Abstract
When human DNA(cytosine-5)methyltransferase was used to methylate a series of snapback oligodeoxy-nucleotides of differing stem lengths, each containing a centrally located CG dinucleotide recognition site, the enzyme required a minimum of 22 base pairs in the stem for maximum activity. Extrahelical cytosines in slipped duplexes that were 30 base pairs in length acted as effective methyl acceptors and were more rapidly methylated than cytosines that were Watson-Crick paired. Duplexes containing hairpins of CCG repeats in cruciform structures in which the enzyme recognition sequence was disrupted by a C.C mispair were also more rapidly methylated than control Watson-Crick-paired duplexes. Since enzymes have higher affinities for their transition states than for their substrates, the results with extrahelical and mispaired cytosines suggest that these structures can be viewed as analogs of the transition state intermediates produced during catalysis by methyltransferases.
Full text
PDF

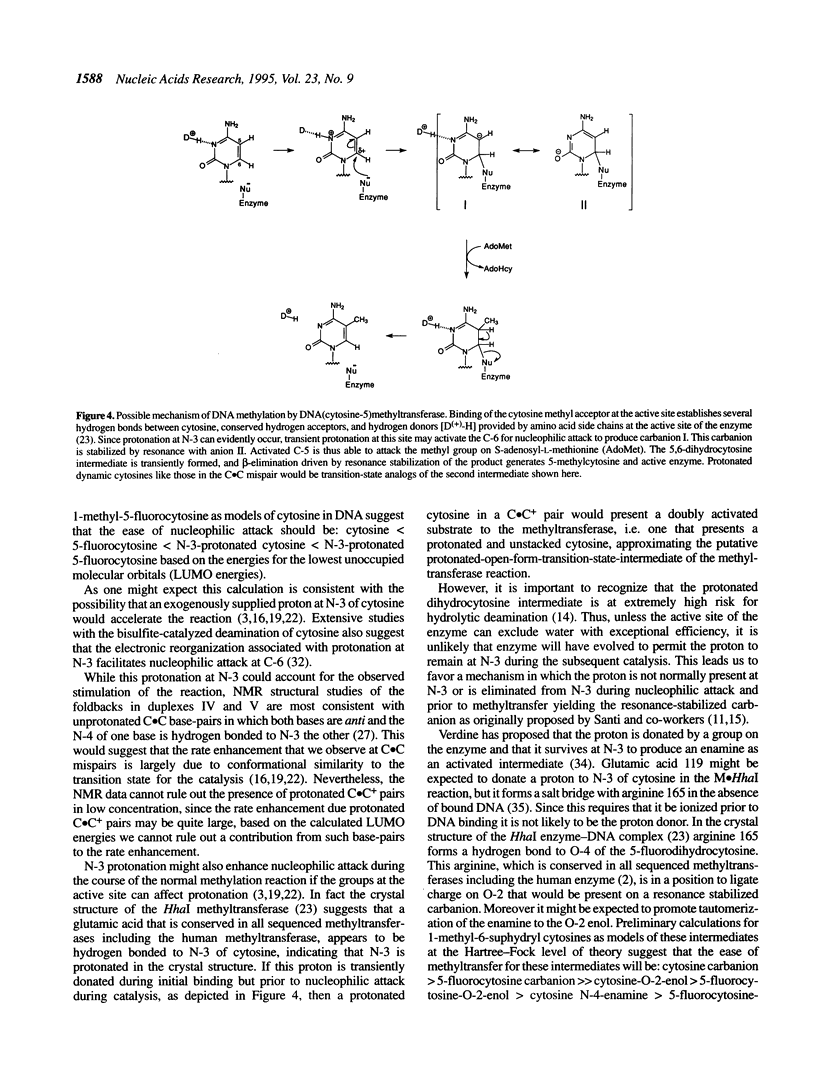

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S., Henderson E. Formation of novel hairpin structures by telomeric C-strand oligonucleotides. Nucleic Acids Res. 1992 Feb 11;20(3):507–511. doi: 10.1093/nar/20.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D. J., Kan J. L., Smith S. S. Recognition of structural perturbations in DNA by human DNA(cytosine-5)methyltransferase. Gene. 1988 Dec 25;74(1):207–210. doi: 10.1016/0378-1119(88)90288-0. [DOI] [PubMed] [Google Scholar]
- Baker D. J., Laayoun A., Smith S. S. Transition state analogs as affinity labels for human DNA methyltransferases. Biochem Biophys Res Commun. 1993 Oct 29;196(2):864–871. doi: 10.1006/bbrc.1993.2329. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Craig I. Human genetics. Methylation and the fragile X. Nature. 1991 Feb 28;349(6312):742–743. doi: 10.1038/349742a0. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Toth M., Kochanek S., Achten S., Freisem-Rabien U., Behn-Krappa A., Orend G. Eukaryotic DNA methylation: facts and problems. FEBS Lett. 1990 Aug 1;268(2):329–333. doi: 10.1016/0014-5793(90)81280-2. [DOI] [PubMed] [Google Scholar]
- Duckett D. R., Murchie A. I., Diekmann S., von Kitzing E., Kemper B., Lilley D. M. The structure of the Holliday junction, and its resolution. Cell. 1988 Oct 7;55(1):79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Catalysis, binding and enzyme-substrate complementarity. Proc R Soc Lond B Biol Sci. 1974 Nov 19;187(1089):397–407. doi: 10.1098/rspb.1974.0084. [DOI] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M., Scott C. R., Chen S. H., Laird C. D. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992 Nov;1(8):571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- Hayatsu H. Bisulfite modification of nucleic acids and their constituents. Prog Nucleic Acid Res Mol Biol. 1976;16:75–124. doi: 10.1016/s0079-6603(08)60756-4. [DOI] [PubMed] [Google Scholar]
- Hepburn P. A., Margison G. P., Tisdale M. J. Enzymatic methylation of cytosine in DNA is prevented by adjacent O6-methylguanine residues. J Biol Chem. 1991 May 5;266(13):7985–7987. [PubMed] [Google Scholar]
- Hwu W. L., Lee Y. M., Lee S. C., Wang T. R. In vitro DNA methylation inhibits FMR-1 promoter. Biochem Biophys Res Commun. 1993 May 28;193(1):324–329. doi: 10.1006/bbrc.1993.1627. [DOI] [PubMed] [Google Scholar]
- Jones P. A., Buckley J. D. The role of DNA methylation in cancer. Adv Cancer Res. 1990;54:1–23. doi: 10.1016/s0065-230x(08)60806-4. [DOI] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laayoun A., Baker D. J., Riley J., Smith S. S. The response of M.HpaII to heteroduplexes. Gene. 1994 Dec 2;150(1):195–196. doi: 10.1016/0378-1119(94)90884-2. [DOI] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Smith S. S. Biological implications of the mechanism of action of human DNA (cytosine-5)methyltransferase. Prog Nucleic Acid Res Mol Biol. 1994;49:65–111. doi: 10.1016/s0079-6603(08)60048-3. [DOI] [PubMed] [Google Scholar]
- Smith S. S. DNA methylation in eukaryotic chromosome stability. Mol Carcinog. 1991;4(2):91–92. doi: 10.1002/mc.2940040202. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Hardy T. A., Baker D. J. Human DNA (cytosine-5)methyltransferase selectively methylates duplex DNA containing mispairs. Nucleic Acids Res. 1987 Sep 11;15(17):6899–6916. doi: 10.1093/nar/15.17.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kan J. L., Baker D. J., Kaplan B. E., Dembek P. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991 Jan 5;217(1):39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Laayoun A., Lingeman R. G., Baker D. J., Riley J. Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J Mol Biol. 1994 Oct 21;243(2):143–151. doi: 10.1006/jmbi.1994.1640. [DOI] [PubMed] [Google Scholar]
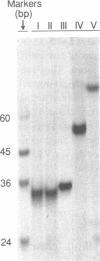
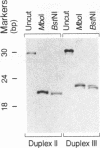
- Smith S. S., Lingeman R. G., Kaplan B. E. Recognition of foldback DNA by the human DNA (cytosine-5-)-methyltransferase. Biochemistry. 1992 Jan 28;31(3):850–854. doi: 10.1021/bi00118a030. [DOI] [PubMed] [Google Scholar]
- Som S., Friedman S. Inhibition of transcription in vitro by binding of DNA (cytosine-5)-methylases to DNA templates containing cytosine analogs. J Biol Chem. 1994 Oct 21;269(42):25986–25991. [PubMed] [Google Scholar]
- Tan N. W., Li B. F. Interaction of oligonucleotides containing 6-O-methylguanine with human DNA (cytosine-5-)-methyltransferase [published erratumm appears in Biochemistry 1992 Aug 4;31(30):7008]. Biochemistry. 1990 Oct 2;29(39):9234–9240. doi: 10.1021/bi00491a018. [DOI] [PubMed] [Google Scholar]
- Verdine G. L. The flip side of DNA methylation. Cell. 1994 Jan 28;76(2):197–200. doi: 10.1016/0092-8674(94)90326-3. [DOI] [PubMed] [Google Scholar]
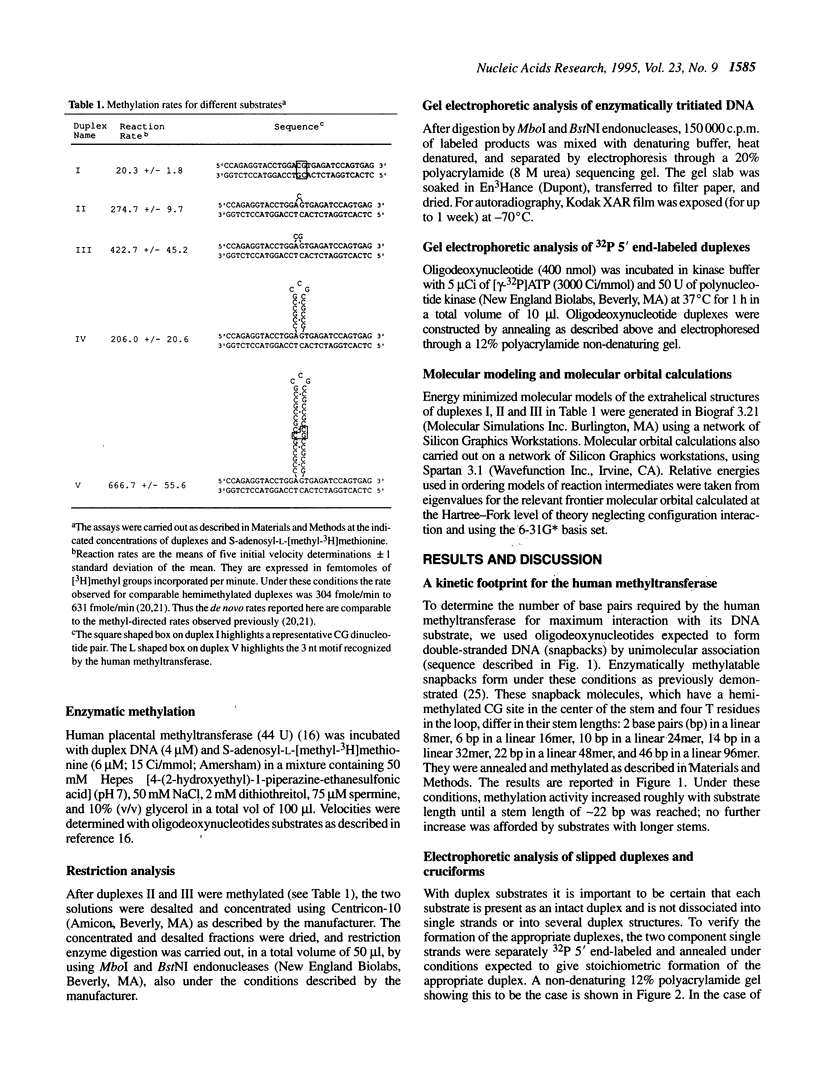
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wurdeman R. L., Douskey M. C., Gold B. DNA methylation by N-methyl-N-nitrosourea: methylation pattern changes in single- and double-stranded DNA, and in DNA with mismatched or bulged guanines. Nucleic Acids Res. 1993 Oct 25;21(21):4975–4980. doi: 10.1093/nar/21.21.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]