Gilley et al., 1988) of DNA containing a mixture of the plasmid of interest (5 μg/μl) and of plasmid DNA directing the expression of the ND7 gene. Transformants were screened for their ability to discharge their trichocysts and further analyzed.
Gene silencing
Gene silencing was performed by the feeding method (Galvani and Sperling, 2002). Sequences of interest were amplified by PCR and cloned into the feeding vector, L4440, between two T7 promoters (Timmons and Fire, 1998): Regions 132–565 of PtSAS4a, 63–398 of PtSAS4b, 1553–2313 of PtSAS4c, and 80–830 of γ-tubulin were used for the cloning. The plasmid containing the IFT172 sequence was a gift from C. Laligné. These constructs were used to transform HT115 bacteria, allowing the production of double-stranded RNAi. WT cells were fed with these bacteria and transferred daily into fresh medium at 18°C. Control cells were fed with HT115 bacteria carrying the complete coding region of the ND7 gene. Control and RNAi cells were fixed together to avoid any differences in labeling due to the experimental procedure. India ink was added to the medium of the control cells. India ink particles are nontoxic for the cells, but they are absorbed in the feeding vacuoles, which allows the control cells, having dark vacuoles by phase contrast, to be distinguished from cells silenced for PtSas4, γ-tubulin, or IFT172.
PtSAS4 overexpression
GFP-PtSAS4 plasmid was injected at 5 μg/μl into Paramecium nuclei. GFP-positive cells were selected and allowed to divide for 15 d in conditions in which a cell cycle takes around 6 h (22°C). Analysis of GFP-PtSAS4 cells in these conditions guarantees that all duplicating basal bodies of a given cell have been assembled in the presence of an excess of PtSas-4.
Fluorescence microscopy
Immunostaining of cells was carried out as previously described in Beisson et al. (2001) with the addition of 0.5% saponin to all buffers. The monoclonal anti–tubulin ID5 (1:100) recognizes polyglutamylation of α-tubulin (Wehland and Weber, 1987; Rudiger et al., 1999), a gift from J. Wehland; the polyclonal anti-GFP antibody (1:100, Interchim, Montluçon, France) and the anti-tubulin decorating the cilia (1:400) (Cohen et al., 1982) were used with the appropriate secondary antibodies from Jackson ImmunoResearch Labs (West Grove, PA) at a dilution of 1:500. Cells were observed under a Zeiss Axioskop 2 Plus epifluorescence microscope and three-dimensional microscope Nikon Eclipse 90i upright equipped with a Piezo stage.
Electron microscopy
For transmission EM, cells were fixed in 1.5% glutaraldehyde and 2% paraformaldehyde in cacodylate buffer for 2 h on ice, washed three times in the buffer, and then postfixed in 1% OsO4 at 4°C. Dividing cells were selected and embedded into 2% agarose. To characterize Sas-4 depletion, we chose cells at their first division following feeding while γ-tubulin depletion was analyzed after three to four divisions following feeding.
Agar blocks were then dehydrated in graded series of ethanol and propylene oxide and embedded in Epon 812 (TAAB, Aldermaston, Bershire, UK). Thin-sectioned samples of 60–70 nm were poststained with 2% uranyl acetate in methanol for 4 min and lead citrate for 1min. The observations were made with a Philips CM120 electron microscope (FEI, Eindhoven, Netherlands). Image acquisition with a KeenView camera (SIS, Munich, Germany) and measurements were made with the iTEM software (Olympus France SA, Rungis, France).
We also attempted to characterize PtSas4 localization at the ultrastructural level by immunolabeling. Unfortunately, although we tried several different conditions and antibodies, we never succeeded in obtaining a clear answer.
Deciliation experiments
Paramecia were transferred several times in the deciliation buffer (10 mM Tris–1 mM Ca2+, 5% EtOH) to allow the cilia to detach from the cell membrane. When immobile, cells were transferred in feeding medium to allow reciliation.
Supplementary Material
Acknowledgments
We are very grateful to Carole Pennetier for technical help, and we thank C. Laligné (Center de Génétique Moléculaire, Centre National de la Recherche Scientifique [CNRS]), who provided the IFT172 construct. We are grateful to J. Beisson for valuable discussions throughout the realization of this project. We thank J. Beisson, M. Jerka-Dziadosz, V. Marthiens, T. Maia, D. Sabino, and M. Rujano for critical reading of the manuscript. We acknowledge the Nikon Imaging Center at Institut Curie and the Imaging Platform team. This work was supported by the Institut Curie, an Action Thématique et Incitative sur Programme grant from the CNRS to the lab and to D.G., and an installation grant from Fondation pour la Recherche Medicale.
Abbreviations used:
- Cr
ciliary rootlet
- EM
electron microscopy
- GFP
green fluorescent protein
- MT
microtubule
- RFP
red fluorescent protein
- RNAi
RNA interference
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0901) on February 2, 2011.
REFERENCES
- Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. In: The centrosome in evolution. In: Centrosome in Development and Disease. Nigg E, editor. Weinheim, Germany: Wiley-VCH; 2004. pp. 93–116. [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Beisson J, Clerot JC, Fleury-Aubusson A, Garreau de Loubresse N, Ruiz F, Klotz C. Basal body-associated nucleation center for the centrin-based cortical cytoskeletal network in Paramecium. Protist. 2001;152:339–354. doi: 10.1078/1434-4610-00072. [DOI] [PubMed] [Google Scholar]
- Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr Opin Cell Biol. 2003;15:96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, Avidor-Reiss T. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cho JH, Chang CJ, Chen CY, Tang TK. Depletion of CPAP by RNAi disrupts centrosome integrity and induces multipolar spindles. Biochem Biophys Res Commun. 2006;339:742–747. doi: 10.1016/j.bbrc.2005.11.074. [DOI] [PubMed] [Google Scholar]
- Cohen J, Adoutte A, Grandchamp S, Houdebine L, Beisson J. Immunocytochemical study of microtubular structures throughout the cell cycle of Paramecium. Biol Cell. 1982;44:35–44. [Google Scholar]
- Culver BP, Meehl JB, Giddings TH, Jr, Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol Biol Cell. 2009;20:1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin–mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Pemble H, Mitchell BJ, McLeod I, Yates JR, III, Kintner C, Desai AB, Oegema K. The hydrolethalus syndrome protein HYLS-1 links core centriole structure to cilia formation. Genes Dev. 2009;23:2046–2059. doi: 10.1101/gad.1810409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in Paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Williams P, Fleury-Aubusson A, de Loubresse NG, Geoffroy H, Vayssie L, Galvani A, Espigat A, Rossier J. Functional role of ε-tubulin in the assembly of the centriolar microtubule scaffold. J Cell Biol. 2002;158:1183–1193. doi: 10.1083/jcb.200205028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani A, Sperling L. RNA interference by feeding in Paramecium. Trends Genet. 2002;18:11–12. doi: 10.1016/s0168-9525(01)02548-3. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2001;2:4. doi: 10.1186/1471-2121-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Preer JR, Jr, Aufderheide KJ, Polisky B. Autonomous replication and addition of telomerelike sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol Cell Biol. 1988;8:4765–4772. doi: 10.1128/mcb.8.11.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the ninefold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftode F, Fleury A, Adoutte A. Development of surface pattern during division in Paramecium. III. Study of stomatogenesis in the wild type using antitubulin antibodies and confocal microscopy. Europ J Protistol. 1997;33:145–167. [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton. 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- Keller LC, Geimer S, Romijn E, Yates J, III, Zamora I, Marshall WF. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol Biol Cell. 2009;20:1150–1166. doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laligne C, Klotz C, de Loubresse NG, Lemullois M, Hori M, Laurent FX, Papon JF, Louis B, Cohen J, Koll F. Bug22p, a conserved centrosomal/ciliary protein also present in higher plants, is required for an effective ciliary stroke in Paramecium. Eukaryot Cell. 2010;9:645–655. doi: 10.1128/EC.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Gull K. Structure and function of the centriole in animal cells: progress and questions. Trends Cell Biol. 1996;6:348–352. doi: 10.1016/0962-8924(96)10033-7. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the ninefold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Oakley BR. γ-Tubulin: the microtubule organizer? Trends Cell Biol. 1992;2:1–5. doi: 10.1016/0962-8924(92)90125-7. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Osborn DP, Giddings TH, Jr, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J Cell Biol. 2009;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Rudiger AH, Rudiger M, Wehland J, Weber K. Monoclonal antibody ID5: epitope characterization and minimal requirements for the recognition of polyglutamylated α- and β-tubulin. Eur J Cell Biol. 1999;78:15–20. doi: 10.1016/s0171-9335(99)80003-x. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Beisson J, Rossier J, Dupuis-Williams P. Basal body duplication in Paramecium requires γ-tubulin. Curr Biol. 1999;9:43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Ruiz F, Krzywicka A, Klotz C, Keller A, Cohen J, Koll F, Balavoine G, Beisson J. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, η-tubulin. Curr Biol. 2000;10:1451–1454. doi: 10.1016/s0960-9822(00)00804-6. [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Skouri F, Cohen J. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol Biol Cell. 1997;8:1063–1071. doi: 10.1091/mbc.8.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. Methods in Paramecium research. Methods Cell Physiol. 1970;4:241–339. [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395(6705):854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehland J, Weber K. Turnover of the carboxy-terminal tyrosine of α-tubulin and means of reaching elevated levels of detyrosination in living cells. J Cell Sci. 1987;88:185–203. doi: 10.1242/jcs.88.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.