The retinoblastoma tumor suppressor (RB) is functionally inactivated at high frequency in nearly all tumor types. Herein the acute deletion of RB in the liver reveals an immediate and profound dysregulation of spatiotemporal coordination in cell-cycle phases, resulting in robust DNA damage and aneuploidy that is not found in other tissues.
Abstract
The integrity of the retinoblastoma tumor suppressor (RB) pathway is critical for restraining inappropriate proliferation and suppressing tumor development in a plethora of tissues. Here adenovirus-mediated RB deletion in the liver of adult mice led to DNA replication in the absence of productive mitotic condensation. The replication induced by RB loss was E2F-mediated and associated with the induction of DNA damage and a nontranscriptional G2/M checkpoint that targeted the accumulation of Cyclin B1. In the context of RB deletion or E2F activation, there was an increase in hepatocyte ploidy that was accompanied by hyperphysiological assembly of prereplication complexes. In keeping with this dysregulation, initiation of DNA replication was readily observed in hepatocytes that were phenotypically in G2/M. Under such conditions, uncoupling of replication initiation from mitotic progression led to altered genome ploidy in the liver. Interestingly, these findings in hepatocytes were not recapitulated in the basally proliferative tissues of the gastrointestinal tract, where RB deletion, while increasing DNA replication, did not lead to a profound uncoupling from mitosis. Combined, these findings demonstrate the critical role of RB in controlling cell-cycle transitions and underscore the importance of intrinsic tissue environments in resultant phenotypes.
INTRODUCTION
The retinoblastoma tumor suppressor (RB) plays a vital role in a number of tumor suppressive processes, and is particularly critical for coordinating entry into the cell cycle. The loss of RB function is a frequently observed event in many cancer types (Burkhart and Sage, 2008), and disruption of the RB pathway in some manner has been suggested to be a requisite event for tumor development (Sellers and Kaelin, 1997; Hahn and Weinberg, 2002; Sherr and McCormick, 2002). Despite these findings, the impact of RB inactivation is highly tissue-specific, and can be clearly masked by compensatory processes that limit the observed phenotype of RB loss (Sage et al., 2003). As a result, in many tissues, it is difficult to decipher the underlying impact of RB loss that would relate to tumor development.
A primary mechanism by which RB exerts proliferative control is by binding to, and inactivating, the E2F family of transcription factors (Weinberg, 1995; Harbor and Dean, 2000; Blais and Dynlacht, 2004). The subsequent recruitment of repressive complexes by RB converts E2F target gene promoters into sites of active transcriptional repression, thus inhibiting cell-cycle entry. By contrast, active E2F transcription factors stimulate the expression of a gene expression program that includes multiple genes that play critical roles in the control of DNA replication and mitosis (Ishida et al., 2001; Markey et al., 2002; Ren et al., 2002). Importantly, this gene expression program both encompasses key drivers (e.g., Cyclin A, Cdc6, Cyclin B1) and inhibitors (e.g., Geminin, Mad2) of replication and mitotic progression. Thus, in the absence of directed functional models, it is impossible to predict the explicit response to RB inactivation and subsequent deregulation of E2F activity with reference to DNA replication and mitotic progression.
The coordination between DNA replication and mitosis is critical for the maintenance of genome integrity and is maintained via complex feedback mechanisms that control the timing and execution of each cell-cycle phase. As cells progress into G1, the assembly of prereplication complexes (preRCs) enables the initiation of DNA replication at specific origins of replication (Bell and Dutta, 2002; Diffley and Labib, 2002). In contrast, S phase inhibits preRC assembly to limit DNA replication to a single round per cell cycle (Blow and Hodgson, 2002; Dimitrova et al., 2002). Thus, in G2, replication reinitiation is inhibited due to the absence of preRCs. These control mechanisms serve as the primary means to restrict replication licensing to the G1 phase of the cell cycle (Blow and Hodgson, 2002). Conversely, the accumulation of key activities for the transition into mitosis is controlled by checkpoint mechanisms that are mitigated only with the completion of S phase and the absence of DNA damage (Sullivan and Morgan, 2007). This combinatorial control ensures that the genome is completely and accurately replicated only once per cell cycle and that mitotic progression proceeds only in the presence of high-fidelity duplication of chromosomes.
Here an acute model of RB deletion in adult tissue was used to interrogate the impact of loss of RB function on cell-cycle control in vivo. It is demonstrated that RB plays an unique role in controlling cell-cycle entry, and serves as an important barrier in regulating the initiation of DNA synthesis with DNA damage response and mitotic progression. These findings underscore RB as a central determinant of S-phase control and replication licensing in vivo.
RESULTS
RB-deleted liver tissue exhibits a specific deficit in mitotic entry
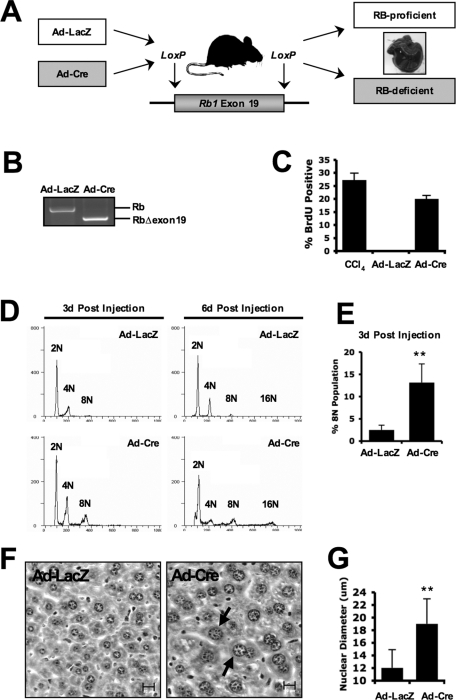
It is well established that, in genetic models of RB loss, compensation can occlude the impact of deleting this key tumor suppressor. To precisely determine the influence of RB status on hepatocyte biology in vivo, an acute model of RB deletion was used. Specifically, intravenous delivery of adenoviruses encoding either β-galactosidase (LacZ) or Cre-recombinase (Cre) was used in mice of Rbf/f genotype (Figure 1A). The adenoviruses efficiently and specifically transduce hepatic cells (Wood et al., 1999) yielding matched adult RB-proficient and -deficient liver tissue (Figure 1A). The adenoviral introduction of Cre-recombinase produced an efficient deletion of exon 19 of the Rb1 gene in liver tissue, as demonstrated by genomic PCR (Figure 1B); this resulted in a complete attenuation of RB protein, as has been previously reported (Mayhew et al., 2005), and a highly significant increase in DNA synthesis at 3 d postinjection, as indicated by the incorporation of BrdU (Figure 1C). Populations in these RB-deficient livers displayed significantly higher levels of 4N and 8N DNA content than did control hepatocytes (Figure 1, D, left, and E). The accumulation of aberrant ploidy continued at 6 d postinjection, where RB-deficient livers harbored a population of 16N hepatocytes, in distinct contrast to control liver cells (Figure 1D, right). In conjunction with the increased DNA ploidy, RB-deficient livers also displayed enlarged nuclear morphology and significantly increased average nuclear size (Figure 1, F and G). Together, these data suggest a critical role for RB in maintaining cell-cycle suppression and ploidy control in adult liver tissue.
FIGURE 1:
Liver-specific RB deletion results in aberrant ploidy and abnormal nuclear morphology. (A) Model of RB deletion in the murine liver wherein Cre-mediated recombination of LoxP sites flanking exon 19 of Rb1 causes loss of RB protein. (B) Liver-specific genomic PCR amplifying the region surrounding exon 19 of Rb1 in mice treated with Ad-LacZ and Ad-Cre for 72 h. (C) Percentage of BrdU-positive hepatocyte nuclei in each treatment condition was counted and graphed. (D) Hepatocyte nuclei from livers harvested either 3 or 6 d postadenoviral delivery were isolated and subjected to propidium iodide flow cytometry to determine cellular ploidy. (E) The 8N population of hepatocytes was counted via flow cytometry and graphed from livers harvested 3 d postadenoviral delivery. (F) Paraffin-embedded liver sections were stained with hematoxylin and eosin (H&E) and visualized via light microscopy. Scale bars = 20 μm. (G) Average nuclear diameter was measured from H&E-stained sections and graphed.
Deregulated E2F activity leads to a DNA damage–induced G2/M checkpoint targeting Cyclin B1 protein levels
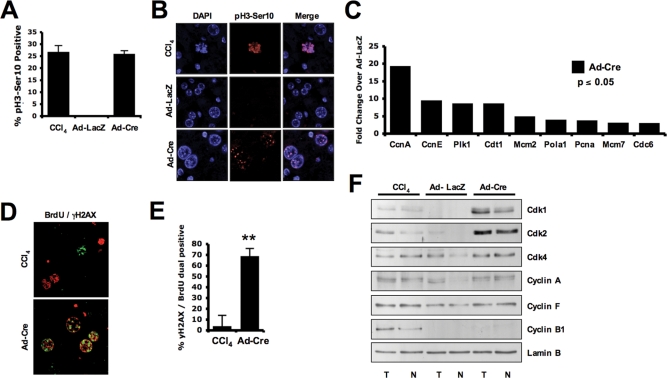
Whereas RB loss has been shown to be associated with altered ploidy or chromosomal instability in specific tissues and cell culture models, the mechanisms surrounding this phenomenon have been attributed to suppression of mitotic progression (Hernando et al., 2004; Sotillo et al., 2007). For example, elevated Mad2 levels, in response to increased E2F activity, have been hypothesized to contribute to mitotic failure. To gain an initial understanding of the basis for altered ploidy in the RB-deficient liver model in vivo, we compared mitotic control in the context of RB deletion against the same process within physiological regeneration induced by the hepatonecrotic agent carbon tetrachloride (CCl4). In this context, phosphorylated serine 10 of histone H3 (pH3-Ser10) was used as an established biochemical marker for mitotic entry. Strikingly, in both regenerating and RB-deleted livers, a dramatic increase in reactivity against this specific modification was observed (Figure 2A). Whereas robust staining for pH3-Ser10 was observed in RB-deleted liver tissues, no mitotic figures were observed in histological sections. In contrast, livers treated with CCl4 exhibited productive mitotic condensation (Figure 2B). These data indicate that although RB loss is sufficient to promote DNA replication in the liver of adult mice, it is not capable of advancing mitotic progression. Mining gene expression array data from these liver tissues, however, revealed that RB deletion resulted in robust induction of genes of critical importance in mitotic entry, suggesting that specific deficits in gene expression are likely not responsible for the observed failure to enter mitosis (Figure 2C). It has been previously shown that loss of RB in cell culture models can lead to replication stresses that induce DNA damage (Bartkova et al., 2005; Tort et al., 2006). To determine whether such a phenomenon could be occurring in vivo and subsequently leading to the induction of a G2/M checkpoint, replicating hepatocytes were labeled with BrdU, and the induction of DNA damage–related stress was detected by Ser-139 phosphorylation of histone H2AX (γH2AX). In regenerating livers, BrdU and γH2AX positivity are largely mutually exclusive, suggesting that DNA damage induces cell-cycle arrest in these hepatocytes (Figure 2D). In contrast, in RB-deficient livers, the majority of BrdU-positive hepatocytes were also strongly positive for γH2AX (Figure 2, D and E). Importantly, administration of Ad-Cre to Rb+/f mice does not recapitulate these results, as <2% of hepatocytes from RB-heterozygous livers were positive for γH2AX (unpublished data), suggesting an absence of Cre-specific effects on DNA damage. Thus these findings suggest that the aberrant replication induced by RB deletion in this model could be producing DNA damage that effectively limits mitotic entry. To delineate the mechanism underlying these observations, the expression of key proteins involved in coordination of mitosis was evaluated in the liver. These analyses revealed that both RB deletion and regeneration provided robust induction of CDK1 and CDK2, which normally contributes to the G2/M transition. RB deletion alone, however, resulted in the absence of Cyclin B1 accumulation (Figure 2F). This finding is critical, as Cyclin B1 is required for mitotic entry. Although it is degraded in response to specific forms of DNA damage (Gillis et al., 2009), the loss of Cyclin B1 can also directly contribute to DNA rereplication (Hook et al., 2007). These combined findings indicate that hepatic RB loss leads to specific breakdown in the coupling of S phase and mitosis in vivo.
FIGURE 2:
Acute RB deletion results in G2/M checkpoint targeting Cyclin B1 protein levels. (A) Sections were stained for presence of phosphorylated Ser-10 of histone H3 (pH3-Ser10). Percentage of positive hepatocytes was graphed. (B) Sections stained for pH3-Ser10 were visualized via fluorescence microscopy for presence of mitotic figures (visually condensed chromosomes) in each condition. (C) Fold change in relevant DNA replication–associated RNA was assessed via microarray. Ad-Cre–treated liver data were graphed as the fold change over control (Ad-LacZ) to assess deregulation of this critical gene expression program with deletion of RB. (D) Sections were dual-stained for incorporation of BrdU and presence of γH2AX and visualized by fluorescence microscopy. Representative images display colabeling only in Ad-Cre–treated livers. (E) Percentage of hepatocytes displaying dual positivity for γH2AX and BrdU is graphed. (F) Total (T) and nuclear (N) protein was resolved via SDS–PAGE, and indicated proteins were detected via immunoblotting.
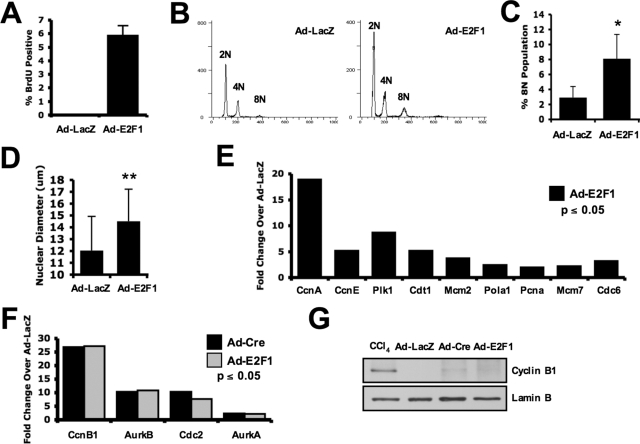
Because RB deletion failed to result in accumulation of Cyclin B1, it is possible that although RB-mediated transcriptional control drives a program for S-phase entry, additional factors are necessary for the transcription of G2/M-phase targets. To determine the specific impact of RB-regulated transcriptional mechanisms on complete cell-cycle control, E2F1 was overexpressed in the liver via intravenous injection of adenovirus encoding E2F1. In this context, E2F1 expression induced robust DNA synthesis (Figure 3A), which was associated with increased DNA ploidy (Figure 3, B and C) and nuclear size (Figure 3D). Importantly, E2F1 overexpression, similar to loss of RB, resulted in a robust increase in target gene transcription (Figure 3E) that included Cyclin B1 transcript (Figure 3F). These livers also exhibited a corresponding failure to accumulate Cyclin B1 protein (Figure 3G), resulting in an absence of mitosis (unpublished data). Thus deregulation of E2F activity is likely the proximal mechanism underlying the observed response to RB deletion. Furthermore it is likely that although RB/E2F-mediated transcriptional programs control multiple facets of the cell cycle, additional antiproliferative feedback mechanisms limit mitotic potential via Cyclin B1 protein levels.
FIGURE 3:
Hyperphysiological E2F activity phenocopies RB deletion in the liver. (A) Percentage of BrdU-positive hepatocyte nuclei are graphed from livers harvested 3 d postadenoviral delivery of Ad-LacZ or Ad-E2F1. (B) Hepatocyte nuclei from livers harvested at 3 d postadenoviral delivery were isolated and subjected to propidium iodide flow cytometry to determine cellular ploidy. (C) The 8N population of hepatocytes was counted via flow cytometry and graphed from livers harvested 3 d postadenoviral delivery. (D) Average nuclear diameter was measured from H&E-stained sections and graphed. (E) Fold change in relevant DNA replication–associated RNA was assessed via microarray. Ad-E2F1–treated liver data were graphed as the fold change over control (Ad-LacZ) to assess deregulation of this critical gene expression program with activation of E2F. (F) Fold change in relevant mitosis-associated RNA was assessed via microarray. Ad-Cre– and Ad-E2F1–treated liver data were graphed as the fold change over control (Ad-LacZ) to assess deregulation of this mitotic gene expression program. (G) Presence of Cyclin B1 protein levels were assessed via immunoblot in total protein lysates for each treatment. Representative blot is displayed.
RB loss deregulates the replication licensing machinery leading to high-density preRC assembly
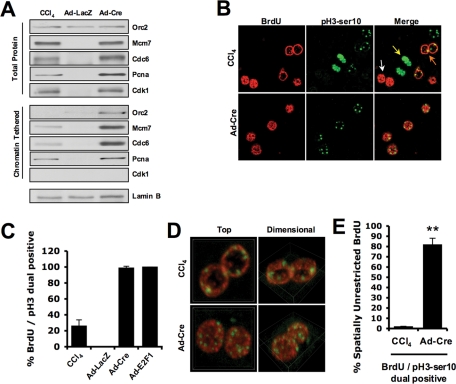
Previous studies suggest that increased levels of mini-chromosome maintenance proteins (MCMs) result in high-density formation of preRCs that are capable of initiating aberrant replication (Woodward et al., 2006). Similarly, inappropriate expression of key replication licensing proteins during G2 can lead to DNA rereplication (Yanow et al., 2001). To determine whether this mechanism was the basis for excessive altered ploidy accumulation following acute RB deletion in the liver, levels of chromatin-tethered replication factors were analyzed (Figure 4A). Whereas increased replication protein levels were observed in both regenerating and RB-deleted livers, conditions of RB deficiency resulted in excessive chromatin loading of these factors. Thus RB loss elicits distinct dysregulation of replication factor loading that likely promotes accumulation of aberrant ploidy in vivo.
FIGURE 4:
RB loss promotes E2F-mediated hyperphysiological preRC assembly and disrupts spatiotemporal replication control in the liver. (A) Total and exclusively chromatin-tethered protein fractions were resolved via SDS–PAGE, and indicated proteins were detected via immunoblotting. Cdk1 served as a negative control for tethering. (B) Liver sections were dual-stained for incorporation of BrdU and presence of pH3-Ser10 and visualized by fluorescence microscopy. Representative images display an aberrant pattern of staining in Ad-Cre–treated liver sections. In contrast, regenerating livers displayed physiological patterns indicative of early/mid-S phase (white arrows), mitotic condensed pH3-Ser10 staining (yellow arrows), and late DNA replication (orange arrows). (C) Percentage of BrdU-positive hepatocyte nuclei that were also pH3-Ser10 positive was counted and graphed. (D) Three-dimensional reconstructions of serial sectioning, imaged via confocal microscopy, demonstrate spatially unrestricted presence of BrdU in Ad-Cre–treated hepatocytes. (E) The total percentage of nuclei displaying the spatially unrestricted pattern of BrdU incorporation was graphed as a percentage of all BrdU/pH3-Ser10 dual-positive hepatocyte nuclei.
To explicitly determine the role of RB/E2F function in maintenance of genomic ploidy, a dual labeling approach was used to assess states of DNA replication and mitosis. Although the deletion of RB was quantitatively more robust at inducing DNA synthesis and pH3-Ser10 positivity, both conditions resulted in a large number of hepatocytes that were dual-positive for these markers (Figure 4, B and C). Because the BrdU pulse exhibits a relatively small observational window, it was possible that the overlap of S phase and mitotic markers could reflect late DNA replication occurring just prior to entry into G2 and mitosis. Consistent with this concept, the majority of hepatocytes in regenerating livers displayed either robust punctate BrdU incorporation across the entire nucleoplasm, indicative of early/mid-S-phase (Figure 4B, white arrows), or mitotic condensed pH3-Ser10 staining (Figure 4B, yellow arrows). The few dual-positive hepatocytes exhibited DNA replication specifically at the nuclear periphery, indicative of late DNA replication (Figure 4B, orange arrows). In stark contrast, RB-deficient liver tissue was characterized by nuclei undergoing early/mid-S-phase replication patterns while being positive for pH3-Ser10 (Figure 4B). Similar results were observed in conditions of E2F1 overexpression (unpublished data). To confirm that these results were truly indicative of spatial organization, optical sectioning was used to define the three-dimensional relationship of BrdU incorporation in pH3-Ser10–positive nuclei. These analyses confirmed that this aberrant pattern of BrdU incorporation was present throughout the entire three-dimensional structure of the nucleus, and appeared in >80% of dual-positive hepatocytes in RB-deficient livers (Figure 4, D and E). In contrast, this biochemically inappropriate pattern occurred in <2% of CCl4-treated hepatocytes (Figure 4E). These findings indicate that RB loss leads to an inappropriate form of DNA replication that underlies alterations in genome integrity, and suggests a unique means through which RB functions to control tumor susceptibility in the liver.
RB loss deregulates DNA replication but fails to uncouple the S and G2 phases of the cell cycle throughout the gastrointestinal tract
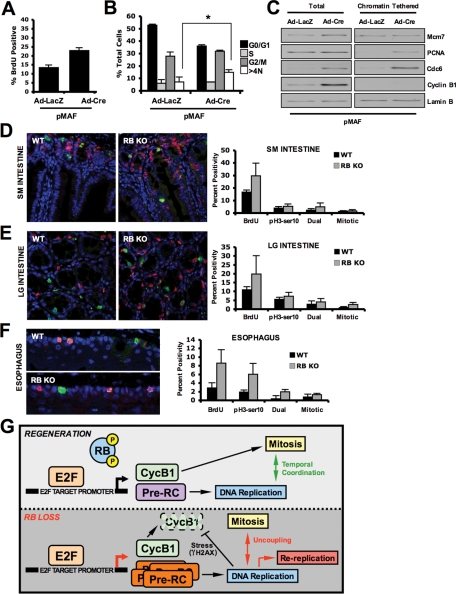
To investigate whether the consequences of RB loss on cell-cycle coordination were present in other model systems or naturally proliferative tissues, we first acutely deleted RB in primary murine adult fibroblast (pMAF) culture. In this context, RB deletion resulted in a modest deregulation of the cell cycle as previously reported (Figure 5, A and B). Importantly, while increased loading of replication factors was observed as in the liver, robust accumulation of Cyclin B1 protein occurred (Figure 5C). Furthermore although RB-deficient cultures harbored increased proportions of 8N cells (Figure 5B), these populations were ultimately proliferative and expanded in culture (unpublished data). These results suggest that the specific conditions that facilitate cell-cycle uncoupling in RB-deficient hepatocytes are not present within this fibroblast model system.
FIGURE 5:
Acute RB deletion abrogates DNA replication control but maintains cell-cycle coordination in other model systems. (A) Percentage of BrdU-positive Ad-LacZ or Ad-Cre treated pMAFs was calculated via bivariate flow cytometry. (B) The total percentage of cells in each condition with indicated DNA content was calculated via bivariate flow cytometry and graphed. (C) Total and exclusively chromatin-tethered protein fractions were resolved via SDS–PAGE, and indicated proteins were detected via immunoblotting for each condition. (D) Sections of murine small intestine taken from Rbf/f;Rosa26+/CreER mice treated with corn oil (RB-proficient) and Rbf/f;Rosa26+/CreER mice treated with tamoxifen (RB-deficient) were dual-stained for incorporation of BrdU and presence of pH3-Ser10 and visualized by fluorescence microscopy. Positive cells were scored and graphed. (E) Sections of murine large intestine taken from RB-proficient and RB-deficient animals and dual-stained for incorporation of BrdU and presence of pH3-Ser10 were visualized by fluorescence microscopy. Positive cells were scored and graphed. (F) Sections of murine esophagus were taken from RB-proficient and RB-deficient animals, dual-stained for incorporation of BrdU and presence of pH3-Ser10, and visualized by fluorescence microscopy. Positive cells were scored and graphed. (G) Modeled interpretation of RB/E2F-mediated cell-cycle control in conditions of natural regeneration and RB deficiency in the liver.
Acute deletion of RB in other naturally proliferative tissues of the gastrointestinal tract provided the opportunity to further interrogate the response of these tissues to the loss of this important tumor suppressor in vivo. For these studies, an Rbf/f;Rosa26+/CreER model was used, where treatment with tamoxifen elicits deletion of RB in multiple tissues. Consistent with prior studies, the acute ablation of RB in the small intestine resulted in increased BrdU that extended into the villi (Figure 5D), consistent with previous reports (Yang and Hinds, 2007). Interestingly, there was only a modest increase in pH3-Ser10 reactivity, and, although a larger proportion of cells stained dual-positive for BrdU/pH3-Ser10 in conditions of RB deletion, this clearly represented a modest fraction of the total labeled cells (Figure 5D). Similar results were found in the large intestine, and both intestinal types harbored mitotic figures, a feature not observed in RB-deficient livers (Figure 5E). In contrast with intestinal tissue, the esophagus was less proliferative, and RB deletion significantly increased the number of BrdU, pH3-Ser10, and dual-positive cells (Figure 5F). Similar to the observation in the intestinal tissues, however, the extent of dual-positive staining was considerably less than that observed in the liver. Together, these findings indicate that RB serves to constrain DNA replication in all tissues investigated; however, among these tissue types, the resultant uncoupling from mitotic progression during RB deficiency was most pronounced in the liver.
DISCUSSION
Although RB is a well-studied tumor suppressor, the response of adult tissues to the deletion of RB is surprisingly poorly understood. Because the vast majority of tumors arising with RB dysfunction are due to sporadic somatic inactivation, such analyses are critical for uncovering how deficiency of this single tumor suppressor can contribute to tumor development (Burkhart and Sage, 2008; Knudsen and Knudsen, 2008). To begin to address this issue, we specifically focused on the liver, as the RB pathway is frequently inactivated in corresponding human disease (Knudsen and Knudsen, 2008). Previous studies have demonstrated the ability of RB deletion to facilitate S-phase reentry in quiescent or differentiated cell populations (Sage et al., 2003; Blais et al., 2007; Pajcini et al., 2010); however, prior work from our laboratory and others has also shown that RB loss can be compensated in the liver (Rivadeneira et al., 2010). Specifically, in a model of neonatal RB deletion, as mouse development proceeds, quiescence is established as indicated by functional analyses and molecular markers. One of the key underlying phenotypes in RB deficient liver tissue, however, is altered hepatic ploidy. Although we have previously observed that such changes in ploidy can be manifest through acute or chronic deletion (Mayhew et al., 2005), the underlying mechanisms that drive this biology have not been elucidated.
Work herein demonstrates that RB serves as a brake for the entry into DNA replication in multiple adult tissues of the gastrointestinal tract. RB deletion mediated through inducible means generated robust DNA replication in gastrointestinal and esophageal tissue. Correspondingly, adenoviral transduction of Cre-recombinase and acute deletion of RB in the liver induced substantial DNA synthesis, similar to that of regenerating livers. It is well appreciated that adenovirus can induce liver damage and inflammatory responses that lead to cell-cycle entry; however, we specifically used low-titer administration of adenovirus, which has been shown to mitigate this caveat. Minimal inflammation was observed in the liver following adenoviral transduction, there was no evidence of necrosis or apoptosis in the infected livers, and control viral transduction did not result in appreciable DNA synthesis. Further solidifying RB as the downstream regulator of DNA replication in the liver, the replicative response to RB deletion is phenocopied by E2F overexpression, suggesting that any lesion capable of derepressing E2F should be sufficient to lead to DNA replication in this tissue.
The dysfunction of the RB pathway has been previously attributed to induction of replication stress–induced DNA damage, which has also been shown to promote specific checkpoints that prompt the termination of replication and cessation of cell-cycle progression (Bartkova et al., 2010). Because RB loss compromises DNA damage–associated G1/S arrest (Knudsen et al., 2000), any such stresses impinge directly on mitotic checkpoints. In this manner, RB-deficient livers failed to progress into mitosis. Importantly, this is a feature which is completely distinct from adenovirus-mediated toxicity, as overinfection with adenoviruses results in mitotic progression similar to that observed in regenerating livers (unpublished data). Although previous studies have demonstrated mitotic arrest following deletion of RB, this phenomenon has been attributed to failure of chromosomal condensation via Mad2- (Hernando et al., 2004; Sotillo et al., 2007) or condensin-related (Longworth et al., 2008) mechanisms. Most recently it was demonstrated that RB-deficient cells in vitro were able to arrest in G2/M due to excessive genomic instability (van Harn et al., 2010), and this arrest was mediated via effects on chromosomal condensation and segregation (Coschi et al., 2010; Manning et al., 2010). Our data in vivo indicate a failure to enter mitosis that functions upstream of chromosomal condensation. In contrast with previous studies wherein defects were observed within condensed chromatin, the RB-deficient livers were devoid of hepatocytes with condensed chromatin. In agreement with this finding, RB-deficient livers failed to accumulate significant levels of Cyclin B1, in stark contrast to regenerative tissue. Importantly, Ccnb1 transcript was up-regulated >26-fold with RB loss, suggesting a protein degradation mechanism preventing Cyclin B1 accumulation in conditions of RB deficiency. While these findings suggest that RB-independent mechanisms control the G2/M transition under such conditions, provocative studies indicate that the RB-related proteins p130 and p107 coordinately contribute to such a transition checkpoint in vitro (Blais et al., 2007). In vivo deletion of either p107 or p130 alone, however, does not yield a significant phenotype in the liver (Sage, unpublished data), suggesting that RB is uniquely involved in coordination of cell-cycle entry in this tissue. Further investigation of this unique mechanism of Cyclin B1 degradation in other model systems revealed relative tissue specificity. Deletion of RB in pMAFs resulted in a robust increase in Cyclin B1 protein. Additionally, acute RB deletion in the naturally proliferative environment of the intestinal tract failed to yield mitotic uncoupling, suggesting that intrinsically proliferative tissues differ in their response to RB deletion. Consistent with this concept, although not as robust as the cell-cycle deregulation observed in liver tissue, a significant increase in dual BrdU/p-H3-Ser10 cells was observed in the esophagus, and deletion of RB in fibroblasts and other models has been shown to be accompanied by increased ploidy. Thus, although clearly observed in the liver, cell-cycle uncoupling is manifest in other cell types via distinct mechanisms, or is perhaps inoperable due to intrinsic tissue differences.
In the context of RB deficiency, hepatocytes continue to aberrantly license replication, even in cells positive for markers of excessive DNA damage. The functional uncoupling of replication licensing and mitotic progression in these livers resulted in the accumulation of aberrant ploidy, a phenotype often regarded as a precursor to tumorigenesis (Pellman, 2007). Replication factors (e.g., Cdc6, Orc2) in these hepatocytes were highly active and tethered to chromatin. Importantly, recruitment of Orc and Cdc6 is sufficient to form artificial origins of replication (Takeda et al., 2005), and inhibition of rereplication is elicited through a Cdt1-independent mechanism (Kerns et al., 2007), suggesting that the increased chromatin tethering of these proteins is sufficient to support the observed rereplication. Moreover, excess levels of MCMs results in nonphysiological formation of preRCs that are capable of initiating aberrant replication (Woodward et al., 2006), and, in similar manner, overexpression of replication licensing proteins during interphase promotes initiation of DNA rereplication (Yanow et al., 2001). Combined, these studies suggest that the loss of RB deregulates the recruitment of various replication factors and formation of the preRC, resulting in unconstrained DNA replication that leads to aberrant ploidy, regardless of cellular checkpoints or inhibitory mechanisms.
Taken together, these studies shed new light on key, previously elusive mechanisms through which RB loss influences cell biology in vivo. The dysfunction associated with RB loss in normally quiescent adult livers directly impinges on physiological replication licensing. These observed phenotypes, especially in the presence of DNA damage, result in mitotic failure and could contribute to the generation of initiated cells that subsequently give rise to tumors. In adult tissues, the acute deregulation of cell-cycle control associated with the loss of such a key tumor suppressor could underlie the mechanism through which RB loss contributes to tumor development in vivo.
MATERIALS AND METHODS
Ethics statement
All mouse care, treatment, and killing were conducted using the highest standards for humane animal care in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals.
Mice
Mice (Rbf/f) containing LoxP sites flanking exon 19 of Rb1 have been previously reported (Marino et al., 2000; Mayhew et al., 2005). Generation and maintenance of Rbf/f;Rosa26+/CreER mice were previously described (Ventura et al., 2007; Burkhart et al., 2008).
Genotyping
Genomic DNA was isolated using phenol/chloroform extraction. Determination of Cre-mediated recombination of exon 19 of Rb1 in Rbf/f mice was done via PCR. Primers used: 5′-GGCGTGTGCATCAATG-3′, and 5′-GAAAGGAAAGTCAGGGACATTGGG-3′.
Adenoviruses, CCl4, and treatment delivery
Adenoviral delivery was performed on male mice anesthetized with isoflurane (2-chloro-2-(difluoromethoxy)-1,1,1-trifluoro-ethane) prior to intravenous injection of 1 × 109 plaque-forming units. Adenovirus expressing Cre-recombinase (Ad-CMV-Cre) was obtained from the University of Iowa Gene Transfer Core Facility. Adenovirus expressing lacZ (Ad-CMV-LacZ) was obtained from Vector Core Laboratory at the University of North Carolina; these were previously described (Mayhew et al., 2005). Adenovirus expressing human E2F1 (Ad-CMV-hE2F1) was obtained from the Joseph Nevins laboratory at Duke University.
CCl4-treated mice were administered a single intraperitoneal injection of 10% CCl4 dissolved in corn oil (Sigma, St. Louis, MO). Mice were killed 48 h postinjection, as previously described (Reed et al.).
Mouse killing and tissue harvest
Mice were given an intraperitoneal injection of BrdU (150 mg/kg in 0.9% saline) ∼90 min prior to killing. Livers were promptly excised, and the left lobe was fixed in 10% neutral buffered formalin for 48 h prior to mounting in paraffin for histological analysis. Remaining liver tissue was snap frozen in liquid nitrogen and stored at –80°C until later use.
Hepatocyte nuclear or total cell isolation
Nuclear isolation was previously described (Mayhew et al., 2005). Briefly, frozen liver tissue was dissociated, and nuclei were released with a Dounce and pestle homogenizer. Samples were then centrifuged at 13,000 rpm to pellet nuclei, and the nuclear pellet was either (immunoblot) subjected to lysis with RIPA buffer or (flow cytometry) resuspended in RNase A (80 μg/ml) and propidium iodide (50 μg/ml) in phosphate-buffered saline (PBS).
For total hepatocyte protein isolation, frozen liver tissue was dissociated, and cells were centrifuged at 8000 rpm. The resulting cell pellet was subjected to lysis in RIPA buffer supplemented with NaF at 50 mmol/l, ß-glycerophosphate at 13 mg/mL, and phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors at 1 mmol/l.
Gene expression array and analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s suggested protocol. Concentration was quantified using the Nanodrop ND-100 spectrophotometer, and quality was determined using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA). Two micrograms of total RNA from each liver was used for Affymetrix one-cycle target labeling as recommended by the manufacturer (Affymetrix, Santa Clara, CA). Each Affymetrix GeneChip for Mouse Genome 430 2.0 was hybridized for 16 h with biotin-labeled fragmented cRNA (10 μg) in 200 μl of hybridization cocktail according to Affymetrix protocol. Arrays were washed and stained using GeneChip Fluidic Station 450, and hybridization signals were amplified using antibody amplification with goat immunoglobulin G (Sigma-Aldrich, St. Louis, MO) and anti-streptavidin biotinylated antibody. Chips were scanned on an Affymetrix GeneChip Scanner 3000 using GeneChip Operating Software version 3.0. Normalization was performed by computing the Robust Multichip Average (RMA) expression measure [PMID: 12582260], and statistically significant gene changes were determined using significance analysis of microrarrays [PMID: 11309499] in the TM4 MultiExperiment Viewer software package [PMID: 12613259]. Significant changes in gene expression were determined using a 1.5-fold cutoff in expression change and an FDR of <25%.
Immunoblotting
Protein concentrations of each lysate were determined via Bio-Rad Protein Assay (Hercules, CA). Equal total protein for each sample was resolved by SDS–PAGE and transferred to Immobilon-P membrane (Millipore, Billerica, MA). Membranes were incubated with the following primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA): Lamin B (sc-6217), Cdk1 (sc-54), Cdk2 (sc-163), Cdk4 (sc-260), Cyclin A (sc-596), Cyclin F (sc-952), Orc2 (sc-28742), Mcm7 (sc-9966), Cdc6 (sc-9964), PCNA (sc-56), and Cyclin B1 (gift from P. Kaldis, NCI, Bethesda, MD).
Cisplatin treatment and bivariate flow cytometry
pMAFs were transduced with either Ad-LacZ or Ad-Cre for 48 h. Subsequent RB-proficient and RB-deficient populations were treated with either dimethyl sulfoxide (DMSO) (control) or 8 μM cisplatin (CDDP) for 24 h. One hour prior to harvest, cells were incubated with BrdU reagent (Amersham Biosciences, Piscataway, NJ) and then fixed in 70% ethanol overnight at 4°C. Fixed cells were centrifuged to a pellet and then resuspended in 2N HCl supplemented with pepsin at 0.5 mg/ml for 30 min. The HCl was then neutralized with 0.1 M sodium tetraborate pH 8.5, and then incubated with anti-BrdU-fluorescein isothiocyanate (BD PharMingen, San Diego, CA) at 37°C for 90 min. Cells were then resuspended in propidium iodide (50 μg/ml) and RNase A (80 μg/ml) in PBS for 30 min before flow cytometric analysis. Histograms were analyzed with FlowJo software, version 8.7 (Tree Star, Ashland, OR).
Liver processing and immunohistochemistry
Paraffin-embedded liver tissues were cut into 5-μm-thick sections and mounted on glass microscope slides. Sections were deparaffinized in xylene and then rehydrated through an ethanol/water gradient. Liver sections were then boiled in Antigen Retrieval Solution (DakoCytomation, Carpinteria, CA) using a microwave for 5 min at 100% power followed by 20 min at 30% power. Sections were permeabilized in 0.4% Triton X-100 for 20 min at 37°C and then washed in PBS. For BrdU detection, sections were incubated in rat anti-BrdU (1:200; AbD Serotec, Raleigh, NC), MgCl2, and DNase for 60 min at 37°C, followed by donkey anti–rat rhodamine (1:1000; Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 min at 37°C. For pH3-Ser10 detection, sections were incubated in rabbit anti–pH3-Ser10 (1:500; Upstate, now Millipore) for 60 min at 37°C, followed by goat anti-rabbit Alexa Fluor 488 (1:1000; Invitrogen) for 30 min at 37°C. For γH2AX detection, sections were incubated in mouse anti-γH2AX (1:200; Millipore) for 60 min at 37°C, followed by goat anti–mouse Alexa Fluor 488 (1:1000; Invitrogen) for 30 min at 37°C. After antibody incubations, slides were washed 3× in PBS and incubated in 1:1000 DAPI in PBS for 5 min before mounting.
Chromatin-tethered fractionation
Isolation of the chromatin-tethered protein fraction has been previously described (Braden et al., 2006). A single cell suspension of hepatocytes or cultured cells was centrifuged to pellet cells, and soluble proteins were then extracted with ice-cold CSK buffer (10 mM PIPES [piperazine-N,N’-bis(2-ethanesulfonic acid)], pH 6.8; 100 mM NaCl; 300 mM sucrose; 1 mM MgCl2; 1 mM EGTA; 1 mM dithiothreitol; 1 mM PMSF) supplemented with 0.1% Triton X-100 for 15 min at 4°C. Extracted cells were centrifuged to a pellet and reextracted in ice-cold CSK buffer for another 10 min at 4°C. The chromatin-tethered fraction was centrifuged at 14,000 rpm to a pellet. Lysates were resolved via SDS–PAGE.
RB Ablation in intestinal/esophageal tissue
Rosa26CreERT2 and control mice received injections with tamoxifen as previously described (Burkhart et al., 2010) and killed 72 h postinjection. All procedures were carried out in the laboratory of J. Sage. One male and two female mice, 8 wk of age, were used for each of the experimental and control genotypes. All images used were adjusted for contrast or brightness without misrepresenting original image and ensuring integrity of raw data.
Acknowledgments
We recognize the members of the Erik S. and Karen E. Knudsen laboratories for insightful scientific feedback and discussion. A special thanks goes to Christopher N. Mayhew for critical instruction in several techniques and procedures. E.S.K. and J.S are supported by grants from the National Cancer Institute.
Abbreviations used:
- DMSO
dimethyl sulfoxide
- γH2AX
histone H2AX
- H&E
hematoxylin and eosin
- HCC
hepatocellular carcinoma
- MCM
mini-chromosome maintenance protein
- PBS
phosphate-buffered saline
- pMAF
primary murine adult fibroblast
- PMSF
phenylmethylsulfonyl fluoride
- preRC
prereplication complex
- RB
retinoblastoma tumor suppressor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0895) on February 2, 2011.
REFERENCES
- Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev. 2004;14:527–532. doi: 10.1016/j.gde.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Hodgson B. Replication licensing–defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braden WA, Lenihan JM, Lan Z, Luce KS, Zagorski W, Bosco E, Reed MF, Cook JG, Knudsen ES. Distinct action of the retinoblastoma pathway on the DNA replication machinery defines specific roles for cyclin-dependent kinase complexes in prereplication complex assembly and S-phase progression. Mol Cell Biol. 2006;26:7667–7681. doi: 10.1128/MCB.00045-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burkhart DL, Ngai LK, Roake CM, Viatour P, Thangavel C, Ho VM, Knudsen ES, Sage J. Regulation of RB transcription in vivo by RB family members. Mol Cell Biol. 2010;30:1729–1745. doi: 10.1128/MCB.00952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Viatour P, Ho VM, Sage J. GFP reporter mice for the retinoblastoma-related cell cycle regulator p107. Cell Cycle. 2008;7:2544–2552. doi: 10.4161/cc.7.16.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010 doi: 10.1101/gad.1917610. 24, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Labib K. The chromosome replication cycle. J Cell Sci. 2002;115:869–872. doi: 10.1242/jcs.115.5.869. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Prokhorova TA, Blow JJ, Todorov IT, Gilbert DM. Mammalian nuclei become licensed for DNA replication during late telophase. J Cell Sci. 2002;115:51–59. doi: 10.1242/jcs.115.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis LD, Leidal AM, Hill R, Lee PW. p21Cip1/WAF1 mediates cyclin B1 degradation in response to DNA damage. Cell Cycle. 2009;8:253–256. doi: 10.4161/cc.8.2.7550. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. Modelling the molecular circuitry of cancer. Nat Rev Cancer. 2002;2:331–341. doi: 10.1038/nrc795. [DOI] [PubMed] [Google Scholar]
- Harbor JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Torke SJ, Benjamin JM, McGarry TJ. Geminin prevents rereplication during xenopus development. J Biol Chem. 2007;282:5514–5521. doi: 10.1074/jbc.M609289200. [DOI] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S, Vooijs M, Van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res. 2002;62:6587–6597. [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D. Cell biology: aneuploidy and cancer. Nature. 2007;446:38–39. doi: 10.1038/446038a. [DOI] [PubMed] [Google Scholar]
- Reed CA, Mayhew CN, McClendon AK, Knudsen ES. Unique impact of RB loss on hepatic proliferation: tumorigenic stresses uncover distinct pathways of cell cycle control. J Biol Chem. 285:1089–1096. doi: 10.1074/jbc.M109.043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira DB, Mayhew CN, Thangavel C, Sotillo E, Reed CA, Grana X, Knudsen ES. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle reentry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- Sellers WR, Kaelin WG., Jr Role of the retinoblastoma protein in the pathogenesis of human cancer. J Clin Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol. 2007;8:894–903. doi: 10.1038/nrm2276. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort F, Bartkova J, Sehested M, Orntoft T, Lukas J, Bartek J. Retinoblastoma pathway defects show differential ability to activate the constitutive DNA damage response in human tumorigenesis. Cancer Res. 2006;66:10258–10263. doi: 10.1158/0008-5472.CAN-06-2178. [DOI] [PubMed] [Google Scholar]
- van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, Te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24(13):1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Wood M, Perrotte P, Onishi E, Harper ME, Dinney C, Pagliaro L, Wilson DR. Biodistribution of an adenoviral vector carrying the luciferase reporter gene following intravesical or intravenous administration to a mouse. Cancer Gene Ther. 1999;6:367–372. doi: 10.1038/sj.cgt.7700090. [DOI] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HS, Hinds PW. pRb-mediated control of epithelial cell proliferation and Indian hedgehog expression in mouse intestinal development. BMC Dev Biol. 2007;7:6. doi: 10.1186/1471-213X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]