siRNA-mediated knockdown of 40 putative ciliome genes reveals distinct roles in cilia formation, cilia elongation, ciliary transport of different cargoes, and signaling capability. Depletion of a subset of genes allows cilia transport to be uncoupled from cilia formation, whereas most genes are required for efficient signaling function.
Abstract
Cilia are microtubule-based protrusions from the cell surface that are involved in a number of essential signaling pathways, yet little is known about many of the proteins that regulate their structure and function. A number of putative cilia genes have been identified by proteomics and comparative sequence analyses, but functional data are lacking for the vast majority. We therefore monitored the effects in three cell lines of small interfering RNA (siRNA) knockdown of 40 of these genes by high-content analysis. We assayed cilia number, length, and transport of two different cargoes (membranous serotonin receptor 6-green fluorescent protein [HTR6-GFP] and the endogenous Hedgehog [Hh] pathway transcription factor Gli3) by immunofluorescence microscopy; and cilia function using a Gli-luciferase Hh signaling assay. Hh signaling was most sensitive to perturbations, with or without visible structural cilia defects. Validated hits include Ssa2 and mC21orf2 with ciliation defects; Ift46 with short cilia; Ptpdc1 and Iqub with elongated cilia; and Arl3, Nme7, and Ssna1 with distinct ciliary transport but not length defects. Our data confirm various ciliary roles for several ciliome proteins and show it is possible to uncouple ciliary cargo transport from cilia formation in vertebrates.
INTRODUCTION
Cilia are microtubule-based extensions of the plasma membrane that project from the surface of cells and carry out motility or sensory functions (Rosenbaum and Witman, 2002; Scholey and Anderson, 2006; Satir and Christensen, 2007). Motile cilia are used by unicellular organisms for locomotion or by specialized cell types in multicellular organisms to direct fluid movement, such as respiratory cilia that beat to generate mucosal flow (Ibanez-Tallon et al., 2003). Most eukaryotic cells, by contrast, contain a single nonmotile primary cilium that plays important roles in helping the cell to sense its extracellular environment (Singla and Reiter, 2006; Berbari et al., 2009). Several signal transduction pathways also use primary cilia, including platelet-derived growth factor (PDGF)Rα, Wnt, and Hedgehog (Hh) (Huangfu et al., 2003; Germino, 2005; Schneider et al., 2005). Defects in both motile and primary cilia have been associated with a large number of human diseases, including primary ciliary dyskinesia, hydrocephalus, polycystic kidney disease, Bardet–Biedl syndrome, and neurological disorders (Badano et al., 2006; Lee and Gleeson, 2010), so understanding cilia function is of great medical importance.
Cilia growth and disassembly are controlled by intraflagellar transport (IFT), the movement of IFT particles and their cargoes toward the cilia tip, driven by kinesin-2 motors; and back toward the cilia base, driven by dynein-2 motors (reviewed in Rosenbaum and Witman, 2002; Scholey, 2008), including Dync2h1 in vertebrates (Huangfu and Anderson, 2005; May et al., 2005). Mutations in murine Ift88 (Tg737) lead to premature death due to shortened, defective cilia resulting in polycystic kidney disease (Pazour et al., 2000), and mutations in other IFT subunits similarly disrupt cilia function (Eggenschwiler and Anderson, 2007). Because cilia formation itself is disrupted by defective IFT, it has been difficult to ascribe a role for IFT in cargo transport through vertebrate cilia (Snell’s IFT Uncertainty Principle [Rohatgi and Snell, 2010]), which lack the convenient temperature-sensitive FLA mutations found in Chlamydomonas.
Hh signaling in vertebrates requires intact primary cilia, because mutations in IFT genes or IFT motors in mice lead to loss of cilia and Hh signaling (Huangfu et al., 2003; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005; Eggenschwiler and Anderson, 2007). In the absence of Hh ligand, the inhibitory Hh pathway receptor Patched is localized to primary cilia (Rohatgi et al., 2007). On Hh binding, Patched departs the cilium and several downstream pathway components enter it, including Smoothened, Gli2, and Gli3 (Corbit et al., 2005; Rohatgi et al., 2007; Chen et al., 2009; Wang et al., 2009; Wen et al., 2010). The transcription factor Gli3 has been proposed to require cilia, or at least IFT, for Hh-dependent activation, although it is currently controversial whether the actual activation event occurs in cilia (Wen et al., 2010) or the nucleus (Humke et al., 2010). Gli3 processing into its repressor form is also thought to require IFT (Haycraft et al., 2005; Huangfu and Anderson, 2005), although its dependence on ciliary entry is unclear (Wen et al., 2010). Other proteins, including the G-protein–coupled receptors (GPCRs) somatostatin receptor 3 (SSTR3), serotonin receptor 6 (HTR6), and melanin-concentrating hormone receptor 1 (MCHR1) have been shown to localize to the cilia as well (Berbari et al., 2008; Mukhopadhyay et al., 2010), although the functional importance of this ciliary localization is unclear. Furthermore, it is not yet completely understood how membranous (GPCRs) and cytoplasmic (e.g., Gli) proteins are transported to and from cilia and whether they use distinct transport mechanisms, although it has been proposed that the coat-like BBSome mediates vesicular transport of GPCRs to the cilia through the GTPases RAB8 and ARL6 (Nachury et al., 2007; Berbari et al., 2008) or, alternatively, mediates transport from the cilia (Lechtreck et al., 2009).
Several laboratories have sought to identify cilia genes using proteomic, comparative genomic, expression, and promoter analyses (ciliome databases compiled in Inglis et al., 2006, and Gherman et al., 2006). Proteomic mass spectrometry studies have been conducted on cilia isolated from human bronchial epithelial cells (Ostrowski et al., 2002), mouse photoreceptor cells (Liu et al., 2007), and Tetrahymena (Smith et al., 2005); and on flagella from Chlamydomonas (Pazour et al., 2005) and Trypanosomes (Broadhead et al., 2006). Two homology-based studies compared the genomes of ciliated and nonciliated organisms (Avidor-Reiss et al., 2004; Li et al., 2004), and another compared ciliated and nonciliated cells in the same organism (Caenorhabditis elegans) (Blacque et al., 2005). Other genomics approaches included identification of consensus X-box sequences, believed to be cilia-specific, in the promoters of C. elegans genes (Blacque et al., 2005; Efimenko et al., 2005); and microarray expression analyses of Chlamydomonas undergoing flagellar regeneration (Stolc et al., 2005) or of ciliated versus nonciliated murine tissues or cells (Li et al., 2007; McClintock et al., 2008). Functional and subcellular localization data are lacking, however, for the vast majority of these putative ciliome genes.
To gain a better understanding of cilia biology, we selected 40 candidate cilia genes from a subset of the previously mentioned studies, the functions of which had not been well characterized. Changes in cilia structure and function were measured after knocking down expression of each putative cilia gene by small interfering RNA (siRNA) treatment in three murine cell lines. We observed several distinct classes of cilia phenotypes upon knockdown, including various combinations of defects in cilia formation, cilia elongation, ciliary cargo transport, and signaling through the cilia.
RESULTS
Selection of candidate cilia genes
The ciliome consists of ∼2500 putative cilia genes (Gherman et al., 2006; Inglis et al., 2006) identified by proteomic, comparative genomic, microarray, and promoter studies (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Li et al., 2004; Blacque et al., 2005; Efimenko et al., 2005; Pazour et al., 2005; Smith et al., 2005; Stolc et al., 2005; Broadhead et al., 2006; Liu et al., 2007; McClintock et al., 2008). To maximize quality hits, we selected those genes appearing in at least one proteomic (P1, P2) and one comparative genomic study (H1, H2) that had not been functionally demonstrated to be ciliary at the time, narrowing the list to 40 genes (Table 1). Ift88, known to be required for ciliogenesis (Pazour et al., 2000), served as a positive control. Another ciliation screen was recently published, with top hits including actin regulators (Kim et al., 2010), but they selected mostly nonciliome genes and so had little overlap with ours.
TABLE 1:
Candidate cilia genes.
| Name | Mouse refseq | Mouse ensembl # | Gene ID | Gene description(s) | P1 | P2 | P3 | P4 | P5 | H1 | H2 | M1 | M2 | X1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arl3 | NM_019718 | 25035 | 56350 | ADP-ribosylation factor-like 3 | X | X | X | X | X | |||||
| mC16orf48 | NM_198299 | 13155 | 102124 | C8H16orf48, orthologue of human C16orf48, E130303B06Rik | X | X | X | X | ||||||
| mC21orf2 | NM_026431 | 20284 | 67884 | C10H21orf2, orthologue of human C21orf2 | X | X | X | X | ||||||
| CapsI | NM_029341 | 39676 | 75568 | Calcyphosine-like, 1700028N11Rik | X | X | X | X | ||||||
| Ccdc147 | NM_001163267 | 46585 | 381229 | Coiled-coil domain containing 147, orthologue of human C10orf80, Flj35908 | X | X | ||||||||
| Ccdc37 | NM_173775 | 48794 | 243538 | Coiled-coil domain containing 37, C230069K22Rik | X | X | X | |||||||
| Ccdc40 | NM_175430 | 39963 | 207607 | Coiled-coil domain containing 40, Flj20753, B930008l02Rik | X | X | X | X | X | |||||
| Ccdc65 | NM_153518 | 03354 | 105833 | Coiled-coil domain containing 65, 4933417K04Rik | X | X | X | X | X | |||||
| Ccdc96 | NM_025725 | 50677 | 66717 | Coiled-coil domain containing 96, 4921513E08Rik, Flj90575 | X | X | X | X | X | |||||
| Dnahc5 | NM_133365 | 22262 | 110082 | Dynein, axonemal, heavy chain 5 | X | X | X | X | ||||||
| Dnaic1 | NM_175138 | 061322 | 68922 | Dynein, axonemal, intermediate chain 1 | X | X | X | X | X | X | X | X | X | |
| Dnali1 | NM_175223 | 42707 | 75563 | Dynein, axonemal, light intermediate polypeptide 1 | X | X | X | X | X | X | ||||
| Dpy30 | NM_024428 | 24067 | 66310 | Epeq4460, Aip1, 2810410M20Rik | X | X | X | X | ||||||
| Efhc1 | NM_027974 | 41809 | 71877 | EF-hand domain (C-terminal) containing 1, 1700029F22Rik | X | X | X | X | X | X | X | |||
| Enkurin | NM_027728 | 26679 | 71233 | Enkurin, 4933434l06Rik, human C10orf63 orthologue | X | X | X | X | X* | |||||
| Flj21827 | NM_023831 | 02031 | 76568 | IFT46, 1500035H01Rik | X | X | X | X | X | |||||
| Gucy1b2 | NM_172810 | 21933 | 239134 | Guanylate cyclase 1, soluble, beta 2 | X | X | X | |||||||
| Iqch | NM_030068 | 37801 | 78250 | IQ motif containing H, Flj1 2476, 4921504K03Rik | X | X | X | |||||||
| Iqub | NM_172535 | 46192 | 214704 | IQ motif and ubiquitin domain containing, 4932408B21Rik | X | X | X | X | ||||||
| Lrrc9 | NM_030070 | 21090 | 78257 | Leucine-rich repeat containing 9, 4921529O18Rik | X | X | ||||||||
| Mns1 | NM_008613 | 32221 | 17427 | Meiosis-specific nuclear structural protein 1 | X | X | X | X | X | X | ||||
| Nme7 | NM_138314 | 26575 | 171567 | Nonmetastatic cells 7, nucleoside diphosphate kinase | X | X | X | X | X | X | X | |||
| Pacrg | NM_027032 | 37196 | 69310 | Park2 coregulated | X | X | X | X | X | X | X | X | X | |
| Phki3718 | NM_177673 | 39523 | 230967 | mKIAA0562, BC046331 | X | X | ||||||||
| Ppp4r4 | NM_028980 | 21209 | 74521 | Protein phosphatase 4, regulatory subunit 4, mKiaa1622, 8430415E04Rik | X | X | ||||||||
| Ptpdc1 | NM_207232 | 38042 | 218232 | Protein tyrosine phosphatase domain containing 1, AW456874 | X | X | X | |||||||
| Ribc2 | NM_026357 | 22431 | 67747 | RIB43A domain with coiled-coils 2 | X | X | X | X | X | X | ||||
| Rshl2 | NM_025789 | 23806 | 66832 | Radial spokehead-like 2, Rsph3a | X | X | X | X | X | X | X | |||
| Rshl3 | NM_001162957 | 39552 | 212892 | Radial spokehead-like 3, Rsph4a | X | X | X | X | X | X | ||||
| Rsph9 | NM_029338 | 23966 | 75564 | Radial spokehead 9 homologue, 1700027N10Rik, orthologue of human C6orf206 | X | X | X | X | X | X | ||||
| Ssa2 | NM_013835 | 18199 | 20822 | Sjögren's syndrome antigen A2, Trove2 | X | X | ||||||||
| Ssna1 | NM_023464 | 26966 | 68475 | Sjögren's syndrome nuclear autoantigen 1 | X | X | X | |||||||
| Tppp2 | NM_001128634 | 14846 | 219038 | Tubulin polymerization–promoting protein family member 2 or 3. p25Beta, p25Alpha, 2700055K07Rik, orthologue of hu C14orf8 | X | X | ||||||||
| Ttc18 | NM_001163638 | 39543 | 76670 | Tetratricopeptide repeat domain 18 | X | X | X | X | ||||||
| Ttc21b | NM_001047604 | 32514 | 73668 | Tetratricopeptide repeat domain 21b | X | X | X | X | ||||||
| Ttc26 | NM_153600 | 56832 | 264134 | Tetratricopeptide repeat domain 26, 9430097H08Rik | X | X | X | X | X | X | ||||
| Wdr19 | NM_153391 | 37890 | 213081 | WD repeat domain 19, DYF-2 | X | X | X | X | X | |||||
| Wdr34 | NM_001008498 | 39715 | 71820 | WD repeat domain 34 | X | X | X | X | ||||||
| Wdr66 | XM_915421 | 29442 | 269701 | WD repeat domain 66 | X | X | X | X | X | |||||
| Wdr78 | NM_146254 | 35126 | 242584 | WD repeat domain 78, FIj23129 | X | X | X | X | X | X |
The 40 putative cilia genes screened in this study, showing their prior identification (“X”) in four types of ciliome studies: 1) Proteomic: mass spectrometry of isolated cilia or flagella (P1–P5); 2) Homology: genomic comparison of ciliated and nonciliated species (H1 and H2); 3) Microarray: of flagellar regeneration (M1) or ciliated versus nonciliated tissues (M2); and 4) X-box identification of genes with conserved DAF19-binding (cilia regulating) X-box promoters (X1). None of the genes appeared in a similar X-box study (Efimenko et al., 2005) or in an IMCD3 microarray study (Li et al., 2007). P1, P2, P3, H1, H2, M1, and X1 were annotated based on www.ciliome.com (Inglis et al., 2006), and P4 and P5 were from www.ciliaproteome.org (Gherman et al., 2006; Nielsen et al., 2008). M2 was added from McClintock et al., 2008. Genes were selected based on their presence in studies P1 and/or P2, plus H1 and/or H2 (boldface), with their appearance in other studies shown for comparison.
P1: Ostrowski et al., 2002: Proteomic mass spectrometry of cilia from human bronchial epithelial cells
P2: Pazour et al., 2005: Proteomic mass spectrometry of flagella from Chlamydomonas
P3: Smith et al., 2005: Proteomic mass spectrometry of cilia from Tetrahymena
P4: Liu et al., 2007: Proteomic mass spectrometry of mouse photoreceptor sensory cilia
P5: Broadhead et al., 2006: Proteomic mass spectrometry of Trypanosome flagellae
H1: Avidor-Reiss et al., 2004: Homology-based comparative genomics using genes in six ciliated and three nonciliated species
H2: Li et al., 2004: Homology-based comparative genomics of genes in human and Chlamydomona absent from Arabidopsis
M1: Stolc et al., 2005: Microarray analysis of genes up-regulated during flagellar regeneration in Chlamydomonas
M2: McClintock et al., 2008: Microarray analysis identifying murine genes enriched in ciliated tissues
X1: Blacque et al., 2005: X-box promoter search in C. elegans and enriched in ciliated cells by SAGE
# Only the last 5 digits of the mouse Ensembl # are shown (all are prefixed with ENSMUSG000000)
* Confirmed by in situ hybridization (McClintock et al., 2008)
Development of functional cilia assays
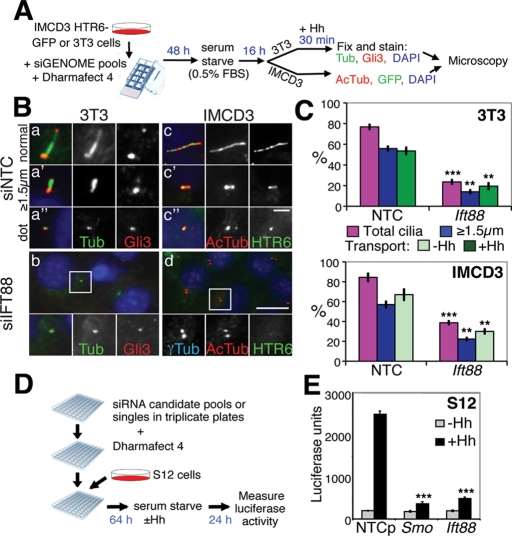
Ideal cell lines for assessing putative ciliary gene function by microscopy would be adherent, readily transfected, and reproducibly ciliate and have ciliary cargo markers to assess ciliary transport. S12 cells (murine C3H10T1/2 osteoblasts) stably express a Hh-inducible Gli-luciferase reporter, an ideal screening cell line for cilia function, but their cilia proved unexpectedly resistant to Ift88 depletion compared with NIH 3T3 (3T3) or intermedullary collecting duct (IMCD3) cells, as assessed by cilia staining with anti-acetylated tubulin, despite efficient knockdown (Supplemental Figure S1, A–C). 3T3 cells afforded a larger ciliation window (+ vs. - siIft88) and are also Hh responsive, accumulating the Hh pathway transcription factor Gli3 at the tips of cilia within 5 min of stimulation, an effect so rapid it likely occurs by IFT (Wen et al., 2010). We hypothesized that induction of stimulated transport after cilia gene knockdown might provide greater sensitivity than a constitutive cargo. We thus knocked down Ift88 in 3T3 cells for 72 h (including 16 h of serum starvation), stimulated them with Hh for 30 min, and stained them for cilia, centrosomes, and Gli3 (Figure 1, A, Ba, and Bb). Cells were scored for % total cilia (% cells with any length cilium), including ciliary “dots” (∼0.5 μm; Figure 1Ba′′); % substantial cilia (≥1.5 μm, i.e., longer than they are wide and clearly distinct from ciliary dots; Figure 1Ba′); and % Gli3 transport (% of ≥1.5-μm-long cilia with strong Gli3 at the tip; Figure 1C, top). As expected, knockdown of Ift88 not only substantially decreased ciliation, but also decreased the number of substantial cilia and the percentage of those with Gli3 accumulation (Figure 1, B and C). Note that, for transport measurements, the cilia had to be ≥1.5 μm long to distinguish the Gli3 staining at the tip from the base, thus transport defects are underestimated in cells with ciliation or elongation defects.
FIGURE 1:
siRNA screen outline and controls. (A) The microscopy-based siRNA screen setup. Transfection of siRNA, serum starvation, fixation, and staining were performed as detailed in Materials and Methods. (B) Microscopy assay controls for cilia structure and transport. 3T3 (a and b) or IMCD3 HTR6-GFP (c and d) cells were transfected with nontargeting control (siNTC, top) or Ift88 (siIft88, bottom) siRNAs then were serum starved for 16 h. 3T3 cells were stimulated with Hh for 30 min immediately before fixation and staining for endogenous Gli3 (red channel), cilia and centrosomes (anti–acetylated tubulin and anti–γ-tubulin, respectively [Tub], green channel), and nuclei (DAPI, blue channel). Note that anti-Gli3 nonspecifically cross-reacts with centrosomes in addition to its specific cilia tip staining (Wen et al., 2010). Three representative cilia types in control cells are shown: a normal ∼2.5-μm-long cilium (a), a substantial but short cilium ≤1.5 μm long (longer than it is wide) (a′), and a ciliary “dot” that is only as long as it is wide (∼0.5 μm) (a′′). IMCD3 HTR6-GFP cells were fixed and stained for cilia (anti–acetylated tubulin [AcTub], red channel) and DAPI (blue channel), and HTR6-GFP fluorescence was detected in the green channel. Again, examples of normal (c), short (c′) and dot (c′′) cilia are shown. Insets at the bottom of b and d show 2× magnification of the boxed regions, with individual and merged channels shown (anti–γ-tubulin [γTub] in the Cy5 channel replaces DAPI in the inset of d). Scale bar represents 2.5 μm in a, c, and insets and 10 μm in the main panel of b and d. (C) Quantification of cilia phenotypes upon siRNA knockdown. Images of 3T3 or IMCD3 HTR6-GFP cells treated as in (B) were scored for ciliation (% total cilia, magenta), cilia length (% ≥1.5-μm-long cilia, blue), and ciliary cargo (% transport, green) of Gli3 (3T3, defined as a Gli3 spot distinct from the centrosome) or HTR6-GFP (IMCD3), scoring as a percentage of the (blue bar) “nondot” cilia (≥1.5 μm long so that the tip and base were clearly distinct) that transport each cargo. The means and standard deviations (SDs) of three independent experiments are shown. Asterisks denote statistical significance between NTC and Ift88 siRNA-treated samples according to the t test (**p ≤ 0.01; ***p ≤ 0.001). (D) Outline of the S12 Gli-luciferase Hh signaling assay. Transfection of siRNA, serum starvation, Hh addition, and luciferase measurements are described in Materials and Methods. (E) Quantification of Hh signaling in the S12 assay with (black) and without (gray) Hh using NTC and positive control siRNAs to Smoothened (Smo) and Ift88. The means and SDs of three independent experiments are shown. ***p ≤ 0.001 vs. siNTC control.
To ensure the screen would not be 3T3- or Hh-specific, we also established a non-Hh-responsive IMCD3 cell line stably expressing green fluorescent protein (GFP)-tagged HTR6, a GPCR that localizes to cilia (HTR6-GFP; Figure 1B, c and d). Because HTR6 and Gli3 differ in membrane association (transmembrane vs. cytoplasm), ciliary localization (shaft vs. tip), and regulation (constitutive vs. Hh-stimulated), we anticipated that these two cargoes may be transported to cilia by different mechanisms (Rohatgi and Snell, 2010).
To monitor cilia function, we also performed the S12 cell Hh signaling assay (Frank-Kamenetsky et al., 2002) (Figure 1D), in which Gli-luciferase levels rise ∼10× upon Hh stimulation. Knockdown of Ift88 reduced Gli-luciferase induction as effectively as that of the essential Hh transduction component Smoothened (Smo) (Evangelista et al., 2008), confirming the critical role of cilia in Hh signaling in these cells (Figure 1E). Combination of all three assays (high-content analysis) thus allows detection of gross structural changes in cilia formation and/or transport as well as signaling and should be more informative than any one assay alone.
Analysis of cilia phenotypes after cilia gene knockdown
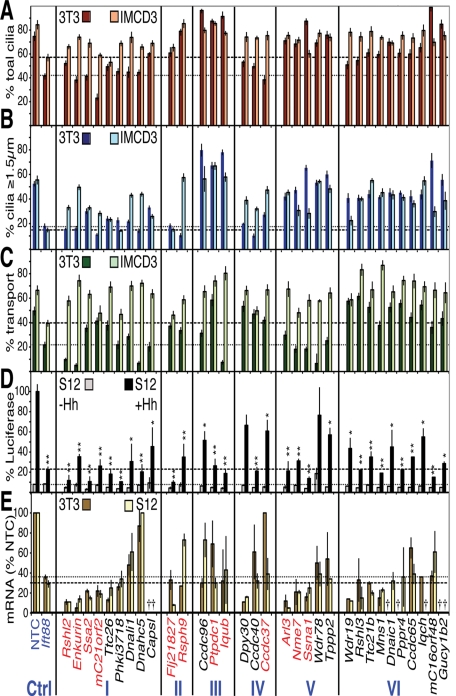
Pools of four siRNAs to each of the 40 candidate cilia genes in Table 1 were transfected and their effects examined using the three assays. The % total cilia (Figure 2A), % ≥1.5 μm cilia (Figure 2B), and % transport of Gli3 in 3T3 cells or HTR6-GFP in IMCD3 cells (Figure 2C) were plotted along with the Gli-luciferase signals in S12 cells (Figure 2D) and the mRNA knockdown efficiencies in 3T3 and S12 cells (Figure 2E). Thirty-two of the 40 genes were successfully depleted by siRNA in at least one cell line and were sorted based on their cilia phenotypes (primarily based on 3T3 and S12 data, as there were differences in transport between Gli3 in 3T3 cells and HTR6 in IMCD3 cells) into the following classes: I) ciliation, transport, and signaling defects; II) short cilia (≤1.5 μm); III) extra long cilia (≥3 μm in 3T3 or ≥5 μm in IMCD3 cells); IV) cilia structure and signaling defects, but normal transport; V) transport and signaling defects, but normal cilia structure; and VI) signaling defects only (Figure 2 and Supplemental Table S1a). Class I (ciliation defects) exhibited the most cilia phenotypes in both IMCD3 and 3T3 cells, with fewer, shorter cilia (Figure 2, A and B) and inhibited transport and signaling (Figure 2, C and D), similar to Ift88 knockdown, suggesting roles for these genes in cilia establishment or maintenance. Classes II and III exhibited relatively normal ciliation, but with shorter or longer cilia, respectively, in 3T3 and/or IMCD3 cells (Figure 2B). Their knockdown led to variable changes in Gli3 and HTR6 transport (Figure 2C), but all strongly reduced Hh signaling (Figure 2D), suggesting that maintenance of proper cilium length is essential for ciliary signaling.
FIGURE 2:
Primary screen results for the 32 candidate siRNAs with ciliary phenotypes. The results of the three functional assays are plotted in categories according to the resulting phenotypes (classes listed at the bottom). (A) Ciliation phenotypes. The percentage of cells bearing any cilium (including ciliary dots) in 3T3 (rusty red) and IMCD3 HTR6-GFP (beige) cells. The means ± standard deviations (SDs) of three different sets of cilia from a single experiment are plotted. (B) Cilia length phenotypes. The percentages of cells with cilia at least ≥1.5 μm in length (i.e., longer than ciliary “dots”) are plotted for 3T3 (dark blue) and IMCD3 HTR6-GFP (light blue) cells. The means and SDs from the same images scored in (A) are shown. (C) Ciliary cargo transport phenotypes. The ≥1.5-μm-long cilia in (B) were scored for the percentage (mean ± SD) with strong cilia tip Gli3 in 3T3 cells (dark green) or ciliary HTR6-GFP in IMCD3 cells (light green). Dotted and dashed lines in A–C highlight the positive control siIft88 3T3 and IMCD3 percentages, respectively. (D) Hh signaling phenotypes. % Gli-luciferase signal remaining after siRNA treatment in S12 cells with (black) or without Hh (gray) normalized to 100% for siNTC + Hh. Dotted and dashed lines indicate the siIft88 luciferase levels – and + Hh, respectively. The means and SDs of three independent experiments are plotted. *p ≤ 0.05; **p ≤ 0.01 vs. siNTC (+Hh data only). (E) Quantitation of mRNA knockdown. The % mRNA remaining relative to siNTC (means and SDs of one triplicate experiment) after siRNA treatment in 3T3 (gold) and S12 (light yellow) cells, with remaining Ift88 mRNA levels marked with dotted and dashed lines, respectively. Crosses indicate genes with low to undetectable levels of mRNA as measured by qRT-PCR TaqMan analysis (Ct > 34.5). Genes listed in red were chosen for further validation; blue are controls.
Class IV had fewer and/or shorter cilia and disrupted signaling, yet apparently normal transport, suggesting that transport and signaling can in fact be uncoupled under certain conditions. This uncoupling was confirmed in Class V, in which transport and signaling were defective in the absence of any obvious cilia structural defects at the light microscopic level. Class VI was the largest (10 genes), which exhibited defective Hh signaling but normal cilia by all other measures. Finally, the eight siRNAs with no significant phenotype (Supplemental Table S1a) could all be attributed to lack of knockdown or undetectable gene expression altogether with two independent probe sets (Supplemental Table S1b). Thus, all but two of the properly depleted genes resulted in impaired Hh signaling (with or without other defects), although off-target siRNA effects cannot be excluded for nonvalidated hits. Control experiments, however, showed that the Hh signaling defects were not simply due to indirect effects on luciferase transcription or stability in SV40-luciferase cells or impaired cell viability (Supplemental Figure S2, B and C).
Validation of selected candidate cilia genes
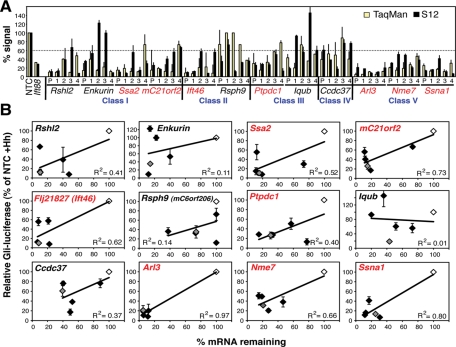
Twelve of the strongest hits (red text in Figure 2) from all classes except signaling only (VI) were chosen for validation. To do this, knockdown of mRNA by the original siRNA pool and its four constituents was correlated with S12 Hh signaling defects because that assay gave the most universal, least labor-intensive readout (Figure 3, A and B). Poor correlation between Hh signaling and mRNA levels (e.g., Enkurin), or signaling defects dominated by siRNAs that do not down-regulate the intended gene (e.g., Rsph9), were considered possible off-target effects, so these genes were rejected. Good correlation was defined as at least three individual siRNAs in addition to the pool showing inhibition of Hh signaling less than 60%, as was the case for Ssa2, mC21orf2, Flj21827 (subsequently identified as Ift46), Ptpdc1, Arl3, Nme7, and Ssna1 in Classes I, II, III, and V, which were considered high-quality hits. These top hits were all confirmed by repeating the immunofluorescence screens twice more and with further experiments (see later) along with Iqub in Class III.
FIGURE 3:
Correlation of S12 phenotype with mRNA knockdown of 12 selected genes. (A) Comparison of mRNA and S12 Hh signaling levels. siRNA knockdown was performed in Hh-treated S12 cells with pooled siRNAs (P) and their four component individual siRNAs (1–4) to the indicated genes. The percentage of mRNA remaining after siRNA knockdown (normalized to NTC as 100%) was measured by TaqMan RT-PCR (yellow), and cilia function was measured using the S12 Gli-luciferase assay + Hh (black). The means and standard deviations (SDs) of three experiments are shown. The dotted line indicates the cutoff value used (60%) for selecting top quality hits in (B). (B) Correlation of cilia function (Hh signaling) and gene knockdown. Same data as in (A) plotted with Gli-luciferase levels (as % siNTC + Hh) on the y-axes and remaining mRNA levels (as % siNTC) on the x-axes. Gray diamonds are pooled siRNAs, black are the four individual siRNAs, and white are the siNTCs. The means and SDs of three independent S12 experiments are shown. R2 is the Spearman’s rho coefficient of correlation. The seven genes in red were considered the top hits due to good correlation.
Class I (ciliation, transport, and signaling defects)
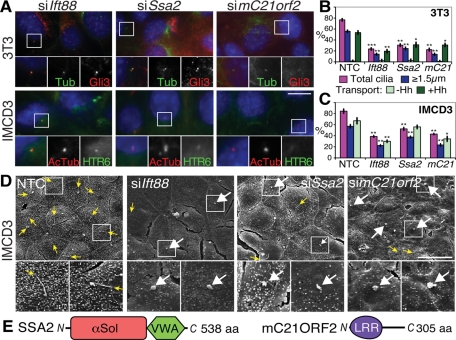
The Class I phenotypes reproduced in repeated knockdown experiments (Figure 4, A–C), suggesting Ssa2 and mC21orf2 may play roles in cilia formation and/or maintenance like Ift88. SSA2, Sjögren’s syndrome antigen A2 (also known as Trove2 or Ro60), is a component of the Ro ribonucleoprotein (Millard et al., 2002) with a von Willebrand factor A (VWA) domain and a so-called TROVE module (found in telomerase and Ro and Vault proteins; Bateman and Kickhoefer, 2003), the crystal structure of which reveals it to actually be a HEAT repeat α-solenoid fold (Stein et al., 2005) (Figure 4E). The HEAT repeat of SSA2 has been implicated in binding to Y RNAs, small noncoding RNAs of unknown function (Perreault et al., 2007), whose connection to cilia is currently unclear. mC21ORF2 (officially C10H21ORF2, the murine orthologue of human chromosome 21 open reading frame 2, on chromosome 10 in mouse) is a conserved leucine-rich repeat containing protein of no known function that was identified by genomic sequencing (Scott et al., 1998) (Figure 4E).
FIGURE 4:
Effects of Class I gene knockdowns (ciliation defects). (A) Representative images of ciliation defects in Ift88, Ssa2, and mC21orf2 siRNA-treated 3T3 (top) and IMCD3 (bottom) cells stained as in Figure 1B. Scale bar is 10 μm; insets show single and merged channel images of the boxed regions at 2× magnification. (B) Quantification of cilia phenotypes upon Class I siRNA knockdown in ≥200 3T3 cells. At least 200 cilia from images as in (A) were scored as before for % total cilia (magenta), % cilia ≥1.5 μm (blue), and % ≥1.5 μm cilia with ciliary Gli3 accumulation (green). The means and standard deviations (SDs) of three independent experiments are shown. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 vs. siNTC. mC21, mC21orf2. (C) Quantification as in (B) except in IMCD3 cells transporting HTR6-GFP instead of Gli3. *p ≤ 0.05, **p ≤ 0.01 vs. siNTC. (D) Representative SEM images of control and Class I–depleted IMCD3 cells. Yellow arrows point to tips of long cilia (note these are rarer in Class I-deficient cells); white arrows point to ciliary stumps. Scale bar is 20 μm in the main panels, and 3× magnifications of the boxed regions are shown underneath. (E) Schematic of SSA2 and mC21ORF2 proteins, drawn approximately to scale. αSol depicts a helical alpha solenoid fold (previously called the TROVE domain; Bateman and Kickhoefer, 2003); LRR, Leu-rich repeat.
Ssa2 or mC21orf2 depletion affected Gli3 more than HTR6 transport, perhaps because HTR6 is constitutively transported to cilia throughout the transfection and serum starvation period, whereas Gli3 transport was measured after only 30 min of Hh stimulation following mRNA depletion. To better understand the structural effects of Class I gene depletion, scanning electron microscopy (SEM) was performed. Cilia were difficult to identify in control 3T3 cells (Supplemental Figure S3A) because they were randomly oriented and usually not emergent (Rohatgi and Snell, 2010), whereas IMCD3 cilia consistently projected from the apical surface of the monolayer (Figure 4D), as recently reported (Molla-Herman et al., 2010). The majority of cilia in Ift88, Ssa2, and mC21orf2 siRNA-treated IMCD3 cells were either completely absent or reduced to stumps (≤1.5 μm), likely representing cilia that have failed to elongate (Figure 4D and Supplemental Figure S3B), with virtually no cilia of the normal mean length of 5 μm. This finding confirms that the structural defects of Class I and Ift88 knockdowns are similar.
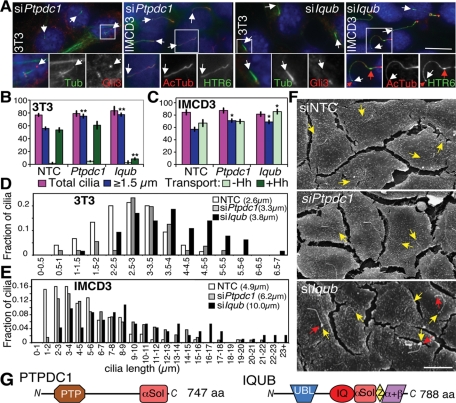
Class III (extra long cilia)
In Class III, Ptpdc1 validated well (Figure 3B), and both Ptpdc1 and Iqub lengthened cilia (confirmed in Figure 5, A–C). 3T3 cilia increased from an average of 2.6 to 3.3 or 3.8 μm upon depletion of Ptpdc1 or Iqub, respectively (Figure 5D), whereas IMCD3 cilia (the length of which was unaltered by HTR6-GFP expression) increased even more dramatically, from 4.9 to 6.2 or 10.0 μm, respectively (Figure 5E). These measurements were confirmed by SEM (Figure 5F and Supplemental Figure S3B). Interestingly, S12 cell cilia lengths increased upon Ptpdc1 depletion, but were resistant to Iqub depletion (Supplemental Figure S5E), perhaps explaining the poor validation in this cell line. This lack of Iqub depletion phenotype in S12 cells may simply be due to fivefold lower Iqub expression than in IMCD3 cells (Supplemental Figure S2A), but because siIft88 also failed to affect S12 cilia length (Supplemental Figure S1A), this peculiarity of S12 cells could suggest that IQ motif and ubiquitin domain–containing protein (IQUB) directly regulates cilia length in other cell types. Three of four Iqub siRNAs did increase cilia lengths in 3T3 and IMCD3 cells (unpublished data), thus validating this hit in these cell types. Iqub knockdown strongly inhibited Gli3 transport, in contrast to Ptpdc1 knockdown, which did not affect the number of Gli3+ cilia (Figure 5B) but significantly increased the amount of Gli3 fluorescence per cilium by 1.7-fold (Supplemental Figure S4A), suggesting that the two proteins negatively regulate cilia length via different mechanisms. Conversely, HTR6-GFP accumulated readily in cilia of Iqub-depleted IMCD3 cells (Figure 5C), where it was detectable in large bulges along the cilia shaft and tips (Figure 5, A and F, red arrows). Because ciliary bulges have been observed only in retrograde mutants (Huangfu and Anderson, 2005; May et al., 2005; Tran et al., 2008), we speculate that Iqub depletion elongates cilia by partially inhibiting retrograde transport (not unique to HTR6) and that Gli3 fails to accumulate due to inability of Patched to exit the cilium and allow Smoothened (SMO) entry and pathway activation. Alternatively, its effects on transport could be secondary to cilia length disruption. By contrast, we speculate that Ptpdc1 depletion may lengthen cilia and accumulate higher levels of Gli3 via increased anterograde transport.
FIGURE 5:
Effects of Class III gene knockdowns (abnormally long cilia). (A) Representative images of 3T3 and IMCD3 HTR6-GFP cells depleted of Ptpdc1 (left) or Iqub (right) and stained as in Figure 1B. White arrows indicate cilia tips, and red arrows indicate bulges in cilia (only seen with siIqub). Scale bar is 10 μm; insets show 2× magnification of the boxed regions for 3T3 cells and 1.5× magnification for IMCD3 cells. (B) 3T3 quantitations as in Figure 4B but with NTC, Ptpdc1, and Iqub siRNAs. **p ≤ 0.01 vs. siNTC. (C) IMCD3 HTR6-GFP quantitations as in Figure 4C with NTC, Ptpdc1, and Iqub siRNAs. *p ≤ 0.05 vs. NTC. (D) Histogram of cilia length distribution in Ptpdc1 and Iqub siRNA-transfected 3T3 cells, with the mean cilia lengths in parentheses. (E) As in (D), but with IMCD3 HTR6-GFP cells. (F) Representative SEM images of Ptpdc1- and Iqub-depleted IMCD3 cells. Yellow arrows indicate cilia tips, and red arrows indicate ciliary bulges in Iqub-deficient cells. Scale bar is 10 μm. (G) Schematic of PTPDC1 and IQUB proteins, drawn approximately to scale. αSol depicts a helical solenoid fold; PTP, protein tyrosine phosphatase; Z, Zn2+-binding module; α+β, novel IQUB C-terminal domain.
Protein tyrosine phosphatase domain containing 1 (PTPDC1) is a homologue of the mitotic phosphatase CDC14 that associates with PLK1 and has been localized to centrosomes (Zineldeen et al., 2009). Its role in ciliary length determination, however, is unlikely directly related to cell cycle control, because we observed no significant defects in viability or cilia number and the cells used for ciliation experiments were not cycling due to serum starvation. IQUB is an orphan protein named for its recognizable calmodulin-binding IQ motif and ubiquitin-like (UBL) domain. We detected three additional C-terminal domains: a helical solenoid segment coupled to a short Cx2Cx20Cx2C Zn2+-binding module (Z), and a novel 120-amino-acid α+β domain (Figure 5G).
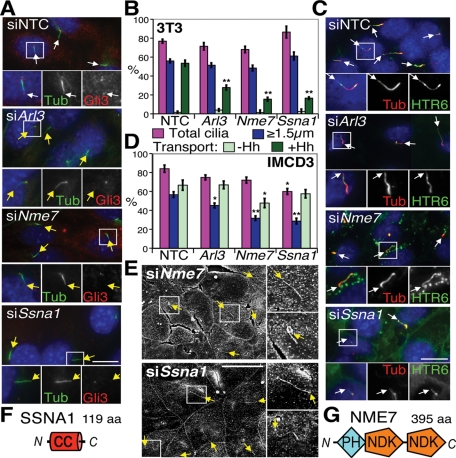
Class V (transport and signaling defects)
Class V hits were particularly interesting in exhibiting defective cytoplasmic cargo (Gli3) transport yet retaining normal-looking cilia, and included Arl3 (ADP-ribosylation factor-like 3), Nme7 (nonmetastatic cells 7, also known as Ndk7 or nucleoside diphosphate kinase 7); and Ssna1 (Sjögren’s syndrome nuclear autoantigen 1), which encodes a small coiled-coil protein (Figure 6F). Knockdown effects were reproducible (Figure 6, A–D), although HTR6 transport was less inhibited than Gli3 transport, with only Nme7 knockdown causing a mild defect in HTR6+ cilia, concomitant with a small decrease in average fluorescence intensity (Supplemental Figure S4C). Ssna1 knockdown had no significant effect on overall HTR6+ cilia numbers, but increased the overall fluorescence per cilium (Supplemental Figure S4C). Nonetheless, the cilia in both cell types were mostly of normal length and appearance by immunofluorescence (Figure 6, A and C), implicating these genes in cargo-specific transport rather than IFT. SEM confirmed both normal length and appearance in IMCD3 cells, bar a few (<5%) bent or kinked cilia (Figure 6E and Supplemental Figure S3B). Although NME7 is a member of the conserved NME/NDK family, it is predicted not to be active as a kinase (Desvignes et al., 2009). As well as two NDK modules, it has an N-terminal DM10/DUF1126 domain of unknown function, but whose pleckstrin homology (PH)-like β6-α fold has been captured in the partial structure of human EFHC2 (PDB ID 2Z13; Figure 6G). Of note, Nme5 is a related inactive kinase also in the ciliome (Ostrowski et al., 2002; Pazour et al., 2005; Stolc et al., 2005) and detectable in ciliated tissues (McClintock et al., 2008).
FIGURE 6:
Effects of Class V gene knockdowns (cilia transport defects). (A) Representative images of NTC, Arl3-, Nme7-, and Ssna1-depleted 3T3 cells stained as in Figure 1B. White and yellow arrows indicate Gli3+ and Gli3− cilia tips, respectively. Scale bar is 10 μm; insets show 2× magnification of the boxed regions with individual and merged channels. (B) 3T3 cilia quantitations following Class V gene depletion. **p ≤ 0.01 vs. NTC. (C) Representative images of Class V–depleted IMCD3 HTR6-GFP cells, with white arrows indicating cilia tips. (D) IMCD3 cilia quantitations. *p ≤ 0.05, **p ≤ 0.01 vs. NTC. (E) SEM of Nme7- and Ssna1-depleted IMCD3 cells. Upper insets on the right of each panel show 2× magnified cilia typical of the majority, whereas lower insets show examples of the few abnormally bent or kinked cilia. Yellow arrows point to cilia tips. Scale bar is 10 μm. Schematics of SSNA1 (F) and NME7 (G) protein domains, drawn approximately to scale. CC, coiled-coil.
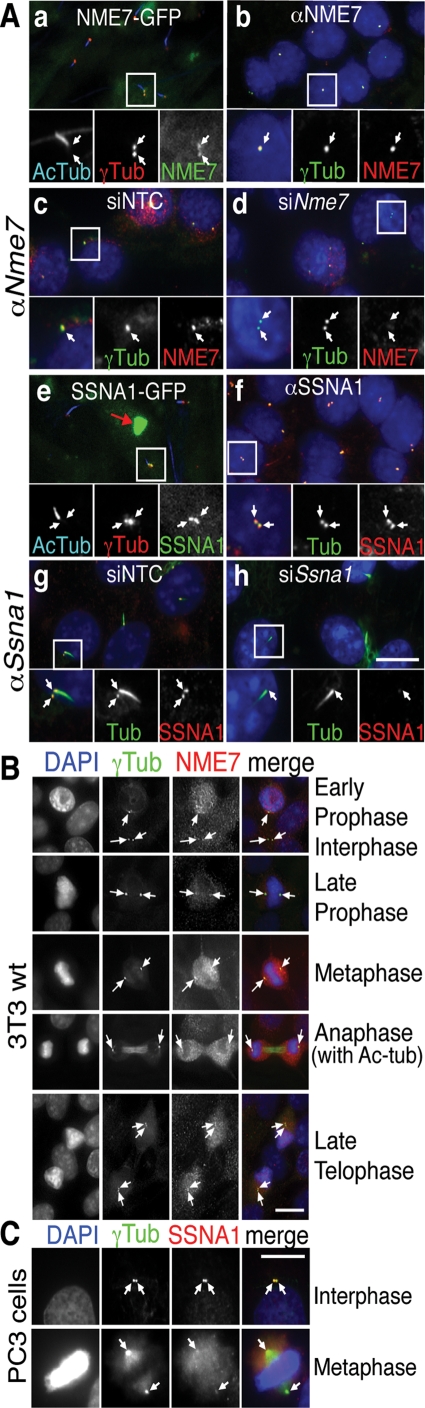
ARL3, a member of the small GTPase Arf family (Kahn et al., 2005), has been localized to centrosomes and cilia (Zhou et al., 2006), so we determined whether this was also the case for NME7 and SSNA1. GFP-tagged NME7 and SSNA1 stably expressed in 3T3 cells both localized to basal bodies, but not cilia, colocalizing well with γ-tubulin (Figure 7A). Endogenous NME7 and SSNA1 (see Supplemental Figure S4 for antibody validation) similarly localized to centrosomes in all cell types tested, ciliated or not, and remained centrosomal throughout mitosis (Figure 7, B and C). Thus all three Class V hits are positioned at the microtubule-organizing center and basal bodies, ideally located for regulating ciliary traffic.
FIGURE 7:
Class V hits localize to centrosomes. (A) Serum-starved 3T3 Nme7-GFP (a) and Ssna1-GFP (e) stable cell lines were stained for cilia (Ac-Tub, Cy5 channel) and centrosomes (γTub, red channel) with GFP fluorescence detected in the green channel. SSNA1 (± GFP tag) aggregated in inclusion bodies (red arrow in e) even when stably expressed, so low SSNA1-GFP expressers were selected by flow cytometry and further siSsna1-transfected to enable centrosome detection. Serum-starved IMCD3 HTR6-GFP cells were stained for nuclei (DAPI, blue), centrosomes (γTub, Cy5 channel, pseudo colored green), and with anti-NME7 (b) or anti-SSNA1 antibodies (f) (Cy3, red channel). 3T3 wild type (wt) cells costained with anti-NME7 (c and d) or anti-SSNA1 (g and h) in red and anti-acetylated and γ-tubulins in green following siNTC (c and g), siNme7 (d), or siSsna1 (h) transfection. Scale bar is 10 μm, and insets show 2× magnification of the boxed region. NME7 staining (red) in nonstarved 3T3 cells (B) and SSNA1 staining in nonciliated PC3 cells (C) during the indicated stages of mitosis, as identified by DAPI (blue) and γ-tubulin (green) staining. Scale bar is 15 μm in (B) and (C). White arrows point to centrosomes. Note the anaphase panel in (B) has both acetylated and γ-tubulin staining.
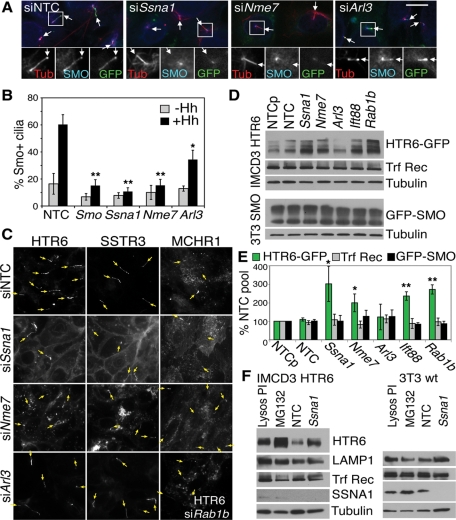
To further investigate Class V gene function, we generated 3T3 cells stably expressing GFP-SMO at low enough levels to retain Hh-dependent ciliary localization, and examined the effect of gene knockdown. Ssna1 and Nme7 depletion inhibited the Hh-stimulated accumulation of GFP-SMO in cilia as effectively as removal of the SMO protein itself, whereas Arl3 depletion had only a partial effect (Figure 8, A and B), mirroring the effects of depleting these genes on Gli3 accumulation (Figure 6B). Because SMO has been proposed to undergo lateral transport from the cell surface to the cilium upon Hh stimulation (Milenkovic et al., 2009), these data potentially implicate NME7 and SSNA1 either in this or the preceding transport step moving SMO from the Golgi to the cell surface. Because SMO was not detectable on the cell surface, we examined Class V knockdown effects on GFP-tagged ciliary GPCRs, SSTR3, and MCHR1 (Mukhopadhyay et al., 2010). Similar to its effects on HTR6 transport, Ssna1 depletion did not prevent ciliary targeting of SSTR3 in IMCD3 cells (Figure 8C) or retinal pigment epithelium 1 (RPE1) cells (unpublished data) or of MCHR1 in RPE1 cells, but did accumulate all three GPCRs at the cell surface (Figure 8C). By contrast, Nme7 depletion accumulated the three GPCRs in transport vesicles, again without abolishing ciliary signals. These vesicles did not colocalize with endocytic markers (transferrin receptor, LAMP1, or EEA1), and were also observed upon knockdown of the secretory transport GTPase Rab1b (Figure 8C) (Hutt and Balch, 2008), suggesting that they could be of secretory origin, consistent with delayed Golgi-to-cell-surface transport and lower ciliary levels. Arl3 knockdown did not cause accumulation of the GPCRs either in vesicles or at the cell surface, suggesting that it may regulate ciliary transport by yet another mechanism.
FIGURE 8:
Class V hits differentially affect GPCR transport. (A) Effect of Class V knockdown on ciliary accumulation of GFP-SMO. GFP-SMO 3T3 stable cells depleted of Class V genes were stimulated with Hh for 16 h, fixed, and stained for acetylated tubulin in red (or bottom left panels) and an unpurified anti-SMO antiserum (the centrosomal reactivity is present in the preimmune serum and not related to SMO) in the cyan Cy5 channel (or bottom middle panels), with GFP fluorescence in green (or bottom right). Scale bar is 10 μm; insets show 2× magnification of the boxed regions; arrows indicate cilia tips. (B) Quantitation of the anti-SMO signal in ∼300 cilia stained as in (A) with (black) or without (gray) Hh treatment. *p ≤ 0.05; **p ≤ 0.01 vs. siNTC + Hh. (C) Differential effects of Class V knockdown on GPCR-GFP localization. IMCD3 HTR6-GFP (left), IMCD3 SSTR3-GFP (middle), and RPE1 MCHR1-GFP (right) cells were transfected with NTC or Class V siRNAs as indicated and were serum starved for 16 h. The intrinsic GFP fluorescence is shown, with arrows pointing to cilia tips identified by Ac-tubulin staining. Bottom right panel shows HTR6-GFP in IMCD3 Rab1b-depleted cells for comparison with siNme7. (D) Western blot of IMCD3 HTR6-GFP (top panels) or GFP-SMO 3T3 cells (bottom panels) following 64-h siRNA transfection and 16-h serum starvation. Lysates (50 μg) were loaded on an 18% Tris–glycine gel and immunoblotted with anti-GFP for GPCRs, tubulin loading control, and anti-transferrin receptor (Trf Rec). NTCp is a second nontargeting pool of siRNAs used as an extra control and for statistical analysis. (E) Quantitation of four (or eight for Ssna1) experiments such as the one shown in (D), normalized to tubulin and NTC pool. *p ≤ 0.05; **p ≤ 0.01 vs. NTC. Knockdown efficiencies of SSNA1 and NME7, for which antibodies are available, are shown in Supplemental Figure S4, F–H. (F) Comparison of SSNA1 knockdown in IMCD3 HTR6-GFP (left) or 3T3 wild-type (wt) cells (right) with 16-h lysosomal protease inhibitor (Lysos PI; leupeptin at 10 μg/ml and 5 μM pepstatin A) or proteasomal inhibitor treatment (MG132; 10 μM). The lysosomal protein LAMP1 and endocytic transferrin receptor (Trf Rec) are shown for comparison, with tubulin as the loading control (50 μg of lysates on 18% gels). SSNA1 (∼14 kDa) was barely detectable in IMCD3 cells (see Supplemental Figure S4F for a higher loaded blot), but is more highly expressed in 3T3 cells. Western blot was performed as in (D).
Western blotting revealed consistently higher levels of HTR6-GFP upon Ssna1 and Nme7, but not Arl3, depletion (Figure 8, D and E, and Supplemental Figure S4, F and H). The increases were posttranslational, because HTR6 mRNA levels were unaltered, and the protein could be stabilized even more by lysosomal protease or proteasome inhibitors (Figure 8F). Inhibition of cell surface transport or ciliogenesis by Rab1b or Ift88 knockdown, respectively, also stabilized HTR6, but none of the knockdowns stabilized the nonciliary endocytic transferrin receptor, raising the possibility that HTR6 is down-regulated, likely by endolysosomal degradation, after passage through the primary cilium. The lateral transport cargo GFP-SMO was not stabilized by any of these knockdowns in the absence of Hh treatment (Figure 8, D and E), although the endoplasmic reticulum pool may obscure any post-Golgi changes. Endogenous SMO in 3T3 cells, however, was similarly unaffected by Ssna1 knockdown with or without Hh treatment (Supplemental Figure S4G), implying a distinct trafficking route for this stimulated cargo and perhaps supporting the lateral transport model (Milenkovic et al., 2009). Consistent with defects in trafficking to or from (rather than within) cilia, IFT88 was distributed normally in cilia of all Class V knockdowns. Taken together, these data imply different roles for the three Class V proteins. We speculate that SSNA1 may regulate postciliary endocytosis of constitutive GPCRs, NME7 (like RAB1B) may regulate their vesicular transport to the cell surface or cilium, and ARL3 appears to have a distinct function that partially affects lateral (SMO) but not constitutive GPCR transport or stability.
Differential effects on Gli3 processing confirms distinct cilia class phenotypes
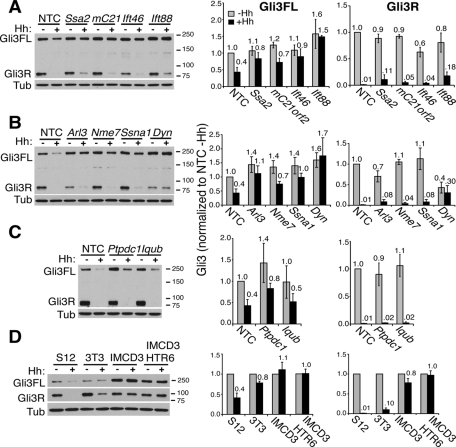
Because all the successfully knocked down genes resulted in defective Hh signaling with variable effects on Gli3 transport, it was of interest to determine whether Gli3 processing or degradation was affected in the different classes. We previously showed that full-length Gli3 becomes unstable following Hh stimulation in S12 cells, which we speculated was due to Gli3 activation (into Gli3A) at cilia tips (Wen et al., 2010). Furthermore, we showed that this instability was prevented upon disrupting cilia by Ift88 or Dync2h1 depletion, whereas Gli3 processing in the absence of Hh (into Gli3R) was only partially affected, suggesting that cilia facilitate but are not absolutely required for processing. We confirmed these results here (Figure 9) and found that none of the cilia hit siRNAs resulted in degradation of Gli3FL, indicating that lack of total Gli3 is not the reason for impaired ciliary Gli3 accumulation. As expected, depletion of the Class I hits, Ssa2 and mC21orf2, consistently resulted in slightly elevated Gli3FL levels, inhibited Hh-mediated Gli3A and Gli3R degradation, but had little effect on Gli3 processing (Figure 9A). This result was similar to that of Ift88 depletion, albeit less pronounced, and was consistent with lack of ciliation and Gli3 transport. Ift46 depletion (with cilia elongation defects) differed from Ift88 depletion in that baseline levels of Gli3FL were less elevated, although Hh-stimulated degradation of Gli3A was similarly impaired (Figure 9A).
FIGURE 9:
Different classes of cilia genes have distinct effects on Gli3 processing and degradation. (A–C, left panels) S12 cells transfected with the indicated siRNAs for a total of 72 h were treated with (+) or without (–) Hh for 16 h prior to lysis and Western blotting with anti-Gli3N antibody 6F5 as described (Wen et al., 2010) to detect both full-length Gli3 (Gli3FL) and the truncated Gli3 repressor (Gli3R). (D) S12, 3T3, IMCD3, and IMCD3 HTR6-GFP cells – or + Hh were blotted as in (A–C). All blots shown are representative of at least three to five experiments. Molecular weight markers in kilodaltons are indicated on the right. Tubulin antibody 1A2 (Tub) served as the loading control. mC21, mC21orf2; Dyn, Dync2h1. (A–D, right panels) Quantitation of Gli3FL (left) and Gli3R (right graphs) from ≥3 blots were quantitated and normalized to the tubulin loading control and then to the NTC siRNA –Hh. The means and standard deviations (SDs) of ≥3 independent experiments for each siRNA are plotted with their values relative to siNTC –Hh above each bar. Gray bars are –Hh; black bars are +Hh.
Knockdowns of Nme7 and Ssna1, which also inhibit Gli3 ciliary accumulation but retain normal length cilia, similarly resulted in impaired Gli3FL degradation (or Gli3A formation, Figure 9B), enhanced baseline Gli3FL levels, and little change in Gli3R. This finding supports our earlier proposal (Wen et al., 2010) that Gli3 transport to cilia tips is essential for Gli3 activation and lability, but not for processing into Gli3R. This phenotype resembles that of the Arl13b hennin mutation (Caspary et al., 2007), although it remains to be determined whether this similarly inhibits Gli3 transport. Knockdown of the other Class V gene, Arl3, similarly affected Gli3A degradation, but also inhibited processing, albeit less strongly than did Dynch21 depletion (Figure 9B).
Ptpdc1 (Class III), the knockdown of which elongates cilia and accumulates Gli3 at their tips, similarly stabilized Gli3FL and inhibited Hh-mediated degradation with no impact on processing, suggesting that the timing of Gli3 activation with respect to transport back out of cilia is also important for regulating Gli3A stability. Iqub depletion had no effect on Gli3 levels in S12 cells, consistent with lack of elongation of cilia in this cell line, although the effect on 3T3 cells (with Gli3− elongated cilia) was also less than expected (Supplemental Figure S5C). Depletion of all the genes gave similar results in 3T3 and S12 cells (Figure 9 and Supplemental Figure S5, A–C), although the extent of Gli3A degradation upon Hh stimulation was much smaller (Figure 9D), perhaps because 3T3 cells are more dependent on serum starvation to ciliate, have fewer robust cilia (Figure 1 and Supplemental Figure S1), and consequently respond less well to Hh (only up-regulating the Hh target gene Gli1 12× compared with ∼100× in S12 cells after 24-h stimulation; Supplemental Figure S5E). By comparison, the non-Hh–responsive IMCD3 (and IMDC3 HTR6-GFP) cells did not destabilize Gli3FL at all (Figure 9D). These data thus support the different phenotypes observed for the various classes of cilia defects, and highlight the need for fully functional cilia to properly regulate Gli3 activation and destabilization.
DISCUSSION
We performed high-content analysis of ciliation phenotypes (number, length, ability to transport two different cargoes, and Hh signaling capability) in three different cell lines after knockdown of 40 putative ciliome genes by siRNA transfection. Although we assayed primary cilia, motile cilia were the main sources of our input gene list (Table 1), thus our hits likely function in both cilia types, despite all exhibiting signaling defects. Indeed, motile cilia have recently been suggested to have sensory as well as motile functions (Quarmby and Leroux, 2010) and may even develop from earlier primary ciliated cells (Jain et al., 2010). Our hits could be divided into six classes based on their ciliary phenotypes: I) ciliation, transport, and signaling defects; II) short cilia (i.e., elongation defects); III) abnormally long cilia; IV) structure and signaling defects; V) transport and signaling defects; and VI) signaling defects only (Figure 2). Although these classifications are based on somewhat arbitrary cutoff values, especially where phenotypes differed in strength among the cell lines, they nonetheless serve as a useful starting point for understanding the different functions of the ciliome proteins.
An advantage of high-content analysis is that it permits such classification of cilia phenotypes, allowing us to uncover (or support previous observations of) several generic aspects of cilia biology:
We found that not only the presence of a cilium, but also its proper length is important for function, because short (Class II) and extra long (Class III) as well as absent cilia (Class I) all signaled defectively. We cannot exclude the possibility, however, that some hits directly inhibited Hh signaling and led to secondary defects in cilia lengths.
Transport defects were generally milder for the three GPCR-GFP cargoes (HTR6, SSTR3, MCHR1) than for Gli3 and GFP-SMO, despite more effective gene knockdown in IMCD3 cells (Supplemental Figure S2A), likely because GPCRs are constitutively transported throughout the experiment rather than stimulated after knockdown. Exceptions include mC21orf2 and Ift46, the knockdown of which impacted HTR6 more strongly than Gli3 transport, perhaps indicative of roles in cilia membrane formation; indeed, both prevented formation of long cilia.
The functions of IFT proteins even in the same subcomplex are not necessarily equivalent, because IFT46 and IFT88 both belong to the IFT-B (anterograde) complex, but had nonidentical phenotypes (Figure 2). Ift88 depletion affected ciliation (Class I), whereas Ift46 knockdown inhibited only elongation (Class II), despite efficient knockdown (Supplemental Figures S2A and S1, D and E). Both affected HTR6 transport similarly, consistent with their direct interaction with each other (Lucker et al., 2010) and the demonstrated association of IFT88 with transport vesicles (Sedmak and Wolfrum, 2010), but Ift88 knockdown had a stronger effect on Gli3 transport and stability, perhaps suggesting differential interactions of distinct IFT particle subunits with various cargoes or adaptors. Indeed Chlamydomonas ODA16 (outer arm dynein 16) has been shown to interact with IFT46 in acting as a cargo adaptor for dynein transport (Ahmed et al., 2008). It will be interesting to determine if IFT-B, like IFT-A, can be dissected into functionally distinct core and regulatory subcomplexes (Mukhopadhyay et al., 2010).
-
Interestingly, Class V genes (Ssna1, Nme7, and Arl3) with defective transport and signaling but grossly normal cilia illustrate that ciliary transport can in fact be uncoupled from cilia formation, at least for certain cargoes, as has been demonstrated for SSTR3 in BBSome-deficient cells (Jin et al., 2010). Cilia formation is thought to be necessarily coupled to IFT, because impaired IFT results in complete lack of or defective cilia, rendering determination of the role of IFT in cargo transport difficult (the “IFT Uncertainty Principle”; Rohatgi and Snell, 2010).
Nme7 depletion in particular may prove a useful tool to get around this problem, although its precise role has yet to be established. Since its depletion accumulates putative secretory transport vesicles and decreases ciliary levels of GPCRs, we speculate NME7 may regulate vesicular transport from the Golgi to the plasma membrane (with a secondary effect on lateral transport of SMO from there to the cilium). Note that the Nme7 GPCR transport defect is much milder than that of Tulp3 or Ift-A depletion (Mukhopadhyay et al., 2010), thus it likely plays a regulatory rather than a critical role. We hypothesize that SSNA1, by contrast, may facilitate endocytosis from the cell surface following transit through cilia, since its depletion caused GPCR accumulation at the plasma membrane and cilia instead of in vesicles. If true, the inhibition of SMO transport to cilia may be an indirect effect of the inability to remove Patched, a possibility we were unable to test due to lack of sensitive antibodies. Since Arl3 depletion only affected GFP-SMO and Gli3 transport, in both cases more modestly than Nme7 or Ssna1, ARL3 likely regulates transport by a distinct mechanism. Indeed, it has been implicated in transport of lipidated cytoplasmic proteins such as the myristoylated retinis pigmentosa RP2 protein to basal bodies (Evans et al., 2010). The presence of all three Class V hits at the centrosome (Figure 7 and Zhou et al., 2006) suggests they are suitably positioned to regulate entry of specific ciliary cargoes either at the ciliary gate or from the Golgi.
The ability to retain long cilia (by depleting Nme7, Ssna1 or Arl3) while inhibiting Gli3 ciliary accumulation provides further support for our hypothesis (Wen et al., 2010) that Gli3 processing does not absolutely require Gli3 transport through cilia, while Gli3 activation (leading to subsequent lability) does (Figure 9).
Finally, the observed accumulation of total HTR6 protein when cilia were absent (siIft88), or when transport to (siRab1b, siNme7) or putatively from (siSsna1) the cilium was inhibited causes us to speculate that not just Gli3, but other ciliary signaling cargoes may be destabilized following transit through the cilium. This possibility is an exciting avenue for further investigation.
In addition to leading to a better understanding of cilia biology, our screen was also successful in identifying high-quality hits, as attested by recent publications showing the involvement of Ift46, Arl3, and Nme7 in ciliary function. Ift46 is now an established component of the IFT complex (Lucker et al., 2005; Gouttenoire et al., 2007; Hou et al., 2007), although it was uncharacterized (gene Flj21827) at the start of our screen; Arl3−/– mice die young with ciliary defects, including polycystic kidney disease and photoreceptor degeneration (Schrick et al., 2006); and Nme7−/– mice have cilia, but exhibit cilia-related defects, including situs inversus and hydrocephalus (Vogel et al., 2009). Other genes that did not pass our stringent criterion of having ≥3 individual siRNA knockdowns correlating with phenotype have also recently been implicated in cilia function, including radial spokehead gene Rsph9 (an elongation gene in Class II), mutated in primary ciliary dyskinesia (Castleman et al., 2009); and Enkurin (in Class I), found in sperm flagellae and proposed to function as a calcium-signaling adaptor (Sutton et al., 2004; McClintock et al., 2008), although, if confirmed, our data would imply a more generic role in cilia formation than in signaling.
Furthermore, our screen assigned ciliary roles to several genes with no other known functional ciliary data: mC21orf2, Ptpdc1, Iqub, Ssna1, and Ssa2. Interestingly, SSA2 and SSNA1 (in Classes I and V) are both targets of autoantibodies in Sjögren’s syndrome, an autoimmune disorder characterized by dry eyes and mouths and defects in nasal mucociliary transport and respiration (Sakakura et al., 1983; Crestani et al., 2007). Depletion of Ssna1, unlike that of Ssa2, did not affect ciliation, although both genes are required for ciliary transport and signaling. Notably, Sjögren’s syndrome scleroderma autoantigen 1 is yet another putative ciliary Sjögren’s autoantigen (Liu et al., 2007). To our knowledge, our data are thus the first to suggest that defective cilia may be the cause of mucociliary defects and respiratory problems seen in Sjögren’s patients, although exactly how the autoantibodies would mediate these ciliary defects remains to be determined.
Overall, we successfully completed a high content analysis of putative ciliome genes, identifying 27 that resulted in at least a signaling defect upon knockdown, with or without additional cilia formation, elongation, or transport defects. Our data thus support the notion (Inglis et al., 2006; Marshall, 2008) that candidate cilia gene lists compiled from a combination of studies, including proteomics, genomic comparisons, and promoter analyses, contain a greater proportion of true ciliome genes than does any one method alone. Furthermore the distinct ciliary phenotypes obtained highlight the advantage of using multiple assays (high-content analysis) over a single output screen for better understanding of cilia biology, and yielded several excellent candidates for further investigation.
MATERIALS AND METHODS
Cell lines
3T3 cells (murine fibroblasts), S12 cells (murine C3H10T1/2 osteoblasts stably transfected with 8× Gli-binding sites driving luciferase expression; Frank-Kamenetsky et al., 2002), and SV40-luc cells (C3H10T1/2 osteoblasts stably transfected with SV40 driving luciferase; Evangelista et al., 2008) were maintained in high-glucose DMEM with 10% fetal bovine serum (FBS), 10 mM HEPES, and 2 mM l-glutamine. IMCD3 cells stably expressing C-terminally localization and affinity purification (LAP)-tagged (Torres et al., 2009) human HTR6-GFP and mouse SSTR3-GFP were made by stable transfection into an IMCD3 Flp-In cell line and maintained in F-12/DMEM high-glucose 50:50 mix with 10% FBS, 10 mM HEPES, 2 mM l-glutamine, and hygromycin B at 750 μg/ml. RPE1 cells stably expressing SSTR3-GFP or MCHR1-GFP were previously described (Mukhopadhyay et al., 2010). GFP-SMO 3T3 cells were made by lentiviral infection of N-terminally GFP-tagged human SMO and selection in puromycin at 1 μg/ml, followed by fluorescence-activated cell sorting sorting for low expressers and screening single clones by microscopy for Hh-dependent cilia translocation. 3T3 and S12 cells were never allowed to become >70% confluent to maintain proper ciliation and Hh signaling.
siRNA transfection and immunofluorescence
Ten thousand 3T3 or IMCD3 HTR6-GFP cells were seeded into eight-well LabTekII microscope slides (Nalge Nunc, Naperville, IL) to achieve 30–40% confluence upon transfection 24 h later. Dharmafect reagent #4 (0.8 μl; Dharmacon, Lafayette, CO) and 24.2 μl of Opti-MEM (Life Technologies, Grand Island, NY) were mixed with 1.25 μl of 20 μM siGENOME siRNA pools or 0.625 μl of 20 μM siRNA singles (Dharmacon) in 25 μl of total Opti-MEM and preincubated at room temperature for 20 min, then added to the cells to final concentrations of 100 nM and 50 nM, respectively. After 48 h, cells were serum-starved in 0.5% FBS for an additional 16 h ± octyl-sonic Hh at 200 ng/ml (Taylor et al., 2001) during the final 30 min as indicated. Cells were fixed and permeabilized at –20°C with 100% methanol, then blocked with 2% FBS and 1% bovine serum albumin in phosphate-buffered saline for 30 min. 3T3 cell cilia were stained with mouse anti-acetylated tubulin 6–11B-1 (0.37 μg/ml; Sigma, St. Louis, MO), centrosomes with mouse anti-γ-tubulin GTU-88 (1 μg/ml; Sigma), and Gli3 with rabbit anti-Gli3 antibody 2676 at 3 μg/ml (Wen et al., 2010), detected with Fluorescein isothiocyanate-anti-mouse and Cy3-anti-rabbit (Jackson ImmunoResearch, West Grove, PA), respectively. IMCD3 HTR6-GFP cell cilia were also stained with 6–11B-1, but detected with Cy3-anti-mouse (Jackson Immuno-Research), and centrosomes were stained with 1:3000 rabbit anti-γ-tubulin (AccuSpecs, Westbury, NY) and Cy5-anti-rabbit (although not shown in all figures). 3T3 GFP-SMO cells were methanol fixed ±16-h Hh treatment on gelatinized slides and stained with unpurified 5928B serum (Wen et al., 2010) and Cy5-anti rabbit to enhance the SMO signal. Endogenous Ssna1 and Nme7 were detected in methanol-fixed IMCD3 HTR6-GFP cells with rabbit anti-NME7 (H278; Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit anti-SSNA1 (11797–1-AP; Proteintech Group, Chicago, IL) and costained with mouse anti-γ-tubulin GTU88 with or without anti–acetylated tubulin 6–11B-1. Slides were coverslipped with ProLong Gold with DAPI (Invitrogen) and imaged by epifluorescence microscopy at room temperature with a 60× 1.42 NA oil objective using a DeltaVision Core (Applied Precision, Issaquah, WA) with an Olympus IX71 microscope, Photometrics CoolSnap HQ2 camera, and SoftWoRx (version 3.7.1) software. Images were processed using Adobe Photoshop CS, adjusting gamma levels in one or more channels where necessary to optimize detection of cilia. Western blots using anti-Gli3 antibody 6F5 were performed after siRNA knockdown in 3T3 cells ±16-h Hh stimulation as reported previously (Wen et al., 2010).
GFP-tagging and immunofluorescence
Murine Ssna1 and Nme7 were PCR amplified using Deep Vent polymerase with CACC 5′ overhangs using the primer sets mSSNA1F, mSSNA1R and mNME7F, mNME7R and the full-length cDNA templates NM_023464 and NM_138314, respectively (Supplemental Table S1b). The PCR products were directionally cloned into the pENTR/D-TOPO vector (Invitrogen) using a TOPO cloning kit (Invitrogen) and subcloned into vectors pGLAP3 and pGLAP5 for N-terminal LAP (GFP)-tagged and C-terminal LAP (GFP)-tagged constructs, respectively (Torres et al., 2009) by Gateway recombination (Invitrogen). GFP-tagged constructs were stably transfected into Flp-In™ 3T3 cells (Invitrogen) and selected with hygromycin B at 150 μg/ml (Clontech, Mountain View, CA). 3T3 cells expressing GFP-tagged genes were fixed with paraformaldehyde followed by methanol permeabilization and stained with mouse anti–acetylated tubulin 6–11B-1 and rabbit anti–γ-tubulin followed by Cy5-anti-mouse and Cy3-anti-rabbit.
Imaging data analysis and quantitation
Approximately 200 cilia were counted for each siRNA. Percent total Cilia was scored as the number of centrosomes with at least a ciliary dot (the length of which was no longer than its width) distinct from the centrosome, as well as longer cilia. A separate category of % cilia ≥1.5 μm was scored as the number of cells bearing a cilium ≥1.5 μm in length, as these were clearly distinct from the ciliary dots. Gli3 transport was assessed as previously described (Wen et al., 2010), and was scored as the percentage of ≥1.5 μm cilia with strong Gli3 signal at the cilia tip, thus transport defects are underestimated due to the exclusion of nonciliated and ciliary-dot-only cells from analysis. Percent HTR6-GFP in IMCD3 cilia was scored as the percentage of ≥1.5-μm-long cilia, the GFP signal of which was stronger than the corresponding acetylated tubulin signal. Nuclei (DAPI staining) were counted to verify even cell density and assess fixation quality.
SEM
After treatment with siRNAs as described earlier in text, IMCD3 HTR6-GFP cell samples were fixed in Karnovsky’s fixative (2% paraformaldehyde/2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) overnight and postfixed in 1% OsO4 for 1 h. The cells were dehydrated through a graded series of EtOH and infiltrated with hexamethyldisilazane. They were then air dried, mounted on stubs, and sputter coated with 10 nm AuPd. The samples were viewed at room temperature on an FEI Electron Optics (Eindhoven, The Netherlands) XL30 environmental scanning electron microscope at 15 kV acceleration voltage, 11 mm WD, spot size 4, and at 500× to 4000× magnification. Images were analyzed using XL Analysis (FEI) software.
Luciferase assays
Pooled siRNAs (12.5 μl of 1 μM; 100 nM final) or 12.5 μl of 500 nM individual siRNAs (50 nM final) were mixed with 0.3 μl of DharmaFECT reagent #4 in a total volume of 25 μl with Opti-MEM (Life Technologies) and added to 96-well plates after 30 min of incubation at room temperature. Reverse-transfection was performed by adding 6000 S12 cells in 100 μl of medium to the siRNA mix for 64 h followed by 24 h in 0.5% FBS medium ± Hh (200 ng/ml). Reverse-transfection of SV40-luc cells was performed the same way but with 9000 cells/well due to slower growth rate. Luciferase assays were done with a steadylite HTS (PerkinElmer, Waltham, MA) following the manufacturer’s instructions and were measured with a TopCount Luminometer (Applied Biosystems, Foster City, CA). Cell viability was measured under the same conditions as the S12 assay with CellTiter-Glo (Promega, Madison, WI) following the manufacturer’s instructions; similar results were obtained when cells were not serum-starved and grown to only ∼80% confluence.
Real-Time quantitative PCR
Gene knockdown for TaqMan analysis was done in 24-well plates by reverse-transfecting pooled siRNAs (100 nM final) or individual siRNAs (50 nM final) with 36,000 cells per well. After 64 h, cells were serum-starved for 24 h in 0.5% FBS medium. Total RNA was extracted from siRNA-treated cells using the RNeasy Mini Kit (Qiagen, Valencia, CA). On-column genomic DNA digestion was performed during the purification using RNAse-Free DNase Set (Qiagen), and cDNA synthesis was done using the High Capacity Reverse Transcription Kit (Applied Biosystems) with the random hexamer primers provided. Quantitative PCR reactions were performed by TaqMan analysis in triplicate, using probes listed in Supplemental Table S1b on an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). The % mRNA remaining after knockdown was calculated according to the 2-Ct method normalized to Rpl19. Spearman’s rho R2 values were calculated for the plots of % mRNA remaining versus the % Gli-luciferase remaining using the Trendline function in Microsoft Excel.
Structural motif determination
Domain architecture and key structural motifs of the top candidate hit proteins were detected by the SMART server (Letunic et al., 2009), in concert with sensitive fold recognition algorithms (Hildebrand et al., 2009).
Supplementary Material
Acknowledgments
We thank James Ernst and Hong Li for octyl Hh, James Lee for assistance with the S12 assays, Peter Dijkgraaf for 3T3 GFP-Smo cells, Saikat Mukhopadhyay for RPE1 MCHR1-GFP and SSTR3-GFP cells, and our colleagues at Curis (Cambridge, MA).
Genentech provided financial support. All authors are employees of Genentech.
Abbreviations used:
- ARL3
ADP-ribosylation factor-like 3
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- GPCR
G protein-coupled receptor
- Hh
hedgehog
- HTR6
serotonin receptor 6
- IFT
intraflagellar transport
- IMCD3
intermedullary collecting duct
- IQUB
IQ motif and ubiquitin domain–containing protein
- MCHR1
melanin-concentrating hormone receptor 1
- NDK7
nucleoside-diphosphate kinase 7
- NME7
nonmetastatic cells 7
- NTC
nontargeting control
- PDGF
platelet-derived growth factor
- PH
pleckstrin homology
- PTPDC1
protein tyrosine phosphatase domain containing 1
- RPE1
retinal pigment epithelium 1
- SEM
scanning electron microscopy
- siRNA
small interfering RNA
- SSA2
Sjögren's syndrome antigen A2
- SSNA1
Sjögren's syndrome nuclear autoantigen 1
- SSTR3
somatostatin receptor 3
- UBL
ubiquitin-like
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0596) on February 2, 2011.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bateman A, Kickhoefer V. The TROVE module: a common element in Telomerase, Ro and Vault ribonucleoproteins. BMC Bioinformatics. 2003;4:49. doi: 10.1186/1471-2105-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, et al. Functional genomics of the cilium, a sensory organelle. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Broadhead R, et al. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Castleman VH, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Crestani B, Schneider S, Adle-Biassette H, Debray MP, Bonay M, Aubier M. [Respiratory manifestations during the course of Sjogren’s syndrome] Rev Mal Respir. 2007;24:535–551. doi: 10.1016/s0761-8425(07)91575-2. [DOI] [PubMed] [Google Scholar]
- Desvignes T, Pontarotti P, Fauvel C, Bobe J. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256. doi: 10.1186/1471-2148-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, Ye W, Davis DP, de Sauvage FJ. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci Signal. 2008;1:ra7. doi: 10.1126/scisignal.1162925. [DOI] [PubMed] [Google Scholar]
- Evans RJ, Schwarz N, Nagel-Wolfrum K, Wolfrum U, Hardcastle AJ, Cheetham ME. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum Mol Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germino GG. Linking cilia to Wnts. Nat Genet. 2005;37:455–457. doi: 10.1038/ng0505-455. [DOI] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Gouttenoire J, Valcourt U, Bougault C, Aubert-Foucher E, Arnaud E, Giraud L, Mallein-Gerin F. Knockdown of the intraflagellar transport protein IFT46 stimulates selective gene expression in mouse chondrocytes and affects early development in zebrafish. J Biol Chem. 2007;282:30960–30973. doi: 10.1074/jbc.M705730200. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt DM, Balch WE. Rab1b silencing using small interfering RNA for analysis of disease-specific function. Methods in Enzymol. 2008;438:1–10. doi: 10.1016/S0076-6879(07)38001-4. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Heintz N, Omran H. To beat or not to beat: roles of cilia in development and disease. Hum Mol Genet. 2003;12(Spec No 1):R27–R35. doi: 10.1093/hmg/ddg061. [DOI] [PubMed] [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Jain R, Pan J, Driscoll JA, Wisner JW, Huang T, Gunsten SP, You Y, Brody SL. Temporal relationship between primary and motile ciliogenesis in airway epithelial cells. Am J Respir Cell Mol Biol. 2010;43:731–739. doi: 10.1165/rcmb.2009-0328OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Roehl White S, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedel syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Volpicelli-Daley L, Bowzard B, Shrivastava-Ranjan P, Li Y, Zhou C, Cunningham L. Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem Soc Trans. 2005;33:1269–1272. doi: 10.1042/BST0331269. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee K, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Gleeson JG. The role of primary cilia in neuronal function. Neurobiol Dis. 2010;38:167–172. doi: 10.1016/j.nbd.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Vega R, Nelms K, Gekakis N, Goodnow C, McNamara P, Wu H, Hong NA, Glynne R. A role for Alstrom syndrome protein, alms1, in kidney ciliogenesis and cellular quiescence. PLoS Genet. 2007;3:e8. doi: 10.1371/journal.pgen.0030008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucker BF, Behal RH, Qin H, Siron LC, Taggart WD, Rosenbaum JL, Cole DG. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J Biol Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- Lucker BF, Miller MS, Dziedzic SA, Blackmarr PT, Cole DG. Direct interactions of intraflagellar transport complex B proteins IFT88, IFT52 and IFT46. J Biol Chem, 2010 doi: 10.1074/jbc.M110.106997. PMID: 20435895 ePub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. Use of transcriptomic data to support organelle proteomic analysis. Methods Mol Biol. 2008;432:403–414. doi: 10.1007/978-1-59745-028-7_27. [DOI] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics. 2008;32:198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TP, Ashton GH, Kondeatis E, Vaughan RW, Hughes GR, Khamashta MA, Hawk JL, McGregor JM, McGrath JA. Human Ro60 (SSA2) genomic organization and sequence alterations, examined in cutaneous lupus erythematosus. Br J Dermatol. 2002;146:210–215. doi: 10.1046/j.1365-2133.2002.04618.x. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, et al. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci. 2010;123:1785–1795. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nielsen SK, Mollgard K, Clement CA, Veland IR, Awan A, Yoder BK, Novak I, Christensen ST. Characterization of primary cilia and Hedgehog signaling during development of the human pancreas and in human pancreatic duct cancer cell lines. Dev Dyn. 2008;237:2039–2052. doi: 10.1002/dvdy.21610. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J, Perreault JP, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol. 2007;24:1678–1689. doi: 10.1093/molbev/msm084. [DOI] [PubMed] [Google Scholar]
- Quarmby LM, Leroux MR. Sensorium: the original raison d’etre of the motile cilium? J Mol Cell Biol. 2010;2:65–67. doi: 10.1093/jmcb/mjp036. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Snell WJ. The ciliary membrane. Curr Opin Cell Biol. 2010;22:541–546. doi: 10.1016/j.ceb.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Sakakura Y, Ukai K, Majima Y, Murai S, Harada T, Miyoshi Y. Nasal mucociliary clearance under various conditions. Acta Otolaryngol. 1983;96:167–173. doi: 10.3109/00016488309132888. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Anderson KV. Intraflagellar transport and cilium-based signaling. Cell. 2006;125:439–442. doi: 10.1016/j.cell.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am J Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HS, Kyriakou DS, Peterson P, Heino M, Tahtinen M, Krohn K, Chen H, Rossier C, Lalioti MD, Antonarakis SE. Characterization of a novel gene, C21orf2, on human chromosome 21q22.3 and its exclusion as the APECED gene by mutation analysis. Genomics. 1998;47:64–70. doi: 10.1006/geno.1997.5066. [DOI] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol. 2010;189:171–186. doi: 10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Smith JC, Northey JG, Garg J, Pearlman RE, Siu KW. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton KA, Jungnickel MK, Wang Y, Cullen K, Lambert S, Florman HM. Enkurin is a novel calmodulin and TRPC channel binding protein in sperm. Dev Biol. 2004;274:426–435. doi: 10.1016/j.ydbio.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Taylor FR, et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry. 2001;40:4359–4371. doi: 10.1021/bi002487u. [DOI] [PubMed] [Google Scholar]
- Torres JZ, Miller JJ, Jackson PK. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 2009;9:2888–2891. doi: 10.1002/pmic.200800873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel P, Read R, Hansen G, Freay L, Zambrowicz B, Sands A. Situs inversus and related ciliopathies in Dpcd-/-, Pkd1l1-/- and Nme7-/- mice. Vet Pathol 47, 120–131. 2009 doi: 10.1177/0300985809353553. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhou Z, Walsh CT, McMahon AP. Selective translocation of intracellular Smoothened to the primary cilium in response to Hedgehog pathway modulation. Proc Natl Acad Sci USA. 2009;106:2623–2628. doi: 10.1073/pnas.0812110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length gli3 accumulation in primary cilia and subsequent degradation. Mol Cell Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zineldeen DH, Shimada M, Niida H, Katsuno Y, Nakanishi M. Ptpcd-1 is a novel cell cycle related phosphatase that regulates centriole duplication and cytokinesis. Biochem Biophys Res Commun. 2009;380:460–466. doi: 10.1016/j.bbrc.2009.01.113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.