Intraflagellar transport (IFT) 20 is required for the formation of photoreceptor outer segments, and deletion of this gene in mature cells leads to opsin accumulation in the cell body. IFT20 interacts with opsin photopigments both in the context of the IFT particle and independent of the particle, suggesting that opsin receptors are cargo for the IFT system.
Abstract
The light-detecting outer segments of vertebrate photoreceptors are cilia. Like other cilia, all materials needed for assembly and maintenance are synthesized in the cell body and transported into the cilium. The highly elaborated nature of the outer segment and its high rate of turnover necessitate unusually high levels of transport into the cilium. In this work, we examine the role of the IFT20 subunit of the intraflagellar transport (IFT) particle in photoreceptor cells. IFT20 was deleted in developing cones by a cone-specific Cre and in mature rods and cones by a tamoxifen-activatable Cre. Loss of IFT20 during cone development leads to opsin accumulation in the inner segment even when the connecting cilium and outer segment are still intact. With time this causes cone cell degeneration. Similarly, deletion of IFT20 in mature rods causes rapid accumulation of rhodopsin in the cell body, where it is concentrated at the Golgi complex. We further show that IFT20, acting both as part of the IFT particle and independent of the particle, binds to rhodopsin and RG-opsin. Since IFT20 dynamically moves between the Golgi complex and the connecting cilium, the current work suggests that rhodopsin and opsins are cargo for IFT transport.
INTRODUCTION
The outer segments of vertebrate rod and cone cells are highly modified cilia and exemplify the paradigm of cilia functioning as cellular antennae to monitor the extracellular environment. In the retina, the cilia or outer segments detect light, but, in other cell types, cilia detect hormones, osmolarity changes, and shear stress (reviewed in Pazour and Bloodgood, 2008). Work over the past 10 years has shown that sensory cilia play key roles in the development of mammals and the maintenance of health. Severe defects in primary cilia cause embryonic lethality whereas moderate defects yield a plethora of phenotypes including cystic kidney disease, obesity, and polydactyly along with defects in every organ system examined (Quinlan et al., 2008). In the eye, ciliary defects cause retinal development and degeneration disorders (Insinna and Besharse, 2008).
All cilia, including photoreceptor rod and cone outer segments, lack capacity for protein synthesis and rely on proteins made in the cytoplasm for their assembly and maintenance (Remillard and Witman, 1982). Because most types of cilia are assembled in small protrusions from the surface of the cell, materials made in the cell body need to be transported into the ciliary compartment for assembly. It is thought that the ciliary proteins and possibly lipids are moved by intraflagellar transport (IFT). During IFT, large protein complexes are transported along ciliary microtubules under the ciliary membrane (Rosenbaum and Witman, 2002). The outward movement is powered by kinesin-2 motors (heterotrimeric Kif3 and homodimeric Osm3/Kif17) (Scholey, 2008), whereas the inward movement is powered by an isoform of cytoplasmic dynein (called Dynein 2, Dynein 1B, or IFT-Dynein). The IFT particles are composed of ∼20 different proteins organized in two subcomplexes called A and B (Cole et al., 1998).
Photoreceptor outer segments have unusually high demands for transport. Approximately 10% of the outer segment disks are shed from the distal tip each day, with this material being replaced by new disks assembled at the proximal end, just beyond the connecting cilium. In mammals, this requires that ∼2000 opsin molecules be transported into the cilium per minute whereas, in fish and frogs, the larger diameter of the cells increases this number to ∼50,000 per minute (Young, 1967; Besharse and Horst, 1990). It is thought that IFT plays a role in this transport as defects in both kinesin motors and IFT complex proteins result in defects in the assembly of the outer segment of photoreceptor cells (Marszalek et al., 2000; Pazour et al., 2002; Tsujikawa and Malicki, 2004; Jimeno et al., 2006; Insinna et al., 2008; Krock and Perkins, 2008). The question remains, however, as to whether the IFT system is responsible for transporting the phototransduction-specific components of the outer segment that make up the disks beyond the connecting cilium. An alternate hypothesis is that IFT is only responsible for building and maintaining the connecting cilium and the effects on the membrane stacks are indirect effects of the connecting cilium defect.
With one known exception, the IFT proteins are predominantly found in a centrosomal pool around the base of the cilium and in the cilia themselves. IFT20, a subunit of the B complex, has an unusual distribution in that, in addition to being found in cilia and centrosome pools, it also is abundantly associated with the Golgi apparatus in vertebrate cells. In previous work, we showed that IFT20 is required for ciliary assembly and that it is highly dynamic in cells, moving from the Golgi apparatus into the cilium and along the length of the cilium. In cultured kidney cells, partial RNAi knockdowns that abolished the Golgi pool without greatly affecting the centrosomal pool blocked ciliary assembly, suggesting that the Golgi pool is critical for ciliary assembly. In addition, less complete knockdowns did not block cilia assembly but reduced the amount of the membrane protein polycystin-2 on cilia. This work suggested that IFT20 played a role in the sorting or trafficking of ciliary membrane proteins from the Golgi to the cilium (Follit et al., 2006).
Although the functions of other IFT proteins have been previously examined in the retina (Pazour et al., 2002; Baker et al., 2003; Krock and Perkins, 2008; Bhowmick et al., 2009), IFT20 has not been characterized. The unique Golgi localization of IFT20 suggests the possibility that IFT20 has functions in addition to transport within the cilium. Furthermore, recent work questioned the identity of IFT20 as a part of the IFT particle in photoreceptor cells. Using high-resolution immuno-electron microscopy (immuno-EM), it was shown that IFT20 was localized at the Golgi and at the base of the cilium but it could not be detected in the cilium. Based on this finding, the authors suggested that IFT20 is not part of the canonical IFT system (Sedmak and Wolfrum, 2010). We show here that IFT20 is fundamentally required for assembly and maintenance of the photoreceptor’s outer segment and is likely functioning both as part of the IFT particle and independent of the canonical IFT system.
RESULTS
In photoreceptor cells, IFT20 is localized to the Golgi complex as well as to the base of the outer segment and the distal end of the connecting cilium (Supplemental Figure S1). This localization places IFT20 in a unique position to couple post-Golgi transport with ciliary transport. Because photoreceptor cells have a large flux of membrane proteins through this pathway, we sought to understand the role of IFT20 in these cells. Germline mutations in Ift20 cause embryonic lethality at midgestation before the development of the retina. Thus, to examine the role of IFT20 in photoreceptor outer segment formation and maintenance, we generated a floxed allele of Ift20 (Jonassen et al., 2008) and used human red/green pigment gene promoter (HRGP)-Cre to delete IFT20 specifically in cone photoreceptors. In the HRGP-Cre mouse, Cre recombinase is expressed from the human red/green pigment promoter. This promoter is highly specific for M opsin-expressing cones and does not appear to cause any cone cell death. Expression initiates with the formation of cone outer segments in the postnatal day (p) 7–10 range (Le et al., 2004). Our Ift20flox allele contains loxP sites in introns flanking exons 2 and 3, which contain the start codon and approximately half of the open reading frame along with all of the ATG codons, and so is expected to be null. Mice homozygous for the floxed Ift20 allele (Ift20flox/flox) were mated to HRGP-Cre, Ift20null/+ animals, and offspring with the genotype HRGP-Cre, Ift20null/flox were used as experimental animals (ΔIFT20), whereas those with the genotype HRGP-Cre, Ift20+/flox were used as controls. The use of Cre-positive animals as controls allowed us to ensure that any phenotypic differences between the control and experimental groups were due to the loss of IFT20 and not the presence of Cre recombinase. It should also be noted that the original HRGP-Cre mice carried the rd (Pde6b) retinal degeneration mutation (Pittler and Baehr, 1991), but this mutation was bred out by backcrossing to C57B6J animals before analysis.
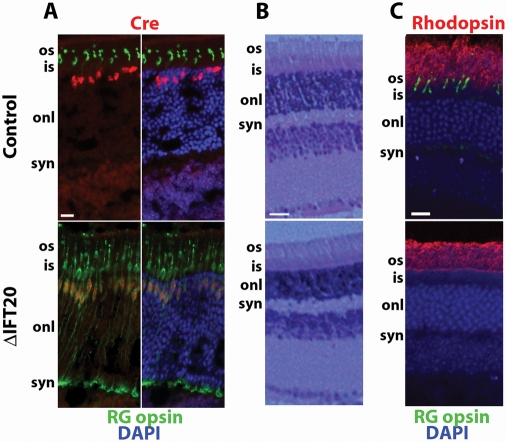
Initially we verified that HRGP-Cre was cone-specific in our genetic background. To do this, we stained p28 retinas with antibodies to Cre and to cone opsin. Cre localizes to the nuclei when the cone opsin labels outer segments. In control animals, the pattern of Cre expression was consistent with cone cells, although the spatial difference between the two markers rules out a definitive conclusion. Cone cell distinction is much clearer, however, in the experimental animals where nearly all Cre-positive cells displayed severe accumulation of cone opsin in the inner segments and cell bodies surrounding the Cre-positive nuclei in addition to the synaptic region (Figure 1A). No mutant animals showed any mislocalization of rod opsin, suggesting that HRGP-Cre is not expressed in rod cells (Figure 1C) and the deletion in cone cells does not adversely affect neighboring rod cells. Overall structure of the retina does not appear to be affected by the deletion of cones (Figure 1, B and C), likely because cone cells constitute only 3% of the photoreceptor cells (Carter-Dawson and LaVail, 1979). The observation that nearly all Cre-positive cell bodies also were positive for cone opsin indicates that HRGP-Cre is efficient at deleting Ift20.
FIGURE 1:
Deletion of IFT20 in mouse cone photoreceptors. (A) Retina cryosections from p28 control and mutant (ΔIFT20) littermates were stained with Cre (red), RG-opsin (green), and DAPI (blue). Scale bar is 20 μm and applies to all panels. (B) Hematoxylin and eosin–stained retina sections from p70 control and mutant littermates demonstrate that rod cell development and overall retina morphology are unaffected by deletion of IFT20 in cone cells. Scale bar is 25 μm and applies to both panels. (C) Specificity of retinal degeneration. Retina sections from p70 control and mutant littermates immunostained with RG-opsin, rhodopsin, and DAPI. Note the absence of cone cells remaining in the mutant whereas rhodopsin is normal. Scale bar is 20 μm and applies to both images. Abbreviations: os, outer segment; is, inner segment; onl, outer nuclear layer; syn, synapse; inl, inner nuclear layer.
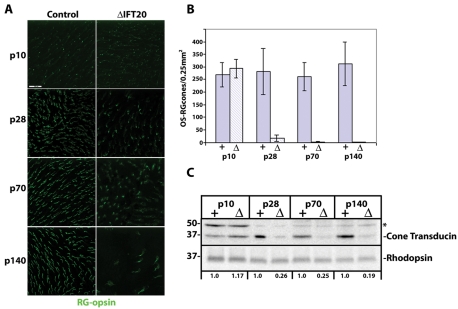
Like rods, the outer segments of cone photoreceptors are modified cilia that extend from the axoneme of the connecting cilium. Because IFT is required for cilia assembly in all other vertebrate cell types examined, we sought to understand the effect of the lack of IFT20 on cone cells. Initially, we used whole mount retinas immunostained for RG-opsin to examine the number of cone outer segments that remained in the mutant animals over time. The advantage of whole mounts is the ability to survey cones across the entire retina, thus eliminating potential artifacts that might result from sections cut from different places in the retina. The expression of HRGP-Cre begins in the p7 to p10 age range (Le et al., 2004) at about the same time that the outer segment is beginning to develop. Development continues until ∼p20, when the outer segments achieve adult proportions (LaVail, 1973; Fei, 2003). Thus, we surveyed cone cells at p10 to examine early development and then at p28, p70, and p140 to characterize degeneration. At p10, approximately equal numbers of cone outer segments were found in the mutant and control animals, and whole mount retinas stained with cone opsin appeared similar in both mutant and control animals (Figure 2, A and B). Even though outer segments were present in p10 animals, higher magnification images showed reduced amounts of opsin in the outer segments and significant mislocalization of opsin in the inner segment and at the synapse (Figure 3A). After p10, the number of cone outer segments in the mutants declined rapidly so that only ∼10% as many as controls were observed at p28, and the number dropped essentially to zero at later time points (Figure 2, A and B). The decline in cone cells over time also can be seen by monitoring the amount of cone transducin that remains in the mutant retinas as compared with the controls. At p10, similar amounts of cone transducin were seen in mutants and controls, but the relative amount declined dramatically in the mutants after that point, with only ∼26% of cone transducin remaining in mutants as compared with controls by p28. The loss of cone outer segments was more dramatic than the loss of cone transducin because some cone cell bodies remained even though their outer segments were gone.
FIGURE 2:
Deletion of IFT20 in cone cells leads to cone cell degeneration. (A) Retina whole mounts from control and mutant littermates of the indicated ages were immunostained with RG-opsin antibody to label red/green opsin cone cells. Images are maximum projections of 10 confocal z-images taken 0.4 μm apart. Scale bar is 50 μm and applies to all images. (B) Quantification of cone cell degeneration using red/green opsin-stained retina whole mounts. Whole retinas from four controls and four mutants were used for each age group. Cone cells with normal outer segments (OS-RG cones) were counted and averaged for three 250 μm2 fields derived from the dorsal, ventral, and middle portions of the retina. Bars depict the mean ± SD of four retinas for each group. (C) Western blot analysis of the cone-specific transducin compared with the rod-specific rhodopsin in protein extracts from control (+) and littermate mutant (Δ) retinas. Antibodies are listed on the right side of Western blots, and molecular weight markers in kilodaltons are listed on the left. Ages and genotypes are indicated above gel lanes. Densitometry for this representative gel is listed at the bottom and indicates the quantitated ratios of transducin to rhodopsin in mutants compared with their age-matched littermate controls. * indicates a nonspecific protein.
FIGURE 3:
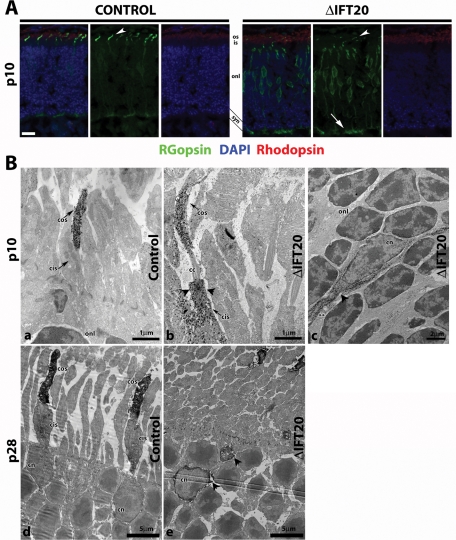
Cone opsin trafficking is impaired in IFT20 mutants. (A) Retina cryosections from p10 control and mutant littermates were immunostained with RG-opsin (green), rhodopsin (red), and DAPI (blue). In the control, RG-opsin is highly concentrated in the cone outer segment (arrowheads) but is severely mislocalized throughout the inner segment, cell body, and synapse (arrows) of mutants. Scale bar is 20 μm and applies to all images. (B) Immuno-EM of retinas stained with anti-RG-opsin. Panels a–c are p10 retinas (a = control; b and c = mutants). Panels d and e are p28 retinas (d = control; e = mutant). Arrowheads designate mislocalized RG-opsin. In c, ** indicates the inner/outer segment side of the cell. At p28, fully grown cone outer segments are seen in controls (d), whereas cone degeneration is seen in mutants (e). Arrowheads in e designate mislocalized RG-opsin throughout the cell body and in aggregated clumps. Abbreviations: cos, cone outer segment; cis, cone inner segment; onl, outer nuclear layer; cc, connecting cilium; cn, cone nuclei.
The most striking phenotype caused by the deletion of IFT20 is the accumulation of cone opsin in the inner segment and at the synapse. This misaccumulation can be observed as early as p10 (Figure 3) and continues until the cone cells are lost from the retina (Figures 1A and 2). An important point to consider is whether the accumulation of opsin in the inner segments is a direct effect of the failure of IFT to transport opsin into the outer segment or if it is an indirect consequence of not having outer segments formed. At p28 and beyond there are few outer segments remaining in the mutant animals, so it is likely that indirect effects play a large role in the mislocalization of opsin. At p10, however, there are approximately equal numbers of outer segments in the mutant and control animals (Figure 2, A and B), so it seems more likely that the mislocalization is a direct effect caused by the failure of IFT to transport opsin to the outer segment. It remains possible, however, that the mislocalization of opsin occurs only in cells lacking outer segments. To directly examine this possibility, we used an immuno-EM technique that allowed us to visualize the localization of opsin and see cell ultrastructural elements such as connecting cilia, inner and outer segments, and nuclei (Maerker et al., 2008). This technique uses a peroxidase detection system to precipitate diaminobenzidine (DAB) in the vicinity of the primary antibody followed by visualization with transmission EM. This has the advantages that the material can be initially lightly fixed to retain epitopes but can be further fixed and processed for traditional EM after staining. In control animals at p10 and p28, opsin is highly abundant in the outer segments and also is seen in the inner segments at a much lower level (Figure 3, Ba and Bd). In p10 mutants, opsin is seen in the outer segments like in controls. Unlike in the control animals, however, opsin also is abundant in the inner segments, with dense accumulations at the basal body of the connecting cilium (Figure 3Bb), suggesting a defect in active transport through the cilium. This mislocalized RG-opsin fills the cell body and continues back toward the synaptic layer (Figure 3Bc).
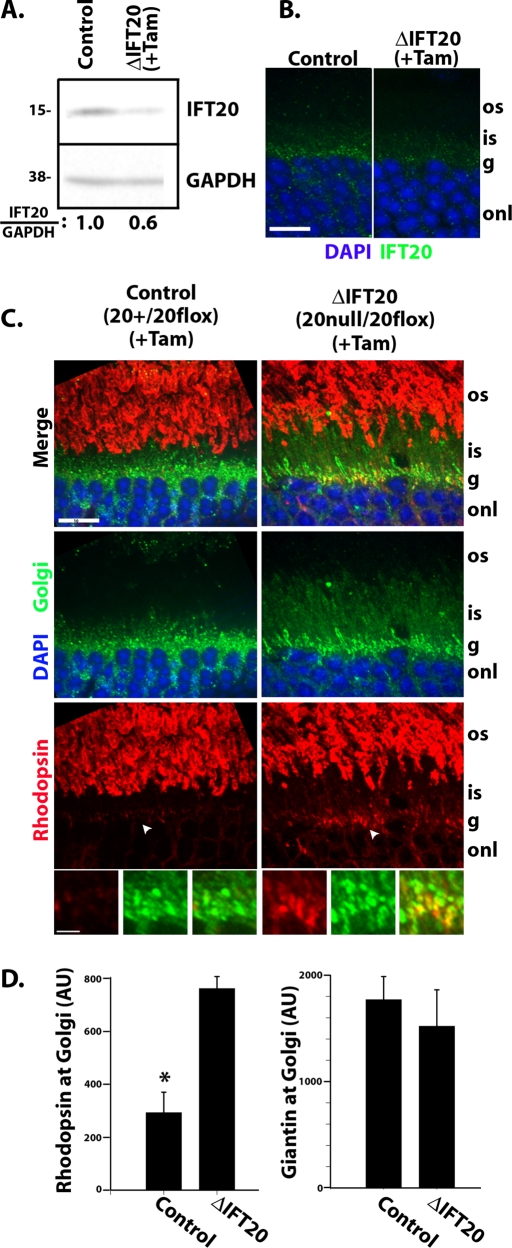
To confirm the results obtained with the RGopsin-Cre and to look at the role of IFT in fully formed photoreceptor cells, we used the tamoxifen-activatable Cag-CreER (Hayashi and McMahon, 2002) to delete Ift20 in adult animals. In this system, Cre is ubiquitously expressed from a hybrid actin/cytomegalovirus (CMV) promoter, but the enzyme is inactive until the animals are treated with tamoxifen. Unlike HRGP-Cre, which only deletes in the cone cells, Cag-CreER drives deletion in all cells of the retina, including rods and cones. Experiments in cell culture indicate that 48 h is the minimal time needed to observe loss of IFT20 protein after administration of tamoxifen (unpublished data). Thus to measure the early effects of the loss of IFT20, we treated 4-wk-old animals with tamoxifen and harvested retinas 48 h later. The treatment with tamoxifen reduced the level of IFT20 as monitored by Western blot (Figure 4A) and by immunofluorescence (Figure 4B). Similar to what was seen at p10 with the HRGP-Cre, rhodopsin was observed in the inner segment, cell body (Figure 4C), and at the synapse (unpublished data). Interestingly, rhodopsin accumulated at the Golgi complex (Figure 4, C and D). This accumulation is best seen in the bottom row of Figure 4C, where enlargements show significant colocalization between rhodopsin and the Golgi protein giantin. Quantification of Golgi-associated rhodopsin indicated that significantly more rhodopsin was found at the Golgi complex in the experimental retinas as compared with the controls, whereas the levels of giantin were not significantly different between the two conditions (Figure 4D). These results suggest that an early defect rising from the loss of IFT20 is an accumulation of this membrane protein in the Golgi complex.
FIGURE 4:
Opsin misaccumulates at the Golgi in fully formed photoreceptors after deletion of IFT20. (A) Western blot analysis of retina extracts from either control or IFT20 flox/null mice 48 h after treatment with tamoxifen. Antibodies are listed on the right side of Western blots, and molecular weight markers in kilodaltons are listed on the left. The quantitated ratios of IFT20 to GAPDH in experimental retina extracts compared with controls are indicated at bottom of gel. (B) Immunofluorescence of IFT20 on retina cryosections from either control or experimental mice demonstrates the loss of IFT20 after tamoxifen treatment in experimental sample. (C) Retina cryosections from control (left panels) or experimental mice (right panels) were immunostained with rhodopsin (red), DAPI (blue), and the Golgi marker giantin (green). The bottom three panels of each row are enlargements of regions around the arrows in the Golgi region. Scale bar in row 1 is 10 μm and applies to the three top rows. Scale bar in the bottom row is 2 μm and applies to that row. (D) Quantification of Golgi-associated rhodopsin and giantin from retinas of three control and three experimental animals treated with tamoxifen. Bars represent mean fluorescence intensity ± SD. * indicates significant difference (p = 0.0048). Abbreviations: g, Golgi layer; os, outer segment; is, inner segment; onl, outer nuclear layer.
Because opsin trafficking was defective in IFT20-deleted cells, we reasoned that IFT20 and the IFT complex might bind cone opsin. Opsin molecules are seven transmembrane receptors with their C-terminal tails projecting into the cytoplasm (Stenkamp et al., 2002). In rod opsin, the cytoplasmic tail is critical for sorting the protein to the outer segment, possibly through interactions with Arf4 (Deretic, 2006) and Tctex1 (Tai et al., 1999). Thus we reasoned that the cytoplasmic tails may be sites for interaction with IFT components. To test this reasoning, we used a coimmunoprecipitation (co-IP) assay to probe for interactions between IFT20 and the tails of either RG-cone opsin or rod opsin (Figure 5). To do this, we cloned the cytoplasmic tails of RG-cone opsin and rod opsin as C-terminal fusions with green fluorescent protein (GFP) and coexpressed these proteins with Flag-IFT20 in mammalian cells. After immunoprecipitating IFT20 with Flag antibodies, the presence of GFP-opsin in the precipitate was determined by Western blotting. This assay indicates that IFT20 interacts robustly with the cytoplasmic tails of both RG-opsin and rod opsin (Figure 5, lanes 3 and 7). This interaction appears to be specific as neither opsin tail precipitated without IFT20 (Figure 5, lanes 2 and 6) nor did they interact with glutathione S-transferase (GST), an unrelated control protein (Figure 5, lanes 1 and 5). Deletion of the last four residues, which contain a VxPx motif (Deretic, 2006), had no effect on the binding to IFT20 (Figure 4, lanes 4 and 8), suggesting that these residues are not critical determinants of IFT20 binding.
FIGURE 5:
IFT20 binds the cytoplasmic tails of RG-opsin and rhodopsin. FLAG-tagged IFT20 or controls were coexpressed in IMCD3 cells with the cytoplasmic tails of either RG-opsin or rhodopsin fused to GFP. The DNA constructs are listed vertically on top of each lane. Anti-FLAG immunoprecipitates were eluted with FLAG peptide and immunoblotted with the indicated antibodies. Molecular weights (kDa) are listed on the left side. The top panel shows the input of GFP fusion proteins. The middle panel shows the GFP fusion proteins that coimmunoprecipitated with IFT20. The bottom panel shows the immunoprecipitated FLAG baits (indicated by arrowheads). The asterisk indicates a nonspecific band recognized by the FLAG antibody.
Because IFT20 is a component of the multiprotein IFT particle, the possibility exists that opsin was pulled down in our co-IP assay by an assembled IFT particle containing Flag-IFT20. If this is occurring, other IFT B complex proteins besides IFT20 should be able to interact with opsin. We tested this idea with a GST pulldown assay (Figure 6). To do this, IFT20, 57, 52, and 54 were expressed in bacteria as fusions to GST, purified, and then incubated with mouse retinal extracts. After incubation, the proteins were reisolated using glutathione resin, and the ability of the fusions to interact with rod opsin was analyzed by Western blot of the bound material. We were unable to test for interactions with RG-cone opsin as the RG-cone opsin antibody was made against a GST-fusion protein and it bound to both the target and the baits. Confirming the coIP result, IFT20-GST was able to pull down endogenous rod opsin in this assay, and the interaction appears to be specific as beads bound with GST alone did not bring down any rod opsin. Interestingly, IFT57, 52, and 54 also were able to bring down rod opsin (Figure 6A). This finding suggests that opsin can interact with the IFT particle and at least part of the IFT20-opsin interaction that we detect is due to interactions with the assembled particle. An alternative explanation is that GST-IFT57, 52, and 54 may be binding to free IFT20 rather than to IFT particles. Previous work reported that IFT20 bound IFT57 in a yeast two-hybrid assay (Baker et al., 2003) and IFT54 in a co-IP assay (Follit et al., 2008; Omori et al., 2008). To test if direct binding could explain the results, we tested for the interaction of purified IFT20 with purified GST-IFT57, 52, and 54 along with GST alone as a negative control and the IFT20 binding domain of GMAP210 (Follit et al., 2008) as a positive control. As expected, IFT20 bound strongly to the IFT20 binding domain of GMAP210 and also bound strongly to IFT54. Detectable, but much less IFT20 bound to IFT57, and no IFT20 binding to IFT52 was detected. The binding appears specific as no IFT20 bound to GST or the resin alone. The observation that IFT52 brings down opsin but does not directly bind IFT20 suggests that at least part of the opsin binding is occurring with the intact particle. To test whether this explains all of the interaction between IFT20 and opsin, we tested whether the IFT20 binding domain of GMAP210 is capable of bringing down opsin. Since GMAP interacts with IFT20 outside of the IFT particle, the presence of opsin in a GMAP pulldown would suggest that IFT20 can interact with opsin independent of the particle. Retinal extracts were incubated with purified GST or GST-GMAP210IFT20BD, and the bound proteins were examined by Western blot (Figure 6C). The IFT20 binding domain of GMAP210 was able to precipitate opsin, but no opsin was brought down by the GST alone. As predicted, GST-GMAP210IFT20BD was able to bring down IFT20 but not IFT88 or IFT52. Importantly, however, the presence of IFT20 and the absence of IFT88 or IFT52 in the GMAP pulldowns strongly suggest that this opsin/IFT20 complex is distinct from the IFT particle. Taken together, these results indicate that IFT20 can interact with rod opsin both in the context of the IFT particle and while bound to GMAP210, supporting the hypothesis that IFT20 is important for trafficking proteins from the Golgi complex to the cilium.
FIGURE 6:
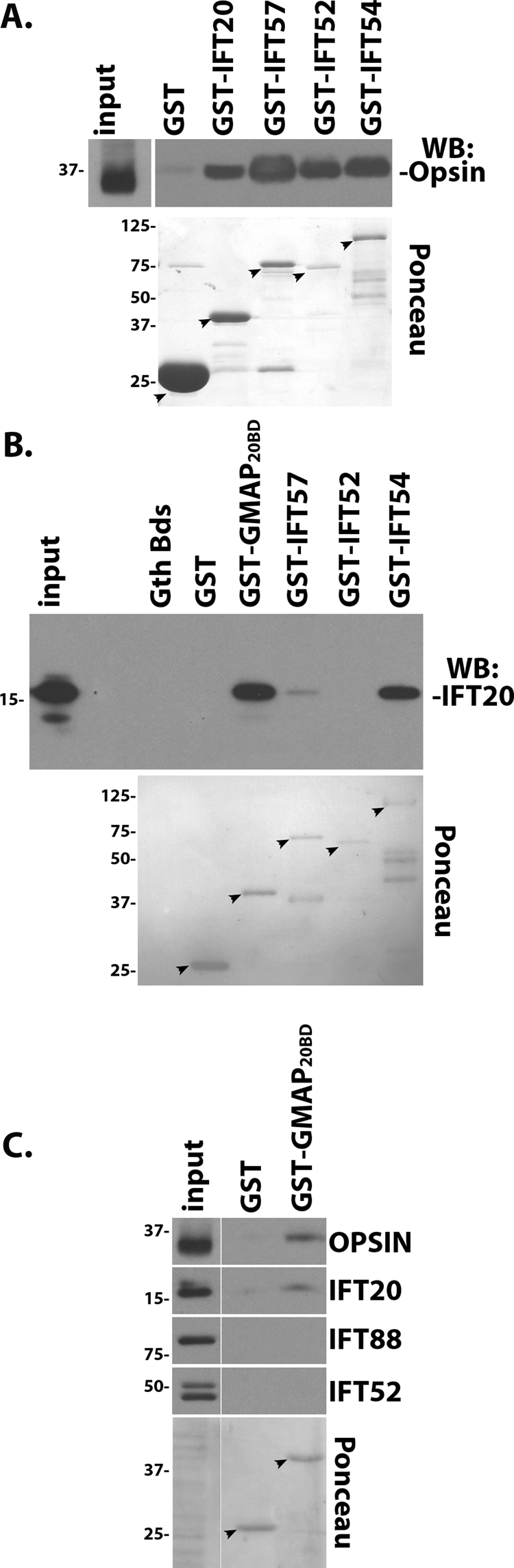
Opsin binds to the IFT particle and also binds to IFT20 outside of the IFT particle. (A) Endogenous opsin is present in a complex with the IFT particle. Mouse retina extracts were incubated with the indicated affinity-purified GST fusion protein. GST pulldowns were then probed for the presence of opsin by immunoblotting (Western blot) with a rhodopsin antibody. The GST baits (arrows) are shown in the bottom panel, which is a Ponceau-stained membrane. Input is equivalent to 6% of the capture. (B) Direct binding assay with purified proteins demonstrates a strong interaction between IFT20-GMAP210 and IFT20-IFT54 and a weak interaction between IFT20-IFT57. The GST tag was cleaved from GST-IFT20, and purified, untagged IFT20 was incubated with affinity-purified GST fusion proteins. GST pulldowns were then probed for the presence of IFT20 by immunoblotting with an IFT20 antibody. The GST baits are shown in the bottom panel of a Ponceau-stained membrane. Input is equivalent to 20% of the capture. (C) Endogenous opsin also binds IFT20 outside of the IFT particle. Mouse retina extracts were incubated with GST or the IFT20 binding domain from GMAP210 fused to GST (GST-GMAP20BD). GST pulldowns were then probed with the indicated antibodies. The GST baits are shown in the bottom panel of a Ponceau-stained membrane. Input is equivalent to 6% of the capture.
DISCUSSION
In this work, we examined the role of IFT20 in the development and maintenance of the outer segments of rod and cone photoreceptor cells. Although rod and cone outer segments share features such as being derived from primary cilia, they have a significantly different membrane topology that has raised questions about their assembly mechanisms (Chuang et al., 2007). In rod outer segments, the light-sensitive membranes are detached from the plasma membrane and form disks that are enclosed within the outer ciliary membrane. In contrast, the light-sensitive membranes of cones do not form detached disks but remain as folds of the ciliary membrane (Eckmiller, 1987). Differences also exist in transport of rod and cone opsins as cone opsins require 11-cis retinal for movement to the outer segment but rod opsin does not (Rohrer et al., 2005). Cre-driven deletions of the IFT motor protein Kif3a in the retina suggested differences between rods and cones in the requirement for heterotrimeric kinesin in photopigment transport to the outer segment (Avasthi et al., 2009). These results, however, are in conflict with other work that found a strong requirement for Kif3a in rod cells (Marszalek et al., 2000; Jimeno et al., 2006; Lopes et al., 2010), and are likely the result of inconsistent recombination by the rod-specific Cre recombinase.
To study the role of IFT20 in cone cells, we used Cre recombinase driven by the human red-green opsin promoter to specifically delete a floxed allele of Ift20 in mouse M-cones. Cre expression from this promoter begins at approximately the time outer segments are being developed from primary cilia (Le et al., 2004). At early time points, outer segments were present in the mutant animals in numbers similar to those seen in controls; however, unlike controls, which have little opsin outside of the outer segment, significant amounts of opsin were seen in the inner segments, perinuclear regions, and synapses of the experimental animals. This accumulation was seen in young animals that still had intact connecting cilia and outer segments. Similarly, if we remove IFT20 from rod cells that are fully developed, we see a rapid accumulation of rhodopsin in the cell body where it is concentrated at the Golgi complex. If opsin and rhodopsin were transported by an IFT-independent mechanism, one would not expect to see this accumulation in cells with intact connecting cilia and outer segments. This finding suggests that the transport of opsin relies on the IFT system.
In vertebrate cells, we find that IFT20 is localized to the Golgi complex as well as the cilium and we proposed that IFT20 was in a unique position to couple the sorting or transport of membrane proteins through the endomembrane system to the IFT system for transport into the cilium. Recent work questioned whether IFT20 was a part of the canonical IFT system within the cilium of photoreceptors and other cells. Wolfrum and colleagues, using immuno-EM and our antibody, were able to confirm that IFT20 was localized at the Golgi complex and at the base of the cilium but not within the cilium itself. From these data, they suggested that IFT20 was only involved in trafficking from the Golgi to the basal body but not within the cilium (Sedmak and Wolfrum, 2010). As discussed previously (Follit et al., 2006), our antibody is epitope blocked at the centrosome and within the cilium in aldehyde-fixed material, likely due to this small protein being obscured within the larger IFT B complex. The ciliary pool can be detected, however, in photoreceptors (Supplemental Figure S1) (Pazour et al., 2002; Luby-Phelps et al., 2008) and in other cells with proper staining conditions. Additional evidence beyond immunolocalization supports a role for IFT20 in IFT within the cilium. This includes biochemistry done on Chlamydomonas cilia that firmly established IFT20 as a subunit of IFT complex B (Cole et al., 1998) and also includes the observation that GFP-tagged IFT20 moves along cilia at IFT rates in mouse (Follit et al., 2006) and Chlamydomonas (Lechtreck et al., 2009). In addition, numerous studies have shown biochemical association between IFT20 and the IFT B complex, which would not occur if the Wolfrum model were correct (Baker et al., 2003; Follit et al., 2006, 2009; Omori et al., 2008). Thus, with the exception of one study, all evidence supports a role for IFT20 contributing to complex B function in transport within the cilium.
In cone cells, the deletion of IFT20 caused M opsin to accumulate in the inner segment and other parts of the cell body, but we did not observe specific accumulation at the Golgi complex. In contrast, when we look quickly after the deletion of Ift20 by the administration of tamoxifen, we see rhodopsin accumulation at the Golgi complex. The kinetics of IFT20 protein loss in this model is dependent on how long it takes for the gene to be deleted and the existing mRNA and protein to decay. We observed significant loss, however, of IFT20 48 h after tamoxifen treatment (Figure 4, A and B). In a cell culture model of fibroblasts derived from these animals, 48 h is the earliest time after tamoxifen treatment that we observed IFT20 loss (Keady, Robinson, and Pazour, unpublished data). This finding suggests that we are looking at cells early after the loss of IFT20. The differences between the cellular distributions of opsin/rhodopsin in the two systems may be that the tamoxifen treatment allows us to look earlier after disrupting the gene than we can analyze in the HRGP-Cre deletions, so the tamoxifen deletion is more likely to reveal primary defects. Alternatively, the results may reflect fundamental differences in the two proteins. Even though M opsin and rhodopsin are homologous, they have differences, including a requirement for 11-cis retinal for outer segment transport of opsin but not rhodopsin (Rohrer et al., 2005).
To further examine the idea that opsin is a direct cargo of IFT, we tested the ability of IFT20 to interact with RG-opsin and rhodopsin in co-IP and GST pulldown assays. By co-IP, IFT20 was able to interact with the cytoplasmic tails of these photopigments. In addition, we found that IFT20 and other IFT complex B proteins were able to pull down rhodopsin in extracts made from mouse photoreceptors. These data indicate that IFT complex B (containing IFT20) can interact with the photopigments; however, the data do not indicate whether IFT20 can interact independently of IFT B. To address this possibility, we tested the ability of the IFT20 binding domain of GMAP210 to pull down rhodopsin from a retinal extract. GMAP210 is a Golgi-associated protein that is responsible for anchoring IFT20 to the Golgi complex (Follit et al., 2008). GMAP210 binds IFT20 independently of IFT complex B, and, as predicted, the GMAP210 pulldown retrieved IFT20 from the retinal extract, but did not recover other IFT B proteins. The observation that rhodopsin is brought down by GMAP210 indicates that IFT20 can interact with rhodopsin independently of IFT B and supports a model in which IFT20 links traffic from the Golgi complex to the canonical IFT system at the cilium.
The photoreceptor outer segment is an extreme example of modifications that can be made to cilia to adapt them to their specialized sensory functions. Because of the extensive elaboration of the membrane compartment, the assembly and maintenance of outer segments require levels of transport not needed in most other cilia. The work presented here and in the literature makes it clear that ciliary assembly mechanisms first identified in single-cell eukaryotes function in the formation of the vertebrate outer segment of both rods and cones. Questions remain about specializations that may have evolved to support the unusually high level of membrane and membrane protein transport that is required for the assembly and maintenance of the outer segment. Our work and the work of Bhowmick et al. (Bhowmick et al., 2009), however, support the idea that IFT proteins can bind to opsin family proteins and direct them through the cytoplasmic compartment and into the outer segment.
MATERIALS AND METHODS
Mouse breeding
The Ift20flox/flox (MGI:Ift20tm1.1Gjp) and Ift20null/+ (MGI:Ift20tm1.2Gjp) mice were generated and genotyped as previously described (Jonassen et al., 2008). The Ift20flox/flox mice have been deposited at The Jackson Laboratory (Bar Harbor, ME) under the stock number 012565. The generation of the HRGP-Cre line was described previously (Le et al., 2004). The rd (Pde6b) mutation was bred out of the HRGP-Cre line by crosses to C57B6J (The Jackson Laboratory) and genotyped as described (Pittler and Baehr, 1991). CAG-CreER line [B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J] (Hayashi and McMahon, 2002) was obtained from The Jackson Laboratory. Tamoxifen (Sigma, St. Louis, MO) was dissolved in peanut oil; 6 mg/20 g mouse was delivered by oral gavage, and the eyes were harvested 48 h later. All mouse work was carried out at the University of Massachusetts Medical School with approval of the Institutional Animal Care and Use Committee.
Histology
Before enucleation, mouse eyes were marked on the dorsal axis to register orientation and then pierced through the cornea to allow penetration of fixative. Eyes were fixed in cold phosphate-buffered saline (PBS)/4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 20 min, followed by overnight incubation at 4°C in methanol. The cornea and lens were removed the next day, and the retina was returned to cold methanol for an additional 1 h. Eye cups were then incubated at 4°C for 1 h in 20% sucrose dissolved in 50% methanol/50% PBS, followed by an overnight incubation in PBS/20% sucrose, and then embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC). Nine-micron cryosections were cut from the central retina near the optic nerve, mounted on Plus slides (Fisher Scientific, Pittsburgh, PA), blocked for 2 h at room temperature in Tris-buffered saline (TBS; 10 mM Tris, pH 7.5, 166 mM NaCl) supplemented with 5% goat serum and 0.1% Triton, and then incubated overnight at 4°C in primary antibody diluted in blocking buffer. After washing in blocking buffer, Alexa Fluor–labeled secondary antibodies (Invitrogen, Carlsbad, CA) were used (goat anti–mouse 594 and goat anti–rabbit 488) to detect the primary antibodies, and specimens were mounted using Prolong Gold with DAPI (Invitrogen). For cryosections, a minimum of five controls and five mutants were examined for each age group. The following primary antibodies were used: cone RG-opsin (AB5405 at 1:1000; Millipore, Billerica, MA), cone transducin (sc-390 at 1:500; Santa Cruz Biotechnology, Santa Cruz, CA), Cre (clone 7–23 at 1:200; Sigma, Natick, MA), IFT20 (1:500) (Follit et al., 2006), rhodopsin (1D4 at 1:3000; Millipore), and Giantin (1:500; a gift from Marvin Fritzler, University of Calgary). Mouse monoclonal rhodopsin (clone B630N) antibody was a gift from Joe Besharse (Medical College of Wisconsin). Immunostaining in Supplemental Figure S1 was done similarly except with the addition of an antigen retrieval step of 0.05% SDS included during primary antibody incubation. Dissociated photoreceptors in Supplemental Figure S1c were prepared as previously described (Liu et al., 2004). Confocal images were acquired as previously described (Jonassen et al., 2008).
Rhodopsin accumulation at the Golgi was quantitated using the measurement tools of OpenLab. Frozen retinal sections were stained with rhodopsin and giantin and imaged. Golgi regions from at least 100 cells within each of three control and three experimental retinas were outlined using giantin as the Golgi marker. The mean fluorescence intensities of rhodopsin and giantin within the outlined Golgi regions were then measured.
Mammalian cell culture and IPs
Culturing of IMCD3 cells (American Type Culture Collection) and IPs were done as previously described (Follit et al., 2008). Briefly, IMCD cells were electroporated with 15 μg of DNA and harvested for IPs 24 h later. Cells were lysed in Cell Lytic (Sigma) supplemented with 0.1% Tween plus Complete Protease Inhibitor (Roche, Basel, Switzerland) and centrifuged at 18,000 × g for 10 min. IPs with FLAG M2 antibody coupled to agarose beads (Sigma) were incubated at 4°C for 1.5 h. Beads were washed four times with wash buffer (50 mM Tris, pH 7.4, 300 mM NaCl, and 0.3% Tween) and eluted with 3× FLAG peptide (Sigma). Western blots were performed as previously described (Follit et al., 2008) with FLAG (F1804; Sigma) and GFP (JL8; Clontech, Mountain View, CA) antibodies.
GST pulldown assay
GST fusion proteins were expressed in BL21 cells (Promega, Madison, WI) by overnight induction at 19ºC with 50 μm IPTG. Harvested cells were resuspended in 50 mM Tris, pH 8, 150 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, lysed by either French Press or sonication, and supplemented with Triton X-100 to a final concentration of 0.5%. Lysates were then centrifuged at 12,000 × g for 15 min, and soluble GST fusion protein was affinity purified using glutathione sepharose 4B (Amersham, Pittsburgh, PA).
For direct binding assays, the GST tag was first cleaved from GST-IFT20 using PreScission Protease (GE Life Sciences, Piscataway, NJ), according to the manufacturer’s protocol. Approximately 10 μg of purified, untagged IFT20 was incubated with an equivalent amount of each purified GST protein for 45 min at 4ºC in binding buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.3% Triton). GST fusion proteins were then affinity purified with equivalent amounts of glutathione sepharose 4B slurry, washed four times in binding buffer, eluted with 20 mM glutathione in binding buffer, and finally placed into SDS–PAGE protein sample loading buffer.
For retina pulldown assays, retinas from adult normal C57B6J mice were homogenized in IP buffer (10 mM Tris, pH 7.5, 250 mM NaCl, 0.5% NP-40, and 0.5 mM EDTA) using a mini-Dounce and then passed through an 18-gauge needle. Homogenates were centrifuged at 18,000 × g for 15 min, and the supernatant from approximately three retinas was incubated with 15 μg each GST protein. Binding reactions were performed at 4ºC for 90 min. GST complexes were affinity purified, washed four times in IP buffer, and resuspended in SDS–PAGE protein sample loading buffer.
Retina extracts
Mouse retina samples for Western blot analysis were prepared by placing eyecups with the lens removed into SDS protein loading buffer and then homogenizing with a mini-Dounce followed by repeated passage through a 22-gauge needle. Images of chemiluminescent Western blots were acquired using the CCD camera of an LAS-3000 imaging system (Fujifilm, Tokyo, Japan) to ensure bands were not oversaturated. Band intensities were quantitated using ImageJ software, and the levels of cone transducin were normalized to the levels of rhodopsin. At least four retinas of each genotype (control and mutant) were analyzed for each age group. The ratios of cone transducin to rhodopsin for the controls in each age group were set to 1 and compared with the same ratios in mutant littermates.
Retina whole mounts
Mouse eyes were fixed and embedded in Tissue Freezing Medium as described earlier in the text and stored at −80ºC. After thawing at room temperature, the fixed eyecups were washed three to four times in PBS. The fixed retina was then marked for orientation, dissected out from the eyecup, and placed in blocking buffer (TBS with 10% goat serum, 5% bovine serum albumin [BSA], and 0.3% Triton) and stained with RG-opsin antibodies as described earlier in the text. To facilitate flat mounting of the retinas onto slides, small cuts were made on the periphery of the retinal specimens. Outer segments were quantitated by determining the average number of RG-opsin–positive outer segments in three separate fields of 250 μm2 for each retina. Four control and four mutant retinas were analyzed for each age group.
Immuno-EM
Methods were similar to those of Maerker et al. (2008) with the following modifications: Mouse eyes were pierced through the cornea to allow penetration of fixative and fixed for 20 min at room temperature with 4% paraformaldehyde in Soerensen phosphate buffer (PB; 0.1 M disodium hydrogen phosphate and 0.1 M potassium dihydrogen phosphate, pH 7.3). The lens and vitreous body were then removed, and the eyecup was placed back in fixative for an additional 60 min. After four washes in PB, the retina was dissected out, incubated sequentially in 10% sucrose/PB for 2 h, 20% sucrose/PB for 1 h, and 30% sucrose/PB overnight. Retinas were then permeabilized by four freeze/thaw cycles and immunostained with RG-opsin primary antibody (1:300 in PBS with 3% goat serum and 1% BSA) followed by a biotinylated secondary antibody (1:200; Invitrogen). The biotin signal intensity was amplified with an ABC Peroxidase staining kit (Pierce, Rockford, IL), and samples were then incubated with diaminobenzidine. After two washes in Tris buffer (50 mM Tris, pH 7.6) followed by two washes in cacodylate buffer (0.1 M, pH 7.4), samples were then postfixed in 2.5% glutaraldehyde/cacodylate buffer for 2 h at 4°C. The samples were then processed for silver enhancement and postfixed in 0.5% OsO4/cacodylate buffer for 40 min. Subsequently the samples were washed, dehydrated, and embedded for transmission EM. Retinas were flat embedded like a sheet, and sections were cut perpendicular to the plane of the sheet to obtain photoreceptors in a longitudinal orientation.
DNA constructs
Flag-IFT20 (JAF134) and Flag-GST (BK35) were generated by PCR amplifying the open reading frames from mouse cDNA and pGex6p1, respectively, and inserting them into JAF113, a slightly modified p3xFLAG-myc-CMV-26 (Sigma) vector (Follit et al., 2008). Similarly, the cytoplasmic C-terminal tails of RG-opsin (residues 320–359) and rhodopsin (residues 308–348) were amplified from mouse cDNA and inserted into the EcoR1 site of pEGFP-C2 (Clontech) to make BK9 and BK32, respectively. GFP-RG-opsin ΔVXPX (BK15) was made by PCR amplifying residues 320–355, and GFP-Rhodopsin ΔVXPX (BK36) was made by PCR amplifying residues 308–344. GST-IFT20 (GP653), GST-IFT57 (GP652), GST-IFT54 (BK34), and GST-IFT52 (GP654) were generated by PCR amplifying the open reading frames and inserting them into either the EcoR1 site (IFT20, IFT57, IFT54) or the BamH1 site (IFT52) of pGex-6p1. GST-GMAP20BD (BK37) was generated by PCR amplifying residues 1157–1319 of mouse GMAP210 from JAF192 (Follit et al., 2008) and inserting it into pGex-6p1.
Accession numbers were as follows: IFT20, NM_018854; RG-opsin, BC014826; rhodopsin, NM_145383; IFT57, NM_028680; IFT54, NM_028718; IFT52, NM_172150; and GMAP210, NM_028446. All genes were obtained from mouse.
Supplementary Material
Acknowledgments
We thank Greg Hendricks and Tracy Levin of the Electron Microscopy Core Facility at the University of Massachusetts Medical School for assistance, Joseph Besharse (Medical College of Wisconsin) and Marvin Fritzler (University of Calgary) for antibodies, and Uwe Wolfrum (Johannes Gutenberg University of Mainz) for advice on immuno-EM. This work was supported by the National Institutes of Health (GM060992). Core resources supported by the Diabetes Endocrinology Research Center grant DK32520 were also used.
Abbreviations used:
- EM
electron microscopy
- GFP
green fluorescent protein
- GST
glutathione S-tranferase
- HRGP
human red/green pigment gene promoter
- IFT
intraflagellar transport
- IP
immunoprecipitation
- MGI
Mouse Genome Informatics
- p
postnatal day
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0792) on February 9, 2011.
REFERENCES
- Avasthi P, Watt CB, Williams DS, Le YZ, Li S, Chen CK, Marc RE, Frederick JM, Baehr W. Trafficking of membrane proteins to cone but not rod outer segments is dependent on heterotrimeric kinesin-II. J Neurosci. 2009;29:14287–14298. doi: 10.1523/JNEUROSCI.3976-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem. 2003;278:34211–34218. doi: 10.1074/jbc.M300156200. [DOI] [PubMed] [Google Scholar]
- Besharse JC, Horst CJ. The photoreceptor connecting cilium: a model for the transition zone. In: Ciliary, Flagellar Membranes., editors. RA Bloodgood, New York: Plenum Press; 1990. pp. 389–417. [Google Scholar]
- Bhowmick R, Li M, Sun J, Baker SA, Insinna C, Besharse JC. Photoreceptor IFT complexes containing chaperones, guanylyl cyclase 1 and rhodopsin. Traffic. 2009;10:648–663. doi: 10.1111/j.1600-0854.2009.00896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I. Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- Chuang JZ, Zhao Y, Sung CH. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell. 2007;130:535–547. doi: 10.1016/j.cell.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D. A role for rhodopsin in a signal transduction cascade that regulates membrane trafficking and photoreceptor polarity. Vision Res. 2006;46:4427–4433. doi: 10.1016/j.visres.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Eckmiller MS. Cone outer segment morphogenesis: taper change and distal invaginations. J Cell Biol. 1987;105:2267–2277. doi: 10.1083/jcb.105.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Y. Development of the cone photoreceptor mosaic in the mouse retina revealed by fluorescent cones in transgenic mice. Mol Vis. 2003;9:31–42. [PubMed] [Google Scholar]
- Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genet. 2008;4:e1000315. doi: 10.1371/journal.pgen.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Xu F, Keady BT, Pazour GJ. Characterization of mouse IFT complex B. Cell Motil Cytoskeleton. 2009;66:457–468. doi: 10.1002/cm.20346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Insinna C, Besharse JC. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno D, Feiner L, Lillo C, Teofilo K, Goldstein LS, Pierce EA, Williams DS. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest Ophthalmol Vis Sci. 2006;47:5039–5046. doi: 10.1167/iovs.06-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. doi: 10.1083/jcb.200808137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krock BL, Perkins BD. The intraflagellar transport protein IFT57 is required for cilia maintenance and regulates IFT-particle-kinesin-II dissociation in vertebrate photoreceptors. J Cell Sci. 2008;121:1907–1915. doi: 10.1242/jcs.029397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM. Kinetics of rod outer segment renewal in the developing mouse retina. J Cell Biol. 1973;58:650–661. doi: 10.1083/jcb.58.3.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le YZ, Ash JD, Al-Ubaidi MR, Chen Y, Ma JX, Anderson RE. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004;10:1011–1018. [PubMed] [Google Scholar]
- Lechtreck KF, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zuo J, Pierce EA. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J Neurosci. 2004;24:6427–6436. doi: 10.1523/JNEUROSCI.1335-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes VS, Jimeno D, Khanobdee K, Song X, Chen B, Nusinowitz S, Williams DS. Dysfunction of heterotrimeric kinesin-2 in rod photoreceptor cells and the role of opsin mislocalization in rapid cell death. Mol Biol Cell. 2010;21:4076–4088. doi: 10.1091/mbc.E10-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K, Fogerty J, Baker SA, Pazour GJ, Besharse JC. Spatial distribution of intraflagellar transport proteins in vertebrate photoreceptors. Vision Res. 2008;48:413–423. doi: 10.1016/j.visres.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerker T, et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Liu X, Roberts EA, Chui D, Marth JD, Williams DS, Goldstein LS. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr Top Dev Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase beta-subunit gene of the rd mouse. Proc Natl Acad Sci USA. 1991;88:8322–8326. doi: 10.1073/pnas.88.19.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan RJ, Tobin JL, Beales PL. Modeling ciliopathies primary cilia in development and disease. Curr Top Dev Biol. 2008;84:249–310. doi: 10.1016/S0070-2153(08)00605-4. [DOI] [PubMed] [Google Scholar]
- Remillard SP, Witman GB. Synthesis, transport, and utilization of specific flagellar proteins during flagellar regeneration in Chlamydomonas. J Cell Biol. 1982;93:615–631. doi: 10.1083/jcb.93.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer B, Lohr HR, Humphries P, Redmond TM, Seeliger MW, Crouch RK. Cone opsin mislocalization in Rpe65-/- mice: a defect that can be corrected by 11-cis retinal. Invest Ophthalmol Vis Sci. 2005;46:3876–3882. doi: 10.1167/iovs.05-0533. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak T, Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J Cell Biol. 2010;189:171–186. doi: 10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp RE, Filipek S, Driessen CA, Teller DC, Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim Biophys Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin’s carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tsujikawa M, Malicki J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron. 2004;42:703–716. doi: 10.1016/s0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- Young RW. The renewal of photoreceptor cell outer segments. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.