This study's findings identify a novel and unexpected function for coilin, independentof its role in snRNP biogenesis, establishing a new link between the DNA damage response and the inhibition of rRNA synthesis.
Abstract
Coilin is a nuclear phosphoprotein that concentrates within Cajal bodies (CBs) and impacts small nuclear ribonucleoprotein (snRNP) biogenesis. Cisplatin and γ-irradiation, which cause distinct types of DNA damage, both trigger the nucleolar accumulation of coilin, and this temporally coincides with the repression of RNA polymerase I (Pol I) activity. Knockdown of endogenous coilin partially overrides the Pol I transcriptional arrest caused by cisplatin, while both ectopically expressed and exogenous coilin accumulate in the nucleolus and suppress rRNA synthesis. In support of this mechanism, we demonstrate that both cisplatin and γ-irradiation induce the colocalization of coilin with RPA-194 (the largest subunit of Pol I), and we further show that coilin can specifically interact with RPA-194 and the key regulator of Pol I activity, upstream binding factor (UBF). Using chromatin immunoprecipitation analysis, we provide evidence that coilin modulates the association of Pol I with ribosomal DNA. Collectively, our data suggest that coilin acts to repress Pol I activity in response to cisplatin-induced DNA damage. Our findings identify a novel and unexpected function for coilin, independent of its role in snRNP biogenesis, establishing a new link between the DNA damage response and the inhibition of rRNA synthesis.
INTRODUCTION
Cajal bodies (CBs) are dynamic nuclear bodies consisting of proteins with diverse functions (Cioce and Lamond, 2005; Nizami et al., 2010). The emerging consensus is that CBs provide locally high concentrations of factors necessary for small nuclear ribonucleoprotein (snRNP) biogenesis (Morris, 2008; Matera et al., 2009). In zebrafish, for example, CBs are required for snRNP assembly and embryogenesis (Strzelecka et al., 2010). In the zebrafish study (Strzelecka et al., 2010), CBs were abolished by depleting coilin, the CB marker protein that plays a crucial role in CB formation and composition (Hebert, 2010). Sm proteins, protein components of snRNPs, and survival of motor neurons protein (SMN), the snRNP assembly protein, have been shown to bind coilin’s C terminus, which contains an atypical Tudor domain (Xu et al., 2005; Shanbhag et al., 2010). Additionally, the C terminus of coilin has been shown to regulate the availability of the N terminus for self-interaction (Hebert and Matera, 2000; Shpargel et al., 2003). Posttranslational modifications of coilin, such as symmetrical arginine dimethylation (Boisvert et al., 2002; Hebert et al., 2002) and phosphorylation (Hearst et al., 2009), play a significant role in mammalian CB formation.
Aside from coilin’s crucial role in CB integrity, little is known about the function of nucleoplasmic coilin, where the vast majority of the protein is located (Lam et al., 2002). Recent evidence indicates that nucleoplasmic coilin may play a part in stress response pathways. For example, coilin interacts with Ku proteins, which are involved in the nonhomologous end-joining (NHEJ) pathway of DNA repair, and inhibits in vitro NHEJ (Velma et al., 2010). Coilin has also been shown to respond to interphase centromere damage induced by herpes simplex virus type 1 infection by accumulating at centromeres (Morency et al., 2007). Additionally, UV-C–induced DNA damage fragments CBs and redistributes coilin to interact with the proteasomal protein PA28γ (Cioce et al., 2006). These findings are significant because they suggest new functions for coilin that appear to be independent of snRNP biogenesis. Although the above-mentioned studies indicate that coilin is responsive to DNA damage, it has not yet been determined what function coilin has in response to this cellular stress.
In this study, we have assessed the role coilin plays in the cellular response to DNA damage induced by cisplatin treatment and γ-irradiation. We demonstrate that coilin localizes in the nucleolus in response to these types of DNA damage. Additionally, we provide evidence suggesting that coilin regulates the activity of Pol I. Altogether, our data suggest that a novel function for coilin in response to cisplatin-induced DNA damage or γ-irradiation is to inhibit Pol I activity.
RESULTS
Cisplatin-induced DNA damage triggers the nucleolar accumulation of coilin
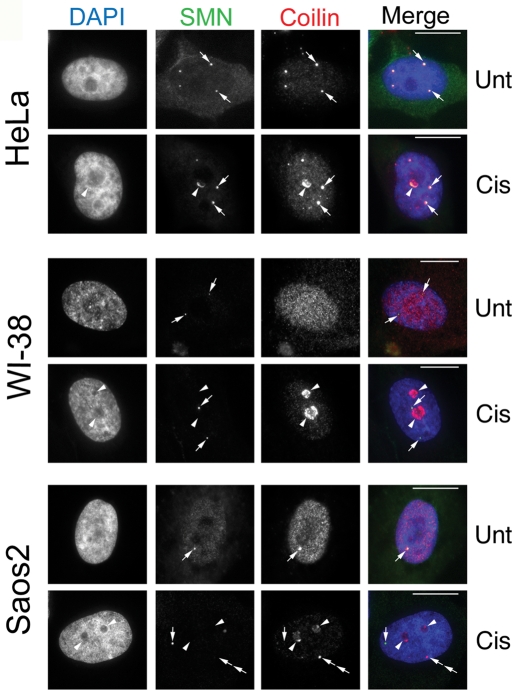
The chemotherapeutic agent cisplatin damages DNA by introducing cross-links that stall the replication fork and induce apoptosis (Bartek et al., 2004). To examine the effect of cisplatin and γ-irradiation (discussed later in this article) on CB and coilin localization, we used four different cell lines (one primary and three transformed) with varying p53 status: Saos2 (human osteosarcoma, p53 null), HeLa (human cervical carcinoma, p53 wild-type; HPV E6 positive), WI-38 (human normal fetal lung fibroblast, p53 wild-type), and H1299 (human non–small cell lung carcinoma, p53 null). In untreated HeLa and Saos2 cells, coilin is localized in the nucleoplasm and CBs, while in untreated WI-38 cells, coilin is distributed diffusely throughout the nucleoplasm (Figure 1). Strikingly, after a 24-h cisplatin treatment, we observed that coilin predominantly accumulated within and around nucleoli in all cell lines tested (Figure 1). These accumulations appear to be redistributions of coilin as coilin protein levels are relatively unchanged in response to cisplatin treatment, despite a twofold increase in message levels (Supplemental Figure 1). SMN, a major coilin interacting partner found in the cytoplasm and in CBs, was also enriched in the periphery of the nucleolus in HeLa cells, but not in the other cell lines (Figure 1). We also observed that 24-h cisplatin treatment induced Gem (SMN foci without coilin) formation in ∼50% of Saos2 cells, which normally have one CB that contains both SMN and coilin (Figure 1). In HeLa cells, coilin predominantly localized in nucleolar cap–like structures, whereas in Saos2 and WI-38 cells, coilin accumulated inside the nucleolus in addition to nucleolar caps. The cisplatin-induced nucleolar cap phenotype of coilin in HeLa and Saos2 cells is similar to that found in response to the transcription inhibitor actinomycin D (ActD) (Carmo-Fonseca et al., 1992; Supplemental Figure 2). However, in cisplatin-treated WI-38 cells (Figure 1), the nucleolar accumulation of coilin was more obvious than that found in ActD-treated cells (Supplemental Figure 2), where coilin localized more exclusively to nucleolar caps. This cisplatin-induced accumulation of coilin within and around the nucleolus has not been described.
FIGURE 1:
Cisplatin-induced DNA damage triggers nucleolar coilin accumulation. HeLa, WI-38, and Saos2 cells were untreated (Unt) or treated (Cis) for 24 h with cisplatin and immunostained for coilin (red) and SMN (green). Nuclei were stained by 4′,6-diamino-2-phenylindole (DAPI) (blue). For HeLa cells, arrows denote CBs containing both SMN and coilin. Arrowheads mark nucleolar accumulation of coilin and SMN. For WI-38 cells, SMN foci lacking coilin (Gems) are indicated by arrows, and arrowheads mark nucleolar coilin. For Saos2 cells, arrows in the untreated cell denote a CB. Arrows in the cisplatin-treated cells mark a Gem while arrowheads show nucleolar coilin. Double arrows represent an SMN-negative coilin focus. Scale bars = 10 μm.
The nucleolar distribution of fibrillarin and Nopp140 is altered in response to cisplatin and γ-irradiation
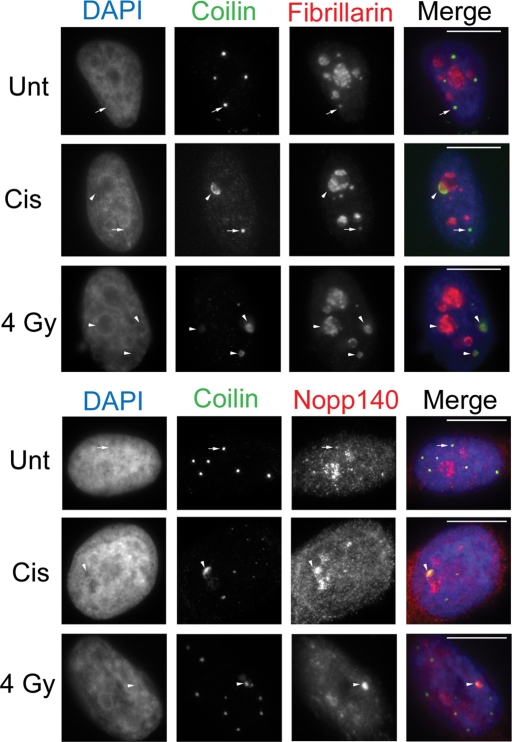
In addition to coilin, we also looked at the CB components fibrillarin and Nopp140 in response to cisplatin and γ-irradiation. Both of these CB components are also normally nucleolar. In HeLa cells, fibrillarin and Nopp140 responded to cisplatin by redistributing to the nucleolar periphery, where they colocalize with coilin (Figure 2). These results were recapitulated in WI-38, Saos2, and H1299 cell lines, although the extent of Nopp140 and fibrillarin nucleolar segregation was less distinct in the H1299 line (Supplemental Figure 3). γ-Irradiation, which causes double-strand breaks, produced similar results in HeLa, Saos2, and H1299 cells as observed for cisplatin treatment in regard to coilin, fibrillarin, and Nopp140 redistribution (Figure 2 and Supplemental Figure 3). Interestingly, in the p53-positive WI-38 primary cell line, γ-irradiation did not result in an appreciable redistribution of coilin, fibrillarin, or Nopp140 (Supplemental Figure 3). To monitor whether the localization of mature snRNPs, which are also enriched in CBs, was altered in response to cisplatin-induced DNA damage, we detected the trimethyl guanosine (TMG) cap of processed small nuclear RNA. The TMG cap epitope colocalized with coilin within and around nucleoli in cisplatin-treated WI-38 cells, but not in HeLa or Saos2 cells (Supplemental Figure 4). These results demonstrate that coilin and other CB components such as fibrillarin, Nopp140, and snRNPs reorganize within and around nucleoli in response to DNA damage caused by cisplatin and γ-irradiation. However, the extent of this reorganization varies among the cell lines studied and the mechanism of DNA damage.
FIGURE 2:
Fibrillarin and Nopp140 redistribute with coilin in response to cisplatin and γ-irradiation. HeLa cells were untreated (Unt) or treated with cisplatin or γ-irradiation (4 Gy) and immunostained for coilin, fibrillarin, and Nopp140 after 24 h. DAPI staining was used to detect the nucleus. Arrows mark CBs. Arrowheads mark coilin and fibrillarin or Nopp140 nucleolar redistributions. Scale bars = 10 μm.
Nucleolar coilin is associated with Pol I suppression
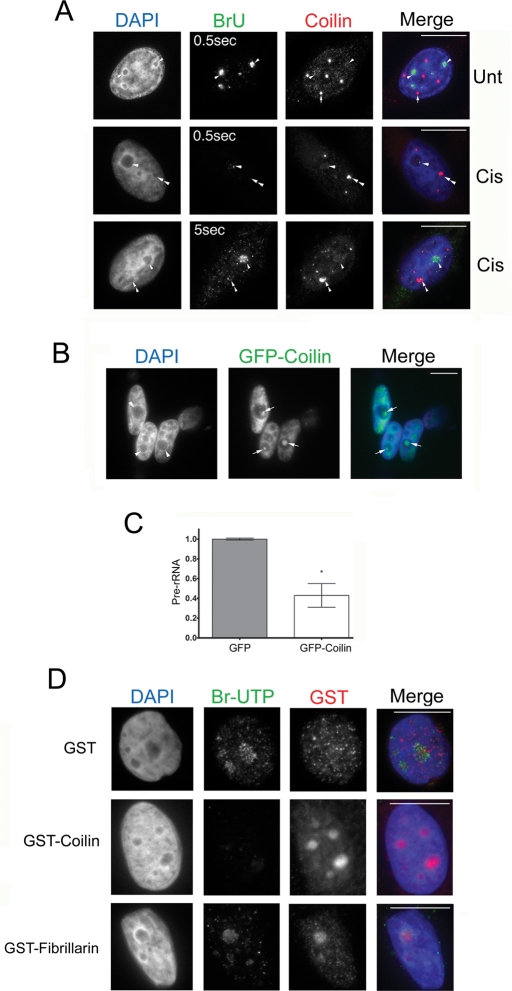
Previous studies have shown that Pol I activity can be disrupted by double-stranded DNA breaks induced by γ-irradiation and by DNA adducts generated from cisplatin treatment (Jordan and Carmo-Fonseca, 1998; Kruhlak et al., 2007). Consequently, we speculated that the nucleolar accumulation of coilin in response to cisplatin treatment and γ-irradiation might be partly responsible for this diminished rRNA synthesis. To test this possibility, we first assessed whether there was a correlation between the accumulation of coilin in the nucleolus and the suppression of rRNA synthesis in cisplatin-treated cells by measuring the incorporation of bromouridine (BrU) into nascent transcripts (Figure 3A). Untreated cells exhibited a strong nucleolar BrU signal (i.e., detectable with a 0.5-s exposure) that is indicative of active Pol I transcription (Figure 3A). In agreement with a previous report (Jordan and Carmo-Fonseca, 1998), cisplatin potently suppressed Pol I activity as indicated by a weak BrU signal (detectable with a 5-s exposure). Interestingly, in cisplatin-treated cells, we observed an inverse relationship between the presence of nucleolar coilin and the extent of BrU incorporation. In nucleoli that lacked coilin staining, we could detect a BrU signal, whereas nucleoli that were positive for coilin staining had a much weaker BrU signal (for the cisplatin-treated cell in the bottom panel of Figure 3A, compare the amount of BrU signal in the nucleolus without coilin [arrowhead] to the nucleolus that contains coilin [double arrowhead]). These data suggest that the presence of nucleolar coilin may inhibit Pol I activity.
FIGURE 3:
Nucleolar coilin suppresses Pol I transcription. (A) Coilin-occupied nucleoli are associated with reduced BrU incorporation. Untreated or cisplatin-treated (24 h) HeLa cells were incubated in 2 mM BrU for 2 h followed by detection of Pol I transcripts with anti-BrdU antibodies (green) and coilin with anti-coilin antibodies (red) and DAPI staining (blue). Note that the BrU signal in the bottom panel was detected using a 5-s exposure. Arrowheads mark BrU-positive nucleoli, while arrows denote CBs. In treated cells, nucleoli with coilin accumulation (double arrowhead) lack BrU signal, yet BrU signal is present in nucleoli that lack intense coilin staining (arrowhead). Scale bars = 10 μm. (B) Transient expression of GFP-coilin results in nucleolar coilin. The nucleolar distribution of GFP-coilin (arrow) is typical of overexpressing cells, which represented ∼20% of transfected cells. Scale bars = 10 μm. (C) Transient expression of GFP-coilin reduces the level of pre-rRNA. RT-PCR was performed on RNA isolated from HeLa cells transfected with GFP-coilin or empty GFP vector. The amount of pre-rRNA was determined relative to GAPDH message levels. Data are normalized to the relative amount of pre-rRNA message obtained from HeLa cells transfected with empty GFP vector (error bars expressed as mean fold induction ± coefficient of variation [CV]; *p < 0.01) and reflect two independent experiments. (D) Exogenous coilin reduces BrUTP incorporation. In situ transcription run-on assay was performed on HeLa cells in the presence of bacterially purified GST-coilin, GST-fibrillarin, or GST. Cells were fixed and stained for BrUTP (green), GST (red), and DAPI (blue). Exposure times were the same for all images. Scale bars = 10 μm.
It has been previously reported (Hebert and Matera, 2000) that cells overexpressing coilin have distinct nucleolar accumulations (Figure 3B). To test whether ectopic coilin expression alone would be sufficient to suppress Pol I activity, we transfected HeLa cells with either green fluorescent protein (GFP) or GFP-coilin plasmids and performed reverse transcription PCR (RT-PCR) analysis of the 5′ external transcribed spacer (ETS) of pre-rRNA, whose levels reflect the rate of Pol I transcription (Grandori et al., 2005; Frescas et al., 2007). RT-PCR analysis of the 5′ETS of pre-rRNA revealed that ectopically expressed coilin suppressed pre-rRNA levels by ∼60% (Figure 3C). These results support the notion that Pol I activity is regulated by coilin, although the possibility exists that ectopically expressed coilin is indirectly regulating rRNA synthesis.
To assess directly whether coilin alone was sufficient to suppress Pol I activity, we conducted an in situ run-on transcription assay in the presence of bacterially generated recombinant coilin, fibrillarin, or GST. Both GST and GST-fibrillarin–treated cells had robust Pol I transcriptional activity, as assessed by detection of BrU triphosphate (BrUTP) within nucleoli (Figure 3D). However, cells treated with GST-coilin had reduced Pol I activity (Figure 3D), although coilin alone was not sufficient to abolish Pol I activity because faint BrUTP signal could still be detected in the nucleolus. Collectively, these results suggest that coilin may be an important factor in the suppression of Pol I activity.
Coilin associates with RPA-194 and upstream binding factor (UBF)
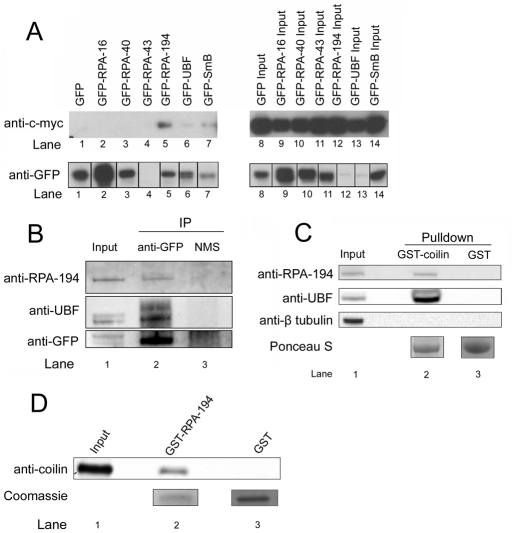
Because cisplatin-induced DNA damage triggers the accumulation of coilin in the nucleolus, and this coincides with the inhibition of Pol I activity, we decided to test whether coilin could interact with Pol I. Active Pol I is a multisubunit protein complex, and thus we performed a screen to determine whether coilin could interact with Pol I–specific subunits and UBF. Myc-tagged coilin was cotransfected with GFP-tagged versions of RPA-16, -40, -43, -194, UBF, and SmB (as a positive control), and coimmunoprecipitation was performed with a GFP antibody (Figure 4A). Among the different Pol I subunits tested, we found that coilin coimmunoprecipitates only with GFP–RPA-194 (lane 5) and GFP-UBF (lane 6). Because GFP–RPA-43 was not sufficiently immunoprecipitated by the GFP antibody (lane 4, bottom), it remains uncertain whether this subunit forms a complex with coilin. Additionally, we performed two separate experiments using a similar approach to confirm that coilin can interact with RPA-194 and UBF. First, transfected GFP-coilin can coimmunoprecipitate endogenous RPA-194 and UBF (Figure 4B) and, second, GST-coilin, but not GST, can recover RPA-194 and UBF from HeLa lysate (Figure 4C). To test for a potential direct interaction between coilin and RPA-194, a GST pull-down assay was performed with recombinant His-coilin and GST–RPA-194 or GST beads. GST–RPA-194 beads specifically recovered His-coilin (Figure 4D).
FIGURE 4:
Coilin interacts with RPA-194 and UBF. (A) Coilin specifically interacts with RPA-194 and UBF. Myc-coilin and various GFP constructs were cotransfected into H1299 cells. Lysate was subjected to IP with a polyclonal GFP antibody, followed by SDS–PAGE, Western transfer, and detection of myc-coilin and the various GFP constructs using monoclonal anti-myc (top) and anti-GFP antibodies (bottom), respectively. The IP samples are shown in lanes 1–7. (B) Ectopically expressed GFP-coilin coimmunoprecipitates endogenous RPA-194 and UBF. Lysate from HeLa cells transfected with GFP-coilin was subjected to IP with anti-GFP antibodies or normal mouse serum (NMS), followed by SDS–PAGE, Western transfer, and detection of RPA-194, UBF, and GFP-coilin using the appropriate antibodies. (C) GST-coilin recovers endogenous RPA-194 and UBF. HeLa lysate was incubated with GST-coilin or GST beads, followed by SDS–PAGE, Western transfer, and detection of RPA-194, UBF, and β-tubulin (negative control) with the appropriate antibodies. Ponceau S verified that approximately equivalent amounts of GST-coilin and GST were used. (D) Coilin directly interacts with RPA-194. Recombinant human GST–RPA-194 or GST was incubated with recombinant human coilin followed by SDS–PAGE, Western transfer, and detection of coilin with a coilin antibody. Coomassie stain shows the amount of GST and GST–RPA-194 used in the assay. Input represents 30% of total protein.
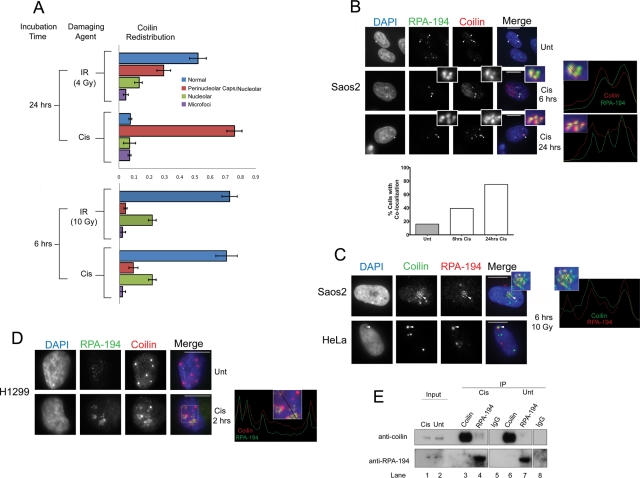
The distribution of coilin within nucleoli in response to cisplatin and γ-irradiation suggested an in vivo interaction between coilin and RPA-194 in response to these types of DNA damage. Before we checked for colocalization between these proteins, we quantified the redistribution of coilin for both types of DNA damage at 6 h and 24 h in HeLa cells. We noted that coilin would often accumulate in perinucleolar caps and within the same nucleolus (Figures 2 and 5C; Supplemental Figure 5) especially after 24 h and in response to cisplatin treatment (Figure 5A). Strictly nucleolar distributions of coilin were more obvious in response to γ-irradiation and at 6 h for both types of DNA damage (Figure 5A). We also occasionally observed coilin distributed in numerous microfoci in response to cisplatin and γ-irradiation (Supplemental Figure 6, quantified in Figure 5A) that appeared similar to the microfoci resulting from UV-C treatment (Cioce et al., 2006).
FIGURE 5:
Coilin colocalizes and associates with RPA-194 in response to cisplatin and γ-irradiation. (A) Quantification of coilin redistribution in response to cisplatin and γ-irradiation. Cisplatin-treated or γ-irradiated HeLa cells were incubated for the indicated time, fixed, and immunostained with coilin. Nuclei were stained with DAPI. The proportion of cells having coilin in CBs (normal), in both perinucleolar caps and inside the nucleolus (Perinucleolar caps/Nucleolar), in the nucleolus exclusively (Nucleolar), or in small punctate foci throughout the nucleoplasm (Microfoci) were quantified (n = 100 cells) for each condition in two separate experiments (error bars represent mean ± CV). (B) Cisplatin induces coilin and RPA-194 colocalization in Saos2 cells. Cells were untreated or treated with cisplatin for 6 h and 24 h and immunostained with RPA-194 (green), coilin (red), and DAPI (blue). In untreated cells (top), arrowheads show normal RPA-194 nucleolar accumulation, and the arrow points to a CB. In treated cells (middle and bottom), arrowheads mark RPA-194 foci that colocalize with coilin after cisplatin treatment (insets). The right two panels are line profiles that represent the RPA-194 (green) and coilin (red) channels for 6 h and 24 h cisplatin treatment using lines shown in the enlarged insets. Scale bars = 10 μm. (Below) Histogram showing the frequency of colocalization between coilin and RPA-194 for 6 and 24 h cisplatin treatment (n = 100). (C) γ-Irradiation induces coilin and RPA-194 colocalization in Saos2 and HeLa cells. Cells were treated with 10 Gy of γ-irradiation, incubated for 6 h, and subsequently fixed and immunostained with coilin (green), RPA-194 (red), and DAPI (blue). Arrowheads mark nucleolar colocalization between coilin and RPA-194. A line profile represents coilin (green) and RPA-194 (red) channels using the line shown in the enlarged inset in the right panel. (D) Cisplatin treatment of H1299 cells results in coilin and RPA-194 colocalization. Cells were untreated or treated with 6 μg/ml cisplatin for 2 h and subsequently fixed and immunostained for RPA-194 (green), coilin (red), and DAPI (blue). A line profile represents coilin and RPA-194 channels using the line shown in the boxed inset in the right panel. (E) The interaction of endogenous coilin with RPA-194 is increased in response to cisplatin treatment. H1299 cells were untreated or treated with 6 μg/ml cisplatin for 2 h, and coimmunoprecipitation was performed using antibodies for coilin, RPA-194, and control rabbit IgG followed by SDS–PAGE and Western transfer. The blot was probed with anti-coilin (top) or anti–RPA-194 (bottom) antibodies. Cisplatin-treated IP lanes are 3, 4, and 5. Inputs represent 1% of the total cell lysate.
We then explored the colocalization of coilin and RPA-194 in Saos2, HeLa, and H1299 after cisplatin treatment and γ-irradiation. In Saos2 cells, cisplatin treatment induced the frequency of coilin and RPA-194 colocalization after both 6 and 24 h (Figure 5B, insets and histogram), although the frequency of colocalized signals was most abundant at 24 h. Extensive colocalization between coilin and RPA-194 was also observed within 6 h of γ-irradiation in Saos2 and in ∼20% of HeLa cells (Figure 5C, inset). Cisplatin treatment of H1299 cells also induced colocalization between coilin and RPA-194 within 2 h (Figure 5D). To verify an induced interaction between endogenous coilin and endogenous RPA-194 in H1299 cells after a 2-h cisplatin treatment, we performed coimmunoprecipitation. A small amount of coilin was coimmunoprecipitated with RPA-194 in untreated cells (Figure 5E, lane 7), and in response to cisplatin treatment this association increased (compare coilin and RPA-194 signals for untreated lane 7 to cisplatin-treated lane 4). Collectively, these experiments demonstrate that coilin associates with RPA-194 and this association increases in response to cisplatin.
Coilin depletion in cisplatin-treated cells partially rescues Pol I transcription
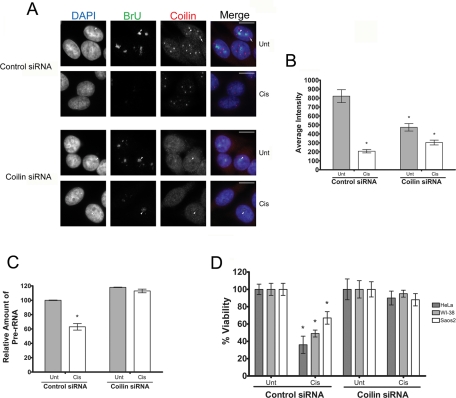
Because we established that ectopically expressed coilin suppressed Pol I activity, we next sought to assess directly whether coilin deficiency impacted the rate of BrU incorporation. Unexpectedly, HeLa cells transfected with coilin small interfering RNA (siRNA) had moderately reduced BrU incorporation (Figure 6A, data quantified in Figure 6B). We suspect that this may reflect the fact that coilin has important roles in other cellular processes such as snRNP assembly. Given that pre-mRNA splicing is perturbed in coilin-deficient cells (Whittom et al., 2008; Strzelecka et al., 2010), it is likely that coilin knockdown may itself generate a stress response in the cell that leads to a decrease in rRNA synthesis. However, in comparison to control siRNA–transfected cells, cisplatin treatment had a much less pronounced effect on Pol I activity in coilin siRNA–transfected cells (Figure 6, A and B). Similarly, coilin knockdown in Saos2 cells partially eliminated the inhibition of Pol I activity caused by cisplatin (Supplemental Figure 7).
FIGURE 6:
Coilin knockdown partially rescues Pol I transcription in the presence of cisplatin. (A) Coilin siRNA–treated cells partially recover Pol I transcription in the presence of cisplatin. HeLa cells transfected with control or coilin siRNA for 48 h were subjected to cisplatin (Cis) for 6 h or were left untreated (Unt). At 2 h before harvest, cells were incubated in 2 mM BrU, followed by detection of BrU (green) and coilin (red). Arrows mark a CB, and arrowheads show some of the BrU-positive nucleoli. Note that all BrU signals were detected using a 0.5-s exposure. (B) BrU pixel intensities from (A) were quantified. At least 49 cells were quantified for each condition, and the values represent the mean pixel intensity ± SD. All data vary from each other with a p value < 0.001. (C) pre-rRNA levels recover in the presence of cisplatin in coilin knockdown cells. RT-PCR using RNA from HeLa cells treated as described in (A) was used to detect the relative abundance of pre-rRNA to GAPDH. Data are normalized to the relative amount of pre-rRNA levels from untreated control knockdown cells (error bars expressed as mean fold induction ± CV; *p < 0.01 and reflects three independent experiments). (D) Cell viability is enhanced in coilin knockdown cells treated with cisplatin. HeLa, WI-38, or Saos2 cells were transfected with control or coilin siRNAs. At 24 h posttransfection, equal numbers of cells were seeded into 96-well dishes. At 48 h posttransfection, cells were untreated (Unt) or treated (Cis) with the appropriate amount of cisplatin for 24 h, followed by cell number determination. Thus all data come from cells 72 h after siRNA transfection. Untreated control siRNA–transfected and untreated coilin siRNA–transfected cell numbers were normalized to 100%, and cisplatin-treated cells are expressed as a percentage of their respective normalized controls (error bars represent the mean ± CV, n = 8; *p < 0.001).
To corroborate these results, we performed RT-PCR on the 5′ETS of pre-rRNA using RNA polymerase II (Pol II)–derived glyceraldehyde-3-phosphate dehydrogenase (GAPDH) message as a normalizer given that it has previously been shown that cisplatin does not affect Pol II activity (Jordan and Carmo-Fonseca, 1998). As expected, cisplatin treatment of control siRNA–transfected cells reduced the relative levels of pre-rRNA (Figure 6C). However, cisplatin treatment of coilin knockdown cells did not significantly reduce pre-rRNA levels as assessed by this method. Hence, coilin is required for the decrease in pre-rRNA message levels in response to cisplatin.
The inhibition of Pol I has previously been suggested to be a key event in DNA damage–induced apoptosis (Kalita et al., 2008; Hetman et al., 2010). We reasoned that because coilin deficiency could override the DNA damage inhibition of Pol I, it may also prevent cells from undergoing apoptosis. Indeed, in all three cell lines tested, coilin knockdown protected cells from cisplatin-induced cell death (Figure 6D). For example, in HeLa cells, a 24-h cisplatin exposure significantly reduced cell number by >60% in control siRNA–transfected cells; however, no significant decrease was observed in coilin knockdown cells. These findings suggest that coilin promotes cell death in response to cisplatin-induced DNA damage. Alternatively, given coilin knockdown has been reported to reduce cell proliferation in HeLa cells 72 h postknockdown (Lemm et al., 2006; Whittom et al., 2008), it was possible that a reduced rate of cell growth could lessen the susceptibility of these cells to cisplatin. However, in untreated cells, we did not detect a significant decrease in proliferation after coilin knockdown for any of the cell lines at the time point examined (48 h postseeding). These data suggest that the protection from cisplatin conferred by coilin knockdown is not likely the result of reduced cell proliferation due to depleted snRNP levels.
Coilin modulates Pol I association with ribosomal DNA (rDNA)
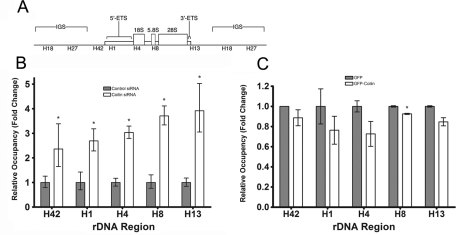
To gain mechanistic insight into how coilin regulates Pol I activity, we tested whether coilin had any effect on the association of Pol I with rDNA. We performed chromatin immunoprecipitation (ChIP) with an RPA-194 antibody to assess the association of Pol I with rDNA loci in control and coilin siRNA–transfected Saos2 cells. A schematic of an rDNA tandem repeat with the amplicons utilized in this experiment is shown in Figure 7A. In control siRNA–transfected cells, we observed that Pol I was present throughout the rDNA transcribed regions represented by amplicons H42, H1, H4, H8, and H13 (Figure 7B). Coilin knockdown resulted in an approximately two- to fourfold increase in the association of Pol I with all of these rDNA regions. In contrast, we did not detect RPA-194 in the intergenic sequences represented by amplicons H18 and H27 in either the control or coilin siRNA–transfected cells. These data indicate that coilin knockdown increases the specific association of Pol I with the transcribed region of the rDNA loci
FIGURE 7:
Coilin affects Pol I occupancy of rDNA. (A) Schematic of rDNA tandem repeat showing amplicons from the transcribed region (H1, H4, H8, H13) as well as amplicons found in the intergenic sequence (IGS) (H18 and H27) used for ChIP analysis. (B) Pol I occupancy of rDNA increases with coilin knockdown. ChIP was performed (Saos2 cells) using either a control or RPA-194 antibody, and real-time PCR was performed using primers for the indicated amplicons. The baseline for Pol I occupancy (obtained from control siRNA–transfected cells) was set as 1, and the fold change in coilin siRNA–transfected cells was calculated relative to that baseline (error bars represent the mean fold change ± CV; *p < 0.05). (C) Ectopic coilin expression displaces Pol I from rDNA. GFP-coilin or GFP was transfected into HeLa cells, and ChIP was performed as in (A) except the baseline for Pol I occupancy was obtained from GFP-transfected cells and set to 1 (error bars represent the mean fold change ± CV; *p < 0.05).
To test the possibility that coilin interferes with the binding of RPA-194 to rDNA, we transfected HeLa cells with GFP alone or GFP-coilin plasmids and performed ChIP analysis to determine the occupancy of RPA-194 on rDNA. Compared with the GFP-transfected cells, GFP-coilin–transfected cells had a significant decrease in RPA-194 occupancy in the H8 amplicon (Figure 7C), indicating that coilin negatively affects the association of Pol I with rDNA. Collectively, these results suggest that coilin can suppress Pol I activity by modulating RPA-194 association with rDNA.
DISCUSSION
Why certain proteins, including coilin, segregate to the nucleolus in some stress conditions has been a long-standing question (Shav-Tal et al., 2005). In this report, we provide three lines of evidence suggesting that coilin responds to cisplatin-induced DNA damage and γ-irradiation by segregating to the nucleolus in order to suppress Pol I activity.
In our first line of evidence, we show that cisplatin causes coilin to translocate within and around nucleoli (Figure 1). It is interesting to note that in the p53-wild-type WI-38 primary cell line, nucleolar coilin accumulation is extensive with cisplatin treatment, despite a diffuse nucleoplasmic coilin distribution in untreated cells (Figure 1). This diffuse pan-nuclear signal in untreated WI-38 cells is diminished in cisplatin-treated WI-38 cells, suggesting that coilin is being recruited to the nucleolus from the nucleoplasm in this line. Moreover, in HeLa cells, despite elevated coilin message levels in response to cisplatin (Supplemental Figure 2A), coilin protein levels remained unchanged following cisplatin treatment (Supplemental Figure 2B). Coilin protein levels also remained constant in response to cisplatin treatment in Saos2 and WI-38 cell lines (our unpublished data). Also, it appears that the nucleolar accumulation of coilin is a generalized response to DNA damage, given that cisplatin and γ-irradiation produce a similar phenotype (Figures 1, 2, and 5A).
Our second line of evidence is that ectopic expression of coilin and recombinant coilin both suppress Pol I activity. Ectopically overexpressed GFP-coilin accumulates in the nucleolus (Figure 3B), reduces pre-rRNA levels (Figure 3C), immunoprecipitates endogenous RPA-194 (Figure 4B), and decreases RPA-194 occupancy at specific sites on rDNA (Figure 7C). Recombinant GST-coilin can also localize to the nucleolus and reduce BrUTP incorporation (Figure 3D) as well as bind to endogenous RPA-194 in a GST pull-down assay (Figure 4C). And recombinant RPA-194 associates with recombinant coilin in vitro (Figure 4D), which suggests a direct interaction between these proteins. Cisplatin treatment and γ-irradiation both induce the colocalization of coilin with RPA-194 (Figure 5 and Supplemental Figure 5). Therefore it is reasonable to suggest that coilin’s role within the cellular response to DNA damage is to mediate the repression of rRNA synthesis.
For our third line of evidence, we show that coilin knockdown in cisplatin-treated cells abrogates the inhibition of Pol I activity as assessed by BrU incorporation (Figure 6A) and RT-PCR analysis of the 5′ETS of pre-rRNA (Figure 6C). Furthermore, coilin knockdown slightly increased pre-rRNA levels by 1.2-fold in HeLa cells (Figure 6C), and this correlated with an increased association of RPA-194 with rDNA (Figure 7B). Moreover, we found that this increased association of RPA-194 was specific and not a generalized increase in DNA binding activity because we detected RPA-194 only on transcribed ribosomal regions and not on the intergenic spacer region. Therefore we speculate that coilin may be involved in the homeostatic regulation of Pol I activity. It is important to note, however, that coilin knockdown alone led to a decrease in BrU incorporation in HeLa cells (Figure 6A), which may reflect a stress response induced by impaired snRNP assembly that leads to reduced rRNA synthesis (Whittom et al., 2008; Strzelecka et al., 2010). Further studies are required to determine whether pre-mRNA splicing defects due to the absence of nucleoplasmic coilin can lead to inhibition of Pol I activity. Importantly, knockdown of coilin in untreated cells appears to slightly increase pre-rRNA levels 1.2-fold in HeLa and 2-fold in Saos2 (Figure 6C and our unpublished data), which we interpret to indicate that coilin may also modulate Pol I activity in unstressed cells. In any case, both methods used to monitor Pol I activity (BrU incorporation and pre-rRNA detection) clearly show that the effect of cisplatin to reduce Pol I is attenuated in the absence of coilin.
Interestingly, the coilin-interacting protein Nopp140 (Isaac et al., 1998), which is found in the nucleolus and the CB, has been shown to associate with RPA-194 (Chen et al., 1999). Full-length and transfected truncations of Nopp140 were shown to reduce Pol I activity (Chen et al., 1999). Likewise, a fragment of coilin consisting of amino acids 94–291 forms aggregates around the nucleolus and reduces Pol I activity (Bohmann et al., 1995). Our data suggest that coilin can directly suppress Pol I activity in response to cisplatin and γ-irradiation. However, given that coilin knockdown failed to completely restore Pol I activity, we acknowledge it is possible that coilin acts indirectly to suppress Pol I activity as well, especially if coilin nucleates the accumulation of other proteins that can suppress Pol I activity, such as Nopp140. We favor this possibility because coilin can effectively nucleate a CB (Kaiser et al., 2008). Indeed, if multiple proteins act to directly suppress Pol I activity, this effect can be cumulative. Future work will be aimed at determining whether Nopp140 and coilin act in either cooperative or antagonistic manners to regulate Pol I, especially during conditions of DNA damage–induced cellular stress.
Cellular stress is not the only impetus for rDNA transcriptional silencing, however. For example, rDNA transcriptional silencing can occur during development (e.g., nucleologenesis) and during the disassembly of the nucleolus during mitosis. It is intriguing to note that CBs have been implicated in nucleologenesis (Zatsepina et al., 2003). Remarkably, a recent electron microscopy study has revealed that coilin itself colocalizes with Pol I on the nucleolar surface at the onset of oocyte nucleologenesis (Pochukalina and Parfenov, 2008). The presence of Pol I components at the surface of the nucleolus have even been shown to correlate with diminished Pol I activity (Pochukalina and Parfenov, 2006). Thus coilin’s role in modulating Pol I activity likely extends beyond the response to cellular stress. In fact, it has been previously proposed that Pol I transcription and processing components undergo preassembly within the CB before shuttling to the nucleolus (Gall et al., 1999).
It remains unresolved how coilin and other proteins are able to translocate to nucleoli upon cisplatin-induced DNA damage and γ-irradiation. Prolonged exposure to cisplatin (48 h) resulted in a clear disruption of the interaction between coilin and SMN in HeLa cells (Supplemental Figure 8). Given that the interaction between SMN and coilin is dependent on the modification of arginines within coilin by symmetrical dimethylation, and disruptions in this coilin modification can induce Gems (Hebert et al., 2001; Hebert et al., 2002; Boisvert et al., 2002), it is tempting to speculate that the induction of Gems upon cisplatin treatment is indicative of alterations in the symmetrical dimethylation of coilin. Recent work shows that hypomethylation of coilin can target coilin to the nucleolus (Tapia et al., 2010). Therefore DNA damage may regulate the methylation status of coilin and thereby impact its cellular distribution.
To date, there are few mechanistic examples of Pol I suppression by a protein. JHDM1B, a histone demethylase, binds rDNA and demethylates lysine 4 on histone H3 in the nucleolus, resulting in the inhibition of Pol I transcription (Frescas et al., 2007). Also, the protein phosphatase cdc14 inhibits Pol I transcription by displacing Pol I subunits from the nucleolus (Clemente-Blanco et al., 2009). Further studies are required to determine whether coilin’s Pol I inhibitory activity involves cross-talk with these pathways.
MATERIALS AND METHODS
Cell lines, cell culture, DNA constructs, and transfection
Saos2 (human osteosarcoma), HeLa (human cervical carcinoma), WI-38 (human normal fetal lung fibroblast), and H1299 (human non–small cell lung carcinoma) were obtained from the American Type Culture Collection (Manassas, VA). All cells were cultured and transfected with siRNA or DNA constructs as previously described (Hebert and Matera, 2000; Sun et al., 2005; Hearst et al., 2009; Toyota et al., 2010). For the Pol I subunit cotransfection experiment, H1299 cells were cotransfected with equivalent amounts of myc-coilin and each respective GFP construct using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. GFP-UBF and GFP–RPA-16, -40, -43, and -194 constructs were used as previously described (Dundr et al., 2002) and obtained from Addgene (Cambridge, MA). His-coilin and GST-coilin constructs were used as previously described (Toyota et al., 2010). Myc-coilin construct was used as previously described (Hebert and Matera, 2000). His-coilin and GST-coilin constructs were used as previously described (Toyota et al., 2010). BL21(DE3)pLysS cells (Invitrogen) were transformed with His or GST fusion constructs. For all recombinant proteins except GST–RPA-194, BL21(DE3)pLysS bearing the respective expression vectors was grown in LB media (Thermo Fisher, Waltham, MA) at 37°C, 250 rpm, until optical density (OD) at 600 nm reached 0.7. The cultures were then induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and incubation was continued at 30°C for an additional 4 h. For GST–RPA-194, when OD at 600 nm reached 1, 0.06 mM IPTG was used for induction at 27°C for an additional 12 h. Cells were harvested and lysed by sonication. Proteins were purified using either Ni-NTA Superflow beads (Qiagen, Hilden, Germany) or glutathione-Sepharose beads (GE Healthcare, Uppsala, Sweden) according to manufacture specification. For all cisplatin treatment studies, unless otherwise indicated, HeLa and H1299 cells were treated with 3 μg/ml cisplatin; WI-38 and Saos2 cells were treated with 7.5 μg/ml cisplatin for 2, 6, 24, or 48 h where indicated. For γ-irradiation, cells were irradiated at the indicated doses with a Cs-137 source (model I43 irradiator, J.L.S. Shepherd, San Fernando, CA).
Construction of recombinant RPA-194 and fibrillarin expression vectors
Human RPA-194 (NM_015425.3) and fibrillarin (NM_001436.3) were cloned into a modified pGEX-2T vector (GE Healthcare). pGEX-2T was used as a template for PCR amplification using Phusion Master Mix with GC (Finnzymes, Espoo, Finland) with primers 5′-GCAATACTCGAGGGATCCACGCGGAACCAGAT-3′ and 5′-GCAATAAAGCTTTGACTGACGATCTGCCTCGC-3′ amplifying the entire vector with the addition of XhoI and HindIII endonuclease recognition sites (underlined sequences), respectively, for the cloning of RPA-194 and FBL. RPA-194 and fibrillarin cDNAs were generated from mRNA isolated from mammalian cells. RPA-194 was amplified with 5′-CAATACTCGAGATGTTGATCTCCAAGAACAT-3′ and 5′-GCAATAAAGCTTCTATCTCAGAGGCTGCTTGA-3′. Fibrillarin was amplified with 5′-GCAATACTCGAGATGAAGCCAGGATTCAGTCC-3′ and 5′-CAATAAAGCTTTCAGTTCTTCACCTTGGGGG-3′. The resulting PCR products were digested with XhoI and HindIII and cloned into the amplified, modified pGEX-2T vector digested with the same endonucleases.
In situ run-on transcription assay
In situ run-on transcription assays were conducted as previously described (Chen et al., 1999), with a few modifications. Briefly, cells were washed in ice-cold phosphate-buffered saline (PBS) and incubated with ice-cold transcription buffer supplemented with Super RNaseIN RNase inhibitor (Ambion, Foster City, CA) and permeabilized with 250 μg/ml digitonin. Cells were then incubated with ∼5 μg each of bacterially prepared recombinant human GST-coilin, GST, or GST-fibrillarin in transcription buffer for 15 min at 4°C. Following incubation, cells were washed again with transcription buffer and incubated at 34°C for 20 min with transcription buffer supplemented with ribonucleotide triphosphates, including BrUTP (Invitrogen). The reaction was stopped with ice-cold PBS, and cells were subsequently fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.5% Triton for 10 min. BrUTP was detected with a bromodeoxyuridine (BrdU) antibody; GST, GST-coilin, and GST-fibrillarin were detected with a GST antibody.
Cell lysis, immunoprecipitation, GST pull-down, and Western blot
For coimmunoprecipitation, a protocol (Toyota et al., 2010) was used, except that cells were harvested and lysed with ice-cold IP buffer (50 mM Tris [pH = 8], 150 mM NaCl, 0.5% NP-40, 10% glycerol, and protease inhibitor cocktail; Roche, Penzberg, Germany). For GST pull-down of HeLa lysate, cells were lysed in ice-cold modified radioimmunoprecipitation assay (mRIPA) buffer (50 mM Tris [pH = 7.6], 150 mM NaCl, 1% NP-40, 1 mM EDTA). Lysate was cleared by syringing with a 25-gauge needle and precleared with 30 μl glutathione-Sepharose beads for 1 h at 4°C. After preclearing, lysate was incubated with GST or GST-coilin beads for 2 h at 4°C, followed by three washes with mRIPA. Recovered protein was suspended in SDS–PAGE loading buffer and subjected to SDS–PAGE and Western transfer. For in vitro GST pull-down of recombinant coilin, GST or GST–RPA-194 beads were incubated with Ni-NTA–purified coilin (Toyota et al., 2010) in mRIPA buffer supplemented with 0.25% sodium deoxycholate and 2 mM dithiothreitol for 1 h at 4°C, followed by four washes with the same buffer. Recovered protein was suspended in SDS–PAGE loading buffer and subjected to SDS–PAGE and Western transfer.
Detection of Pol I transcription
For detection of nascent Pol I transcripts, a protocol modified from a previous study (Pellizzoni et al., 2001) was used. Briefly, cells were cultured for 2 h in 2 mM BrU (Sigma-Aldrich, St. Louis, MO), followed by fixation and extraction by incubation in methanol for 20 min at −20°C and a 30-s room temperature incubation in acetone. The slides were then air-dried and subjected to immunofluorescence (IF). BrU pixel intensities were quantified using MetaVue image software (Molecular Devices, Sunnyvale, CA). For comparative RT-PCR, RNA was isolated using the Ambion RNAqueous kit according to the manufacturer’s protocol. Message levels were quantified using Brilliant II SYBR Green QRT-PCR (Stratagene, Santa Clara, CA) according to the manufacturer’s protocol, and GAPDH was used as a normalizer, as previously described (Hearst et al., 2009). For two-step RT-PCR, cDNA was made using the iScript cDNA kit (Bio-Rad, Hercules, CA). Coilin, GAPDH, and 5′ETS of pre-rRNA primers were as described (Grandori et al., 2005; Hearst et al., 2009).
Antibodies and immunofluorescence
The following antibodies were utilized in this study: rabbit polyclonal RPA-194 (1:500 Western, 1:200 IF) (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal UBF (1:1000) (Santa Cruz), mouse monoclonal β-tubulin (1:2000) (Sigma), mouse monoclonal GFP (1:1000) (Roche), rabbit polyclonal GFP (1:1000) (Santa Cruz), mouse monoclonal c-myc (1:500) (Santa Cruz), mouse monoclonal BrdU (1:200) (Sigma), rabbit polyclonal coilin H300 (1:1000 Western, 1:200 IF) (Santa Cruz), mouse monoclonal coilin clone pdelta (1:500) (Abcam, Cambridge, UK), mouse monoclonal SMN (1:1000 Western, 1:200 IF) (BD Transduction Laboratories, San Jose, CA), rabbit polyclonal GST (1:200 IF) (Abcam), mouse monoclonal actin AC-15 (1:2000) (Sigma), rabbit immunoglobulin G (IgG) (Santa Cruz), Nopp140 rabbit serum RE10 (1:1000) (Meier and Blobel, 1992), and fibrillarin monoclonal antibody 72B9 (1:2000) (gift from E. Chan). Immunofluorescence and image acquisition were performed as previously described (Sun et al., 2005).
Proliferation
For proliferation studies, cells were seeded at a density of 1200 cells/well into 96-well plates 24 h postknockdown. At 48 h postknockdown, cells were untreated or treated with 3 μg/ml cisplatin for HeLa and 7.5 μg/ml cisplatin for WI-38 and Saos2. At 72 h postknockdown, cell titer blue (Promega, Fitchburg, WI) was added according to the manufacturer’s protocol and fluorescence output was read on an FLx800 Spectrophotometer (BioTek, Winooski, VT) using the 490/540 filter set.
ChIP
For the coilin siRNA ChIP experiment, Saos2 cells transfected with either control or coilin siRNA were harvested and processed following the Upstate ChIP (EZCHIP) protocol, but with protein G agarose from KPL Laboratories (Gaithersburg, MD). The antibodies used were control rabbit IgG (Santa Cruz) and RPA-194 (H300; Santa Cruz). For the ectopic coilin ChIP experiment, HeLa cells were transfected with GFP-coilin or GFP and subjected to the same processing as described above. Real-time PCR was performed using primers specific to the ribosomal loci, as has been previously described (Grandori et al., 2005). Similar results were obtained in two independent experiments.
Acknowledgments
We thank Leslie Robinson for assistance with γ-irradiation. This work was supported by National Institutes of Health grant R01-GM-081448 (to M.D.H.), Research Scholar grant RSG-08-084-01 from the American Cancer Society (to L.A.M.), and startup funds from the University of Mississippi Cancer Institute (to L.A.M.).
Abbreviations used:
- ActD
actinomycin D
- BrdU
bromodeoxyuridine
- BrU
bromouridine
- BrUTP
bromouridine triphosphate
- CB
Cajal body
- ChIP
chromatin immunoprecipitation
- CV
coefficient of variation
- DAPI
4′,6-diamino-2-phenylindole
- ETS
external transcribed spacer
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- IF
immunofluorescence
- IP
immunoprecipitation
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- mRIPA
modified radioimmunoprecipitation assay
- NHEJ
nonhomologous end-joining
- OD
optical density
- PBS
phosphate-buffered saline
- Pol
polymerase
- rDNA
ribosomal DNA
- RT-PCR
reverse transcription PCR
- siRNA
small interfering RNA
- SMN
survival of motor neurons protein
- snRNP
small nuclear ribonucleoprotein
- TMG
trimethyl guanosine
- UBF
upstream binding factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0731) on February 2, 2011.
REFERENCES
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Lamond A. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol. 2002;159:957–969. doi: 10.1083/jcb.200207028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HK, Pai CY, Huang JY, Yeh NH. Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol Cell Biol. 1999;19:8536–8546. doi: 10.1128/mcb.19.12.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol. 2006;175:401–413. doi: 10.1083/jcb.200604099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Clemente-Blanco A, Mayan-Santos M, Schneider DA, Machin F, Jarmuz A, Tschochner H, Aragon L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 2009;458:219–222. doi: 10.1038/nature07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys. 2010;496:69–76. doi: 10.1016/j.abb.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, Vashishta A, Rempala G. Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J Neurochem. 2010;114:1537–1549. doi: 10.1111/j.1471-4159.2010.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:407–417. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P, Carmo-Fonseca M. Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res. 1998;26:2831–2836. doi: 10.1093/nar/26.12.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kalita K, Makonchuk D, Gomes C, Zheng JJ, Hetman M. Inhibition of nucleolar transcription as a trigger for neuronal apoptosis. J Neurochem. 2008;105:2286–2299. doi: 10.1111/j.1471-4159.2008.05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Lam YW, Lyon CE, Lamond AI. Large-scale isolation of Cajal bodies from HeLa cells. Mol Biol Cell. 2002;13:2461–2473. doi: 10.1091/mbc.02-03-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- Morency E, Sabra M, Catez F, Texier P, Lomonte P. A novel cell response triggered by interphase centromere structural instability. J Cell Biol. 2007;177:757–768. doi: 10.1083/jcb.200612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochukalina GN, Parfenov VN. The nucleolus in oocytes of multylayer mouse follicles: topography of fibrillarin, RNA polymerase I and coilin [in Russian] Tsitologiia. 2006;48:641–652. [PubMed] [Google Scholar]
- Pochukalina GN, Parfenov VN. Nucleolus transformation in oocytes of mouse antral follicles. Revealing of coilin and RNA polymerase I complex components [in Russian] Tsitologiia. 2008;50:671–680. [PubMed] [Google Scholar]
- Shanbhag R, Kurabi A, Kwan JJ, Donaldson LW. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett. 2010;584:4351–4356. doi: 10.1016/j.febslet.2010.09.034. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci. 2003;116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Sun J, Xu H, Subramony SH, Hebert MD. Interactions between Coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci. 2005;118:4995–5003. doi: 10.1242/jcs.02613. [DOI] [PubMed] [Google Scholar]
- Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma. 2010;119:743–755. doi: 10.1007/s00412-010-0276-7. [DOI] [PubMed] [Google Scholar]
- Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB′. Chromosoma. 2010;119:205–215. doi: 10.1007/s00412-009-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velma V, Carrero ZI, Cosman AM, Hebert MD. Coilin interacts with Ku proteins and inhibits in vitro nonhomologous DNA end joining. FEBS Lett. 2010;584:4735–4739. doi: 10.1016/j.febslet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittom AA, Xu H, Hebert MD. Coilin levels and modifications influence artificial reporter splicing. Cell Mol Life Sci. 2008;65:1256–1271. doi: 10.1007/s00018-008-7587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;114:155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatsepina O, Baly C, Chebrout M, Debey P. The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the Cajal (coiled) body. Dev Biol. 2003;253:66–83. doi: 10.1006/dbio.2002.0865. [DOI] [PubMed] [Google Scholar]