Abstract
Circadian rhythms provide organisms with an adaptive advantage, allowing them to regulate physiological and developmental events so that they occur at the most appropriate time of day. In plants, as in other eukaryotes, multiple transcriptional feedback loops are central to clock function. In one such feedback loop, the Myb-like transcription factors CCA1 and LHY directly repress expression of the pseudoresponse regulator TOC1 by binding to an evening element (EE) in the TOC1 promoter. Another key regulatory circuit involves CCA1 and LHY and the TOC1 homologs PRR5, PRR7, and PRR9. Purification of EE–binding proteins from plant extracts followed by mass spectrometry led to the identification of RVE8, a homolog of CCA1 and LHY. Similar to these well-known clock genes, expression of RVE8 is circadian-regulated with a dawn phase of expression, and RVE8 binds specifically to the EE. However, whereas cca1 and lhy mutants have short period phenotypes and overexpression of either gene causes arrhythmia, rve8 mutants have long-period and RVE8-OX plants have short-period phenotypes. Light input to the clock is normal in rve8, but temperature compensation (a hallmark of circadian rhythms) is perturbed. RVE8 binds to the promoters of both TOC1 and PRR5 in the subjective afternoon, but surprisingly only PRR5 expression is perturbed by overexpression of RVE8. Together, our data indicate that RVE8 promotes expression of a subset of EE–containing clock genes towards the end of the subjective day and forms a negative feedback loop with PRR5. Thus RVE8 and its homologs CCA1 and LHY function close to the circadian oscillator but act via distinct molecular mechanisms.
Author Summary
Circadian clocks help organize 24-hour rhythms in physiology and behavior so that critical organismal functions are optimally timed relative to highly predictable daily changes in the environment. Circadian clocks run at approximately the same pace across a wide range of temperatures, ensuring accurate timekeeping in all seasons. Although molecular components of the circadian clock are not conserved across higher taxa, eukaryotic circadian clocks are composed of analogous interlocked transcriptional feedback loops. In this study, we report the isolation and characterization of a new component of the plant circadian system, REVEILLE 8 (RVE8). RVE8 is a clock-regulated Myb-like transcription factor that binds with high affinity to the evening element (EE) promoter motif and helps to set the pace of the clock in a light- and temperature-dependent manner. RVE8 promotes expression of the clock component PSEUDO-RESPONSE REGULATOR 5 (PRR5), likely via direct action at the PRR5 promoter. RVE8 expression is in turn repressed by PRR5. Thus, RVE8 is a new component of the plant circadian oscillator that takes part in a novel transcriptional feedback loop.
Introduction
We live on a world with prominent and predictable daily changes in the environment. To better anticipate and mitigate these changes, most organisms possess circadian clocks, internal timers that generate roughly 24-hour rhythms in physiology even in the absence of environmental cues. Processes influenced by the circadian clock include once-in-a-lifetime developmental events such as the eclosion of insects from their pupal cases or the transition of plants from vegetative to reproductive growth, to daily events such as changes in activity levels in animals or the opening and closing of flowers [1], [2]. Given this diversity of clock outputs, it is not surprising that the circadian system influences expression of a substantial fraction of the genome, with 30% of plant genes and 10% of mammalian genes estimated to be circadian regulated [3]–[6].
Although the molecular components of the circadian clock are not conserved across higher taxa, basic features of the circadian system are shared. In all organisms that have been investigated, circadian clocks are cell autonomous and can be reset by environmental cues such as changes in light or temperature [7]–[9]. However, circadian clocks are strongly temperature compensated; that is, they run at a similar pace across the physiologically relevant range of temperatures [10], [11]. This allows them to keep accurate time in all seasons. In eukaryotes, transcriptional feedback loops play a crucial role in the circadian oscillator, although post-transcriptional events such as regulated protein degradation are also essential for robust clock function [12].
In recent years, rapid progress has been made in uncovering the mechanisms underlying clock function in many model organisms. For example, molecular genetic and genomic studies in Arabidopsis thaliana (Arabidopsis) have led to the identification of interlocked transcriptional feedback loops that act at the heart of the plant clock [1]. Two Myb-like transcription factors, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), inhibit expression of TIMING OF CAB EXPRESSION 1 (TOC1), which encodes a nuclear-localized protein that indirectly promotes expression of CCA1 and LHY, forming the first transcriptional feedback loop [13], [14]. A second negative feedback loop is formed between CCA1 and LHY and two TOC1 homologs, PSEUDO-RESPONSE REGULATOR 7 and 9 (PRR7 and PRR9). CCA1 and LHY promote expression of PRR7 and PRR9, which in turn negatively regulate CCA1 and LHY via direct binding to their promoters [15], [16]. Another TOC1 homolog, PRR5, has recently been shown to also negatively regulate CCA1 and LHY expression [17], although it is not currently clear how expression of PRR5 itself is regulated. A third negative feedback loop has been proposed, based primarily on mathematical modeling, in which an unknown component forms a negative feedback loop with TOC1 [18].
In addition to the transcriptional feedback loops, post-transcriptional control mechanisms are also indispensible for normal clock function. ZEITLUPE (ZTL), a blue light photoreceptor with an F-box domain, is involved in the regulated degradation of TOC1 [19]. ZTL stability is in turn regulated by its light-dependent interaction with GIGANTEA (GI) [20]. Regulation of clock protein phosphorylation and intracellular localization are also important control mechanisms [21]–[23].
Analysis of circadian regulated genes has led to the identification of several promoter motifs implicated in the phase-specific regulation of gene expression [4], [24]–[26]. One such motif is the evening element (EE, AAAATATCT), which when multimerized is both necessary and sufficient to confer evening-phased expression on a reporter gene [27]. A number of clock genes, including TOC1, GI, and PRR5, have EE sequences in their promoters. CCA1 and LHY bind to the EE-containing region of the TOC1 promoter to directly repress TOC1 expression [13]. We have recently shown that a protein related to CCA1 and LHY, REVEILLE 1 (RVE1), also binds specifically to the EE. However, rather than affecting central clock function, RVE1 controls daily rhythms of auxin production, serving as a node connecting the circadian and auxin signaling networks [28]. Two RVE1 homologs, CIRCADIAN 1 (CIR1)/RVE2 and EARLY-PHYTOCHROME-RESPONSIVE 1 (EPR1)/RVE7, also seem to function primarily as clock outputs via undefined mechanisms [29], [30].
Although a framework describing the plant circadian oscillator is now in place, there are still considerable gaps in our understanding. The interactions between known components are not completely understood, some clock genes have been identified but not yet placed into the clock model, and some components have been predicted but not yet identified on a molecular level [1]. We have therefore taken a biochemical approach to identify new factors that act within the circadian system. Previously, we found a specific EE-binding activity in extracts made from both wild-type and cca1 lhy plants [27]. We now report on the characterization of RVE8, an EE-binding protein identified by liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. RVE8 is a member of a previously uncharacterized clade in the CCA1/LHY/RVE transcription factor family. RVE8 acts not only in setting the pace of the clock, but also plays roles in temperature compensation and light signaling. Although the circadian transcriptional profiles of CCA1, LHY, and RVE8 are very similar, the protein accumulation patterns are quite different: RVE8 protein peaks in the subjective afternoon while CCA1 and LHY proteins peak near subjective dawn. Our data suggest that RVE8 directly promotes expression of PRR5. Microarray data indicate that PRR5, in turn, represses expression of RVE8. Thus RVE8 and PRR5 comprise a negative transcriptional feedback loop that acts within the plant circadian network.
Results
Mass spectrometric identification of RVE8, an EE–binding protein
In previous experiments, we identified an afternoon-phased activity in plant extracts that bound specifically to the EE [27]. To identify the factor(s) responsible, we used wild-type and mutant versions of the EE bound to magnetic beads to isolate DNA binding proteins from plants harvested eight hours after dawn. Purified proteins were subjected to LC-MS/MS and peptide sequences compared to the Arabidopsis proteome. At a peptide false discovery rate of 0.1%, a number of transcription factors were identified, including three Myb-like, two B3-domain, two trihelix, two WHIRLY, one WRKY, and one basic-leucine zipper transcription factors (Table 1). Several of these factors were identified specifically in wild-type but not in mutant EE samples. Only the Myb-like factor RVE8 (At3g09600) was identified in each of three independent experiments; intriguingly, its close homolog RVE4 (At5g02840) was also identified in one of the replicates. In addition to the peptides that could be assigned uniquely to either RVE8 or RVE4 (indicated in Table 1), additional peptides that could have been derived from either protein were identified in each of the three experiments (Table S1). Using more liberal filtering parameters to identify proteins with lower sequence coverage, peptides uniquely derived from RVE5 and RVE6 could also be identified (Table S1). Complete protein and peptide identification data can be found at ProteomeCommons.org and in Table S2.
Table 1. Putative EE–binding factors identified by LC-MS/MS.
| Accession number | Family(Gene name) | Number of protein-specific peptides identified | ||||||
| Replicate 1 | Replicate 2 | Replicate 3 | ||||||
| AGI | Uniprot | EE-wt | EE-mt | EE-wt | EE-mt | EE-wt | EE-mt | |
| At3g09600.1 | Q8RWU3 | Myb-like (RVE8) | 1 | - | 4 | - | 4 | - |
| At5g02840.1 | Q6R0G4 | Myb-like (RVE4) | - | - | - | - | 2 | - |
| At5g47390.1 | Q9LVS0 | Myb-like | 3 | - | - | - | - | - |
| At1g49480.1 | Q9XIB5 | B3 domain(RTV1) | 5 | 2 | - | - | - | - |
| At3g18990.1 | Q8L3W1 | B3 domain (VRN1) | 2 | - | - | - | - | - |
| At4g36730.1 | P42774 | Basic-leucine zipper (GBF1) | 3 | 3 | - | - | - | - |
| At1g76890.1 | Q39117 | Trihelix (GT2) | 2 | - | - | - | - | - |
| At1g76880.1 | Q9C6K3 | Trihelix | - | 2 | - | - | - | - |
| At1g13960.1 | Q9XI90 | WRKY (WRKY4) | - | 4 | - | - | - | - |
| At1G14410.1 | Q9M9S3 | WHIRLY (WHY1) | 14 | 14 | 11 | 9 | 10 | 9 |
| At2g02740.1 | A8MR64 | WHIRLY (WHY3) | 13 | 14 | 9 | 9 | 9 | 6 |
In three independent experiments, plant extracts were incubated with either wild-type or mutant versions of the EE bound to beads. Purified proteins were digested with trypsin and peptides were identified by LC-MS/MS. The number of peptides that could be specifically assigned to a single protein (not shared with any homologous proteins) using a peptide false discovery rate of 0.1% is indicated. Only known and putative transcription factors are listed (169 total proteins were identified). Complete protein identification data and statistics are available from the proteome commons data set as detailed in the methods section. The peptide numbers reported are based on scaffold grouping. For full information see Table S2 or the scaffold file in the proteome commons data repository.
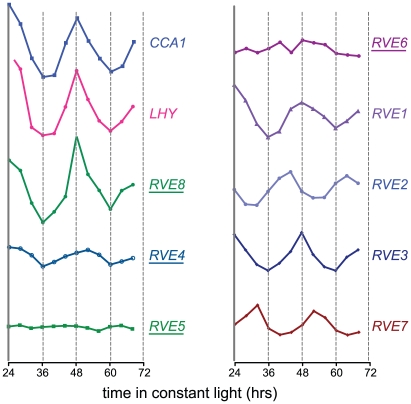
The RVE genes are part of a small family of transcription factors that includes the well-studied clock genes CCA1 and LHY. Many RVE proteins have previously been reported to bind the EE in vitro [31]. One clade consists of RVE1, RVE2/CIR1, and RVE7/EPR1, which likely functional primarily as clock outputs [28]–[30]. RVE3, RVE4, RVE5, RVE6, and RVE8 belong to a separate clade not previously functionally characterized. Expression of RVE3, RVE4 and RVE8, but not RVE5 or RVE6, was clock-regulated in seedlings, with peak message levels occurring near subjective dawn (Figure 1) [4], [32]. RVE8 expression in particular was very similar in amplitude and phase to that of the known EE-binding proteins CCA1, LHY, and RVE1 (Figure 1) [28], [33], [34].
Figure 1. Circadian expression patterns of the CCA1, LHY, and RVE genes.
Expression of the RVE family of genes was assayed in a previously published microarray experiment [83]. Seedlings were entrained in LD 12:12 for 7 days before being transferred to constant white light. Plants were harvested at the indicated times and subjected to microarray analysis [83]. Relative expression levels of the different Myb-like genes are shown; all are graphed using the same scale. Underlined names indicate that peptides from the corresponding gene product were identified by mass spectrometry after affinity purification of EE-binding proteins.
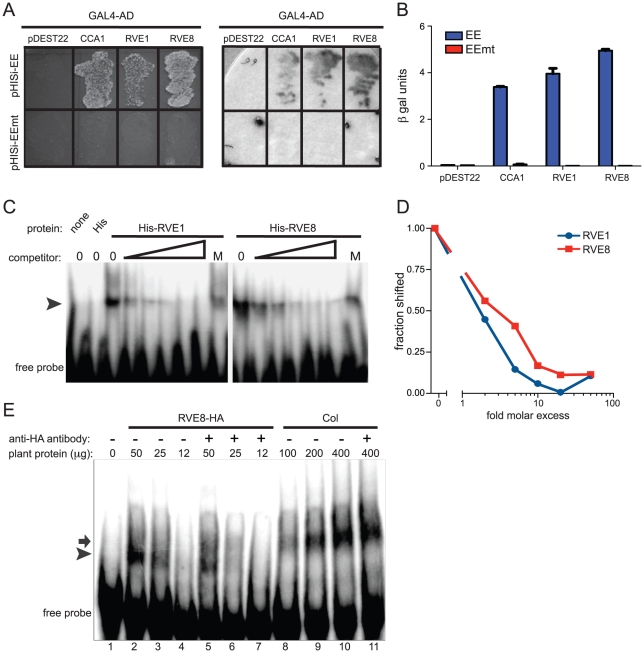
Since RVE8 was the only transcription factor identified as a sequence-specific EE binding protein in all three of our LC-MS/MS experiments, we further characterized its ability to bind to the EE. First, we used yeast one-hybrid experiments to assay RVE8 binding in a heterologous system. RVE8 interacted with wild-type but not mutant EE sequences, showing similar EE binding activity as CCA1 and RVE1 (Figure 2A, 2B). Electrophoretic mobility shift assays (EMSAs) carried out with recombinant RVE8 also demonstrated that RVE8 bound the EE specifically and with similar affinity as RVE1 (Figure 2C–2D).
Figure 2. RVE8 binds specifically to the EE both in vivo and in vitro.
(A, B) Four copies of wild-type or mutant EE were multimerized and used to drive expression of the HIS3 or lacZ genes in S. cerevisiae. The CCA1, RVE1, and RVE8 cDNAs were fused to the GAL4 activation domain, transformed into yeast containing the above bait vectors, and plated on media lacking histidine (A, left panel). They were also assayed for β-galactosidase activity on a filter (A, right panel) and in a liquid assay (B). (C) Recombinant RVE1 or RVE8 was incubated with a multimerized, radiolabeled, EE probe sequence. A 2, 5, 10, 20, or 50-fold molar excess of unlabeled EE or a 50-fold molar excess of unlabeled mutant EE (indicated by the letter M) was added to each reaction as competitor. Protein/DNA complexes were separated on a non-denaturing polyacrylamide gel and the radiolabeled DNA visualized using a phosphorimager. The arrowhead indicates the position of protein/DNA complexes. (D) The fraction of probe shifted in each lane was quantified and normalized to the fraction shifted in the lane with no added competitor. (E) An EMSA assay was performed with protein extracted from wild-type or plants overexpressing HA-tagged RVE8. 1 µg of anti-HA antibody was added to some reactions, as indicated. The arrow indicates the mobility of the shifted DNA/protein complex using extracts made from wild type while the arrowhead indicates the mobility of the complex using extracts made from plants overexpressing RVE8. Data are representative of three independent experiments.
We next investigated whether RVE8 might contribute to the EE binding activity we found in plant extracts, first generating plants overexpressing HA-tagged RVE8 (Figure S1A). A similar amount of EE binding activity was detected in 25 µg of extract made from RVE8-OX plants as in 100 g of wild-type extract (Figure 2E, compare lanes 3 and 8). However, the mobility of DNA/protein complexes was slightly different depending upon whether extracts were made from wild-type (arrow) or RVE8-OX (arrowhead) plants, suggesting that the composition of EE binding complexes was somewhat different in these genotypes.
The increased EE binding activity in RVE8-OX plant indicated that RVE8 directly or indirectly contributed to EE binding activity in these plants. To help distinguish between these possibilities, we added anti-HA antibody to our EMSA reactions. The addition of anti-HA antibody caused a reduction in the amount of shifted probe in the RVE8-HA lanes (Figure 2E; compare lanes 2 and 5, and lanes 3 and 6) but no reduction in binding activity in the wild-type control lanes (Figure 2E, compare lanes 10 and 11). These data suggested that RVE8 can bind directly to the EE in plants, at least when overexpressed, and encouraged us to further examine its role in the circadian system.
Perturbation of RVE8 expression causes growth phenotypes
Plants with circadian clock defects frequently also show changes in light-mediated inhibition of hypocotyl elongation and photoperiodic control of flowering time [35], [36]. Indeed, overexpression or mutation of CCA1, LHY, and several other RVE-family genes affects these processes [29], [30], [33], [34], [37]. We therefore examined light regulation of hypocotyl growth and flowering time responses in both RVE8-OX and SALK-053482, a rve8 loss-of-function mutant obtained from the SALK T-DNA collection [38]. This mutant, designated rve8-1, did not express detectable levels of RVE8 message (Figure S1B, S1C) and is thus likely a null allele.
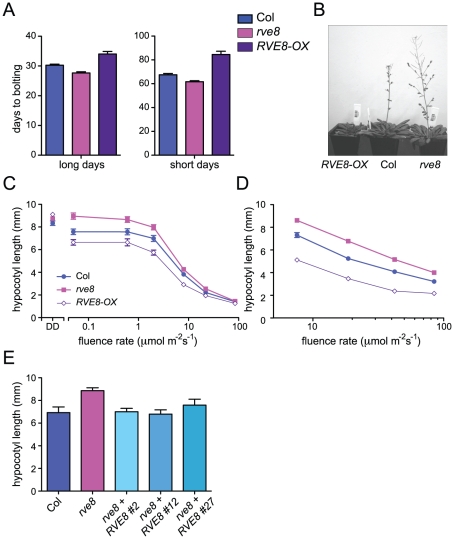
Appropriate photoperiodic control of flowering time requires a functional circadian clock [36]. We therefore examined time to flowering in plants grown in short day (SD; 8 hours light:16 hours dark) or long day (LD; 16 hours light:8 hours dark) conditions. rve8-1 plants flowered slightly earlier than wild type in both LD and SD (2.6 days earlier in LD and 5.8 days earlier in SD) (Figure 3A, 3B). The effects of RVE8-OX overexpression were also relatively modest, but acted to delay rather than promote flowering (RVE8-OX plants flowered 3.8 days later than wild type in LD and 17 days later in SD) (Figure 3A, 3B). Although these effects were small, they were reproducible and highly statistically significant. These moderate differences in time to flowering are quite different from the strong flowering time phenotypes seen in CCA1 and LHY mutant and overexpressing plants [33], [34], [37], but are reminiscent of the relatively subtle effects on flowering time reported in rve2, RVE2-OX, and RVE7-OX plants [29], [30].
Figure 3. RVE8 affects seedling growth and flowering time.
(A, B) Plants were grown in either long-day (16 hr white light/8 hr dark; LD 8:16) or short day conditions (8 hr white light/16 hr dark; SD 8:16). Plants were classified as bolting when a 1 cm bolt was observed; n = 18–25. (C–E) Seedlings were grown in darkness (DD) or constant white light (C) or in SD (D) at the indicated fluence rates for 6 days and hypocotyl lengths were measured using ImageJ. In constant light, hypocotyl lengths of rve8-1 and RVE8-OX were significantly different from Col at all except for the highest fluence rate tested (p<0.05) whereas in short days, hypocotyl lengths of rve8-1 and RVE8-OX were significantly different from Col at all fluences rates tested (p<0.0001). (E) Wild type, rve8-1, and independent rve8-1 lines transformed with a RVE8 gene driven by the native promoter were grown in constant white light at 2 mol m−2 s−1. n = 20–30 for all experiments; means ± SEM are shown. Data are representative of at least two independent experiments; ** indicates p<0.005; Student's two-tail heteroscedastic t test.
We next examined the hypocotyl length of seedlings grown in different light conditions. Hypocotyl lengths of etiolated RVE8-OX and rve8-1 seedlings were very similar to those of wild-type seedlings (Figure 3C). However, when grown in low or medium fluence-rate constant white light, rve8-1 seedlings had longer hypocotyls and RVE8-OX had shorter hypocotyls than wild type (Figure 3C). These differences in phenotype diminished at higher fluence rates of constant white light. When grown in SD conditions, rve8-1 and RVE8-OX displayed long and short hypocotyls, respectively, over a range of fluence rates, although the differences were slightly reduced at the highest light intensity tested (Figure 3D). The rve8-1 long-hypocotyl phenotype was rescued by transformation of these plants with a wild-type genomic copy of the RVE8 locus (Figure 3E), demonstrating that loss of RVE8 expression was indeed responsible for this trait.
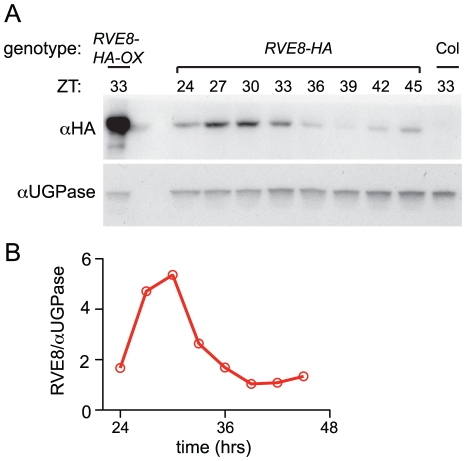
RVE8 protein levels are clock-regulated but lag transcript accumulation
Given the dawn phase of expression of RVE8, it was somewhat surprising that we identified peptides uniquely derived from this protein in extracts from plants harvested eight hours after dawn. To investigate the temporal pattern of RVE8 protein accumulation, we performed immunoblots on samples made from a rve8-1 line rescued with a RVE8::RVE8-HA construct (Figure 3E). We found that peak levels of RVE8-HA protein occurred three to six hours after subjective dawn (ZT27 – ZT30; Figure 4A, 4B). In contrast, CCA1 and LHY protein levels closely track their respective transcript levels [33], [39]. However, post-transcriptional regulation of clock genes is not uncommon; for example, accumulation of many of the pseudo-response regulator proteins that function in the plant clock is also significantly delayed relative to their transcript profiles [21], [40]–[42], as is accumulation of the clock protein PER in animals [43], [44]. This delay helps explain why RVE8 was identified as an afternoon-phased EE-binding protein. The mechanism underlying the sizable delay in RVE8 protein accumulation relative to its transcript is not clear and will be an interesting topic for further research.
Figure 4. RVE8 protein accumulates in the subjective afternoon.
Col, RVE8::RVE8-HA and 35S::RVE8-HA seedlings were entrained in 12 hr light:12 hr dark for 6 days before being released to constant white light (55 mol m−2 s−1) (A) Plants were harvested at the indicated times and extracts were subjected to immunoblot analysis using either an anti-HA antibody (upper panel) or an anti-UGPase antibody (lower panel). (B) Data shown in panel (A) was quantified using ImageQuant software. Data are representative of two independent experiments.
RVE8 helps set the pace of the clock
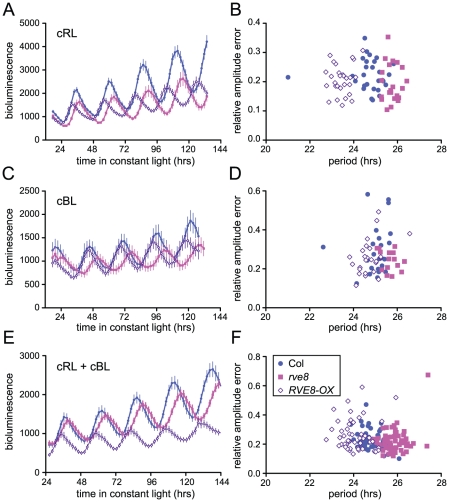
Loss-of-function alleles of CCA1 and LHY cause period shortening whereas overexpression of these genes causes arrhythmicity [33], [34], [37], [45], [46]. Similarly, overexpression of RVE1, RVE2, or RVE7, all members of the same clade, reduces rhythmic amplitude [28]–[30]. However, rve1, rve2, and rve1 rve2 rve7 mutants have no circadian phenotypes [28], [30] suggesting that they normally primarily act as output genes rather than clock components. In contrast, RVE8 overexpression caused an approximately one hour shortening of free-running rhythms of CCR2::LUC activity when plants were maintained in constant red or red + blue light (Figure 5A and 5E). In constant blue light, the average period of RVE8-OX plants was only 0.3 hours shorter than wild type, a difference that was not statistically significant in this experiment (Figure 5C). In contrast, the average free-running period of rve8-1 plants was approximately 1 hour longer than wild type in all three light conditions (Figure 5A, 5C and 5E; see figure legend for period estimates). The robustness of rhythms, as measured by the relative amplitude error (RAE), was not appreciably different in rve8-1 or RVE8-OX compared to Col controls (Figure 5B, 5D, and 5F).
Figure 5. Perturbation of RVE8 expression changes free-running period.
Seedlings were entrained in 12 hr light:12 hr dark for 6 days and then transferred to either constant red (44 mol m−2 s−1) (A–B), blue (19 mol m−2 s−1) (C–D), or red + blue (36 mol m−2 s−1 red and18 mol m−2 s−1 blue) (E–F) light; CCR2::LUC activity rhythms were then monitored. (A, C, E) Average luciferase activity of Col, rve8-1, and RVE8-OX plants expressing CCR2::LUC; each point is the average of 20–25 seedlings and error bars represent ± SEM. (B, D, F) RAE, a measure of rhythmic robustness (with smaller values indicating stronger rhythms) is plotted relative to free-running period. (A, B) In red light, the free-running periods of rve8-1 and RVE8-OX were significantly different from Col (Col = 24.75±0.15 hr; rve8-1 = 25.73±0.07 hr; RVE8-OX = 23.44±0.07 hr; p<0.0001 for both comparisons). (C, D) In blue light, rve8-1 had a significantly longer period than Col (Col = 24.93±0.11 hr; rve8-1 = 25.70±0.09 hr; p<0.005); RVE8-OX had a shorter period (RVE8-OX = 24.60±0.09 hr) than wild type but this was not statistically significant (p = 0.08). (E, F) In red + blue, rve8-1 had a significantly longer period and RVE8-OX had a significantly shorter period than Col (Col = 24.84±0.07 hr; rve8-1 = 25.82±0.06 hr; RVE8-OX = 24.06±0.09 hr; p<.0001 for both; Student's two-tail heteroscedastic t test used for all comparisons). These data are representative of at least two independent experiments.
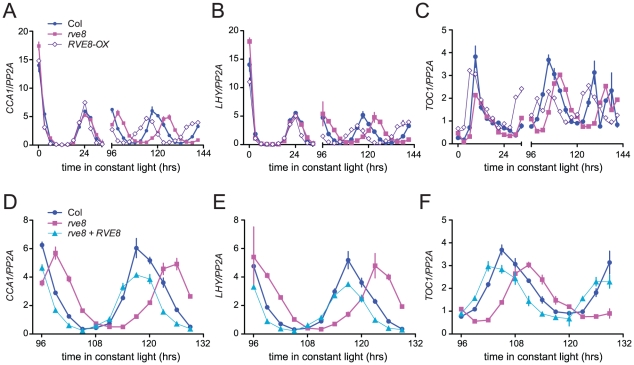
Changes in free-running period are often accompanied by changes in the expression levels of core clock genes. In particular, CCA1, LHY, and other members of the RVE family have previously been reported to regulate each other's expression levels [28], [30], [33], [34], [37]. We therefore used quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) assays to monitor expression of CCA1, LHY, and TOC1 in rve8-1 and RVE8-OX plants. No changes in expression of LHY or CCA1 were observed immediately upon release into constant conditions in either rve8-1 or RVE8-OX; however, the times of peak expression were clearly altered after five days in free running conditions (Figure 6A, 6B). In rve8-1 plants, the time of peak expression was delayed whereas in RVE8-OX the time of peak expression was advanced, consistent with the respective long- and short-period CCR2::LUC phenotypes seen in these genotypes (Figure 5). In contrast, the phase of TOC1 expression was appreciably advanced in RVE8-OX in the first day of free run, although a delay in the peak phase of TOC1 expression in rve8-1 was not seen until the plants had been in free run for five days (Figure 6C). The late phase of expression of all three clock genes was rescued by introduction of a RVE8 transgene expressed under the control of the native promoter (Figure 6D–6F), indicating that the rve8-1 mutation was indeed responsible for the observed long-period phenotype. However, there were no reproducible changes in overall expression levels of CCA1, LHY, or TOC1 in either RVE8-OX or rve8-1. This suggests these core clock genes are not significant targets for RVE8.
Figure 6. Expression of CCA1, LHY, and TOC1 in rve8 and RVE8-OX.
Seedlings were entrained 12 hr light:12 hr dark for 6 days before being transferred to continuous white light and harvested at the indicated times. Expression of CCA1 (A, D), LHY (B, E), and TOC1 (C, F) was determined using qRT-PCR in (A–C) Col, rve8-1, RVE8-OX, and in (D–F) Col, rve8-1, and rve8-1 plants transformed with the RVE8 genomic region. Values are expressed relative to PP2a. Error bars represent ± SE. These data are representative of at least two independent experiments.
RVE8 acts in a light- and temperature-dependent manner
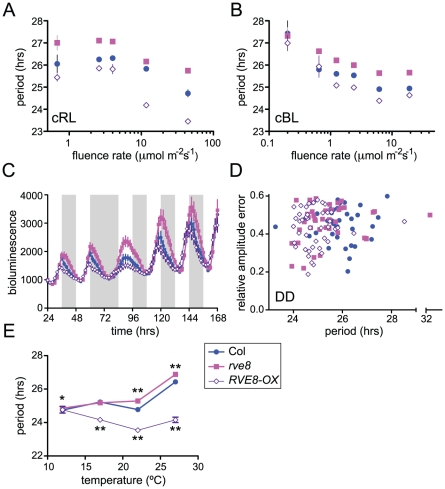
Light signaling pathways affect the circadian clock, both setting clock phase and influencing the pace at which the clock runs. When monitored under constant light conditions, Arabidopsis plants display a shorter free-running period as fluence rates are increased [47]. We speculated that the altered free-running periods in rve8-1 and in RVE8-OX might be due to altered sensitivity to light in these genotypes. To test this, we examined the effects of different fluence rates of continuous red or blue light on CCR2::LUC rhythms in Col, rve8-1, and RVE8-OX. After entrainment in white light/dark cycles, seedlings were moved to free run in continuous red or blue light of different fluence rates. In red light, both the rve8-1 long-period phenotype and the RVE8-OX short-period phenotype were observed across a wide range of fluence rates (Figure 7A). The responsiveness of rve8-1 mutants to red light was very similar to that of wild type, whereas the short-period phenotype of RVE8-OX seedlings was slightly enhanced at higher fluence rates of red light. Two-way ANOVA analysis revealed that only about 3% of the variance could be attributed to a genotype by fluence rate interaction. In contrast, the genotypes and fluence rates each accounted for approximately 30% of the total variance. This indicates that the red light sensitivity of the circadian clock in Col, rve8-1, and RVE8-OX plants is fundamentally similar.
Figure 7. RVE8 clock function is both light- and temperature-dependent.
Seedlings were entrained in 12 hr light:12 hr dark for 6 days at 22°C before being analyzed for CCR2::LUC activity in different environmental conditions. (A–B) Plants were transferred to constant red (A) or blue (B) light of the indicated fluence rates; average free-running period at each fluence rate, ± SEM is indicated. (C) Plants were maintained in the same light/dark regimen as during entrainment and then subjected to one long night before resumption of light/dark cycles. Average luciferase activity, ± SEM, is depicted. White bars represent times lights were on and grey bars times lights were off during imaging. (D) After entrainment, plants were transferred to continuous darkness. The average free-running periods were: Col = 25.87±0.14 hr; rve8-1 = 25.42±0.22 hr; RVE8-OX = 24.94±0.12 hr. The average period of RVE8-OX, but not rve8-1 plants, was significantly different from Col (p = 0.000002 and p = 0.10, respectively). (E) Seedlings were released to constant red light at the indicated temperatures; free-running period ± SEM is shown. (* indicates p<0.05; ** indicates p<0.001; Student's two-tail heteroscedastic t test used for all comparisons). These data are representative of at least two independent experiments. Note that in many cases the error bars are smaller than the symbols in the graphs.
We next examined the effects of different fluence rates of blue light on Col, rve8-1, and RVE8-OX. As is shown in Figure 7B, RVE8-OX plants had a period approximately 0.6 hours shorter than wild type at most fluence rates tested, a weaker effect than seen in constant red light. The long-period phenotype of rve8-1 plants was observed at all but the lowest fluence rate (0.2 mol m−2 s−1) tested; however, it is possible that the relatively poor rhythmicity of all three genotypes in this low-light condition may have masked a period phenotype in rve8-1. Two-way ANOVA analysis indicated there was not a significant genotype by fluence rate interaction. Therefore clock sensitivity to continuous blue light is not altered in plants with perturbed RVE8 expression.
Since mutation of genes involved in light input to the clock can affect resetting of clock phase in response to light [48], we examined how rapidly Col, rve8-1, and RVE8-OX plants re-entrained to new light conditions. Wild-type and mutant plants containing a CCR2::LUC reporter gene were entrained in 12 hrs light: 12 hrs dark and luciferase rhythms were initially monitored under the same photocycles (Figure 7C). Even in light/dark cycles, the early-phase phenotype of RVE8-OX and the late-phase phenotype of rve8-1 plants were evident. We next subjected these three genotypes to an extended night of 24 hours before resuming light/dark cycles. All three genotypes recovered from this 12 hour ‘jet lag’ treatment at approximately the same rate, regaining appropriately evening-phased CCR2::LUC activity within two days of the perturbation. Together, the fluence rate response curve and phase-resetting data strongly suggest that the period phenotypes in rve8-1 and RVE8-OX are not caused by changes in light input to the circadian clock.
To assess the importance of light for the rve8-1 and RVE8-OX period phenotypes, we examined free-running rhythms in constant darkness (DD) after entrainment in light/dark cycles. RVE8-OX plants had a period approximately one hour shorter than that of the Col control, similar to the RVE8-OX phenotype in constant red or red + blue light. In contrast, the rve8-1 free running period was slightly shorter than but not significantly different from wild type (Figure 7D). This is in marked contrast to the long-period phenotype of rve8-1 plants seen in all light conditions tested (Figure 5, Figure 6, and Figure 7A and 7B). Therefore the rve8-1 circadian phenotype is light-dependent, even though light input to the clock is not appreciably altered in this mutant.
Circadian clocks are typically robustly temperature compensated; that is, they maintain approximately the same free-running period over a physiologically relevant range of temperatures [1]. Since mutation of clock genes can specifically affect this process, we examined periodicity of CCR2::LUC activity in Col, rve8-1, and RVE8-OX plants at different ambient temperatures. rve8-1 seedlings had significantly longer free-running periods when assayed at 22°C or 27°C (Figure 7E), but essentially wild-type periodicity at 12°C or 17°C. RVE8-OX plants had significantly shorter free-running rhythms than Col at 17°C, 22°C, and 27°C but had the same period as wild-type controls at 12°C (Figure 7E). Therefore temperature compensation is disrupted in both rve8-1 and RVE8-OX plants, with loss- and gain-of-function RVE8 alleles showing normal rhythmicity at the lowest temperature assayed. This suggests that RVE8 normally functions only at the high end of the range of physiologically relevant temperatures.
RVE8 forms a negative feedback loop with PRR5
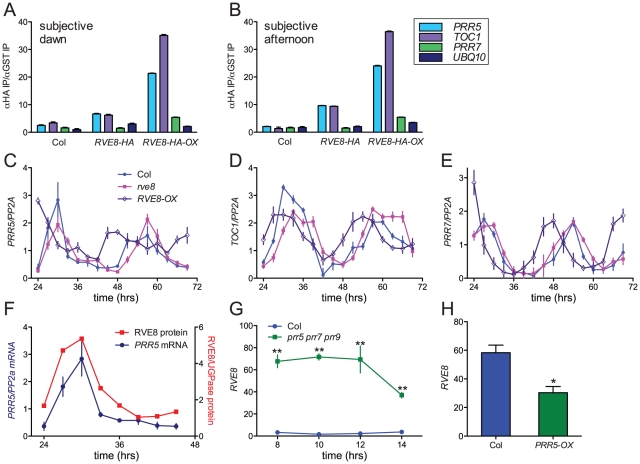
Since RVE8 binds to EE sequences in vitro, we next investigated whether it binds to promoters containing these motifs in vivo. TOC1 is perhaps the best-known clock gene that contains an EE within its regulatory region [13]. However, other genes that function within the circadian network also contain EE sequences in their promoter regions. One, PRR5, has a similar phase of expression as TOC1 and has recently been shown to regulate nuclear accumulation of TOC1 protein and to repress CCA1 and LHY expression [17], [23]. We therefore investigated whether RVE8 could bind to the EE-containing regions of the TOC1 and PRR5 promoters using chromatin immunoprecipitation (ChIP) assays followed by qRT-PCR. We used plants entrained in white light/dark cycles and then transferred to constant red light since the phenotype of RVE8-OX plants was strongest in this condition (Figure 7A, 7B). Wild-type, rve8-1 + RVE8::RVE8-HA, and rve8-1 + 35S::RVE8-HA plants were harvested at subjective dawn (Figure 8A) and in the subjective afternoon (Figure 8B). ChIPs were carried out using anti-HA antibodies (experimental samples) and anti-GST antibodies (negative controls) and the ratio of genomic DNA isolated in each type of IP was determined. In plants harvested at subjective dawn, PRR5 and TOC1 sequences were slightly enriched (∼2 fold) in extracts made from RVE8::RVE8-HA plants compared to wild-type controls. A greater enrichment for these promoter sequences (∼6 fold) was found in RVE8::RVE8-HA plants harvested in the subjective afternoon (Figure 8B). In contrast, a similar strong enrichment for TOC1 and PRR5 promoter sequences was found in extracts made from plants overexpressing RVE8 harvested at either subjective dawn or subjective afternoon (Figure 8A, 8B). These data indicate that RVE8 binds to EE-containing promoter sequences in vivo. Since RVE8 protein levels are higher in the afternoon than in the morning (Figure 4) it is not surprising to find that RVE8 expressed under its endogenous promoter has more EE binding activity at that time.
Figure 8. RVE8 and PRR5 form a negative feedback loop.
(A, B) RVE8 binds to promoter regions containing EE motifs. Col, rve8-1 + RVE8::RVE8-HA and rve8-1 + 35S::RVE8-HA seedlings were entrained in white light/dark cycles for six days before release into continuous red light (35 mol m−2 sec−1). Plants were harvested at ZT48 (A) or ZT 32 (B); chromatin immunoprecipitations (IPs) were carried out using anti-HA and anti-GST antibodies. qRT-PCR was performed using primers that amplify the EE-containing regions of the PRR5 and TOC1 promoters and the CBS-containing region of the PRR7 promoter; primers that amplify a portion of the UBQ10 locus were used as a background control. The ratios of the amount of DNA isolated in the anti-HA IPs vs. the control anti-GST IPs are presented. (C–E) Expression of clock-associated genes in Col, rve8-1 and RVE8-OX. Plants were entrained as described for panels A–B and samples were harvested at the indicated times. Expression of PRR5 (C), TOC1 (D), and PRR7 (E) was determined using qRT-PCR. Values are expressed relative to PP2a. Similar results were obtained in two independent experiments. (F–H) Regulatory interactions between RVE8 and PRR5. (F) Relative abundance of RVE8 protein and PRR5 mRNA (re-plotted from Figure 4B and Figure 8C). (G) RVE8 transcript levels are elevated in prr5 prr7 prr9 mutants; data are derived from previously-published microarray data [84], [85]. (H) RVE8 transcript levels are reduced in plants expressing PRR5 under the control of the strong viral 35S promoter. Data are derived from microarray data available at NASC (submitted by Hitoshi Sakakibara; NASCArrays experiment reference number NASCARRAYS-420). ** indicates p<0.01 and * indicates p<0.05; Student's two-tail heteroscedastic t test. Error bars represent ± SE.
In addition to binding to EE sequences, CCA1 and LHY are thought to regulate clock gene expression by binding to a related motif termed the CCA1 binding site (CBS) [14], [49]; for example, CCA1 and LHY are thought to promote expression of PRR7 by binding to a CBS-containing region of the PRR7 promoter [16]. We therefore examined the ability of RVE8 to bind to this region of the PRR7 promoter by ChIP. However, at both subjective dawn and afternoon there was not appreciable enrichment of the PRR7 promoter region in plants expressing RVE8 under its native promoter, and only a very modest enrichment was seen in plants overexpressing RVE8 (Figure 8A, 8B). These data suggest that RVE8 does not bind to the CBS-containing portion of the PRR7 promoter in vivo.
We next examined expression levels of PRR5, PRR7, and TOC1 in plants grown in the same conditions used for the ChIP assays. PRR5 levels were very similar in Col and rve8-1 two and three days after transfer to constant environmental conditions. However, trough levels of PRR5 were significantly elevated in RVE8-OX plants; this was especially apparent during day three of free run (Figure 8C). In contrast, peak and trough expression levels of TOC1 and PRR7 were very similar in Col, rve8-1, and RVE8-OX (Figure 8D, 8E).
Since RVE8 binds to the PRR5 promoter and PRR5 transcript levels are elevated in plants overexpressing RVE8, we examined the temporal relationship between amounts of RVE8 protein and PRR5 transcript (Figure 8F). Their levels are highly correlated, with both peaking in the subjective afternoon and having low levels during the subjective night. Together with the direct binding of RVE8 to the PRR5 promoter and the increase in PRR5 expression in RVE8-OX plants, this suggests that RVE8 directly and positively regulates PRR5 expression. In contrast, TOC1 levels are not appreciably altered in RVE8 loss- or gain-of-function alleles (Figure 6C and Figure 8D), suggesting that although RVE8 binds to the TOC1 promoter in vivo it may not regulate TOC1 expression.
PRR5, PRR7, and PRR9 have recently been shown to be direct and potent repressors of CCA1 and LHY expression [17]. Since RVE8 has a very similar pattern of gene expression to these homologs (Figure 1), we examined whether RVE8 expression was altered in prr5 prr7 prr9 triple mutants and in PRR5-overexpressing plants. Using publicly accessible microarray data [50], we found that RVE8 levels were significantly increased in prr5 prr7 prr9 triple mutants and decreased in PRR5-overexpressing plants (Figure 8G, 8H). This indicates that PRR5 represses RVE8 expression, either directly or indirectly. RVE8 and PRR5 therefore comprise a novel negative feedback loop within the plant circadian network.
Discussion
RVE8 is a clock-regulated EE–binding protein
We isolated RVE8 from plant extracts based upon its ability to bind to the EE. Further investigation in yeast and in vitro (Figure 2) revealed its EE-binding affinity is similar to that of both CCA1 and RVE1, which is not surprising since these proteins all contain a similar Myb-like/SANT DNA binding domain. The Myb-like domain of RVE8 shares 64% identity and 86% similarity with that of CCA1; its relatedness to LHY and RVE1 is similar. Sequence similarity between these proteins is also seen in short basic regions immediately N-terminal and short proline-rich regions immediately C-terminal to the Myb-like domains. These three regions are conserved among all 11 of the RVE-related proteins [28]. A search of the Eukaryotic Linear Motif resource database [51] suggests a portion of the conserved proline-rich region (PRPKRKAA in RVE8) may act as a nuclear localization signal while part of the conserved basic region (RKPYTIT in RVE8) may be a binding site for 14-3-3- proteins. 14-3-3 proteins often bind to their ligands in a phosphorylation-dependent manner, and this binding may affect client protein activity or intracellular localization [52]. It will be very interesting to determine whether the RVE-related proteins are regulated in this manner.
It is notable that despite the similar affinity of RVE8, RVE1, and CCA1 for the EE (Figure 2B) we identified peptides derived from RVE4, RVE5, RVE6 and RVE8 but not from other RVE-related proteins in our affinity purifications. This may be explained by our finding that RVE8 protein levels are high in the subjective afternoon (Figure 4) whereas CCA1 and LHY proteins are difficult to detect at that time [33], [39]. It will be of interest to determine whether RVE4, RVE5, and RVE6 protein levels are also highest in the subjective afternoon.
RVE8 acts close to the circadian oscillator in a temperature- and light-dependent manner
Mutations in clock genes often affect temperature compensation, the ability of the circadian system to run at a similar pace across a wide range of temperatures. For example, mutation of casein kinase 2 in Neurospora, casein kinase I epsilon in hamster, or PERIOD in Drosophila, affects clock pace and disrupts temperature compensation [53]–[55]. Similarly, mutation of CCA1 or LHY differentially affects circadian period at different temperatures in Arabidopsis. CCA1 function is more important at lower temperatures while LHY function is more important at higher temperatures [56]. We show that RVE8 is also involved in temperature compensation, but in a different manner from its homologs: the period phenotypes of the RVE8 mutant and overexpressing plants completely dissipated at the lowest temperatures tested (Figure 7E). It has recently been reported that prr7 prr9 mutants have a similar temperature compensation phenotype as rve8 mutants, with normal free-running rhythms at low temperature and longer rhythms at higher temperatures [57]. It will be very interesting to determine whether other RVE genes also exhibit temperature-dependent phenotypes and to investigate possible temperature-dependent regulatory relationships between the RVEs and the PRRs.
Light is a potent regulator of most aspects of plant growth and development, not only directly affecting many processes that are also modulated by the circadian clock [35], [58] but also influencing clock function directly. Illustrating the intertwined natures of the clock and light signaling networks, many genes that act close to the plant circadian oscillator, such as TOC1, ZTL, PRR5 and PRR7, also function in light signaling pathways [59]–[62]. It can therefore be difficult to determine whether clock mutant phenotypes are due to alterations in light signaling, the circadian oscillator, or both. To address this point, we examined the light-dependence of developmental and clock phenotypes in rve8-1 and RVE8-OX plants.
Using light inhibition of hypocotyl elongation to investigate the role of RVE8 in light-regulated development, we found that rve8-1 and RVE8-OX seedlings had long and short hypocotyls, respectively, at low fluence rates of continuous white light. Thus RVE8 differs from its characterized homologs in that it represses rather than promotes hypocotyl elongation. Interestingly, the RVE8 phenotypes were less obvious at fluence rates of 8 mol m−2 s−1 or higher, and almost absent at a fluence rate of 85 mol m−2 s−1 (Figure 3C). This type of light-dependent phenotype is reminiscent of mutants in the phyA signaling pathway such as fhy1 [63], and suggests that RVE8 may be a positive mediator of the very low fluence response. None of the other characterized RVE-related genes shows a similar low-light dependent hypocotyl phenotype, indicating they affect hypocotyl growth in a fundamentally different manner from RVE8 [28]–[30], [33], [34], [59].
Unlike the hypocotyl phenotypes, rve8-1 and RVE8-OX displayed similar circadian period phenotypes across a wide range of fluence rates (Figure 7A, 7B). This discrepancy strongly suggests that the primary mechanism by which RVE8 affects the pace of the clock is not via a light input pathway. In support of this conclusion, rve8-1 and RVE8-OX showed similar kinetics of clock re-setting in a jet lag experiment (Figure 7C). Given these results, it was somewhat surprising to discover that rve8-1 mutants did not have a long-period phenotype when assayed in constant darkness (Figure 7D), demonstrating that RVE8 function is light-dependent. However, this is not a unique observation since other clock-associated genes including EARLY FLOWERING 3 (ELF3), JUMONJI DOMAIN-CONTAINING PROTEIN 5 (JMJD5), PRR5, and PRR9 have been reported to have light-dependent period phenotypes [64]–[66]. Together, these data indicate that RVE8 functions in multiple signaling pathways, affecting light signaling and clock pace via different mechanisms.
What is causing the period phenotypes in rve8-1 and RVE8-OX plants? By analogy with CCA1 and LHY, one obvious role for RVE8 might be the direct regulation of TOC1. Indeed, we found that RVE8 binds directly to the TOC1 promoter in vivo (Figure 8A, 8B). However, the overall levels of TOC1 expression were not appreciably altered in either rve8-1 or RVE8-OX (Figure 6C, 6F, and Figure 8D). This suggests that despite the ability of RVE8 to bind the TOC1 promoter, TOC1 is not an important target for RVE8 function. It is not uncommon for transcription factors to bind to the promoters of genes in vivo but not affect their expression levels [67], [68]. This discrepancy may be explained by the requirement at some promoters for specific combinations of transcription factors to activate gene expression [69].
Whereas we found no changes in TOC1 expression levels in plants misexpressing RVE8, trough levels of PRR5 transcript were increased in RVE8-OX plants (Figure 8C). Combined with the ability of RVE8 to bind the PRR5 promoter and the correlation between RVE8 protein and PRR5 message levels (Figure 8A, 8B, 8F), this suggests that PRR5 is a direct target of RVE8. However, PRR5 levels are not affected in rve8 mutants. Given that we isolated not only RVE8 but also its homologs RVE4, RVE5, and RVE6 bound to the EE (Table S1), it may be that these proteins function in a partially redundant manner. The lack of apparent changes in PRR5 levels in rve8-1 (Figure 8C) might be caused by such redundancy. Similar findings have been previously reported; for example, cca1 and lhy single mutants have no apparent change in TOC1 levels but cca1 lhy double mutants have greatly increased levels of TOC1 [37]. CCA1 transcript levels are only slightly altered in prr5, prr7, or prr9 single mutants [16], [62], [66], [70], [71] but are greatly increased in prr7 prr9 double and prr5 prr7 prr9 triple mutants [15], [16]. We are currently generating higher-order rve mutants to investigate whether PRR5 levels are altered in these plants. It seems likely that RVE8 has many targets and that the long-period rve8 phenotype is due to subtle alterations in expression levels of multiple clock genes.
Members of the three clades of Myb-like proteins play distinct roles in the Arabidopsis circadian system
Members of the three separate clades of the CCA1/LHY/RVE family of transcription factors have now been found to fall into separate functional categories. CCA1 and LHY are the best-studied and affect clock pace by repressing TOC1 expression and promoting expression of PRR7 and PRR9 [13], [16]. They also profoundly influence control of hypocotyl elongation and flowering time by regulating expression of the PHYTOCHROME INTERACTING FACTOR4 (PIF4) and PIF5 transcription factors and genes in the photoperiodic pathway, respectively [36], [72]. RVE1 acts primarily as a clock output gene, regulating daily rhythms in auxin production but not playing an important role in clock function [28].
We now show that RVE8 plays a distinct role in the light signaling and circadian networks. RVE8 affects light inhibition of hypocotyl elongation at low light intensities, suggesting it may affect the phytochrome-mediated very low fluence response, a novel phenotype for a clock mutant. RVE8 also acts in temperature compensation, suggesting that its activity is important for fine-tuning clock function in different environmental conditions.
Our findings also provide insight into the architecture of the transcriptional networks that make up the plant circadian clock. PRR5, PRR7, and PRR9 act sequentially to repress CCA1 and LHY expression throughout the day [17]. While CCA1 and LHY promote expression of the morning-phased genes PRR7 and PRR9 [16], the mode of regulation of the afternoon-phased gene PRR5 has not previously been reported. We now provide evidence that RVE8 promotes PRR5 expression and that PRR5 represses RVE8 expression (Figure 8), the same type of regulatory interactions previously reported for their homologs [16], [17]. However, the delay in accumulation of RVE8 protein relative to its transcript likely accounts for the delayed phase of PRR5 accumulation compared to PRR7 and PRR9. The post-transcriptional mechanisms controlling RVE8 protein accumulation may therefore be key to proper functioning of the clock network.
All of the CCA1/LHY/RVE transcription factors that have been characterized to date have similar in vitro binding affinity for the EE [13], [27], [28], leaving open the question as to why their mutant phenotypes are so different. Inspection of the AtGenExpress developmental data set shows that individual family members are expressed throughout plant development and in most organs. In general, expression levels are lower in roots than in aerial tissues and expression is highest in germinating seedlings, flowers, and developing siliques [73]. Since there are no obvious differences in spatial and developmental expression patterns, their contrasting functions in the circadian system are likely due primarily to post-transcriptional differences. Our data suggest at least two such mechanisms are crucial: differential regulation of protein accumulation and differential association of co-regulators at target promoters. The study of these regulatory mechanisms will now be a high priority as we begin to resolve the disparate circadian functions of this fascinating group of transcription factors.
Materials and Methods
Purification of the evening element–binding proteins
Whole-cell extracts were generated from Arabidopsis plants as previously described [27]. 5′-biotinylated oligonucleotides containing four tandem repeats of the EE_wt sequence (AAAATATCT) or EE_mt sequence (AAAATcgag) were purchased from Sigma (St. Louis, MO) and annealed together to obtain double stranded DNA (dsDNA). 18 mg of DYNAL®M-280 Streptavidin (Invitrogen) beads (1.8 ml) were washed twice with wash buffer [10 mM Tris-HCl (pH 7.5), 1 mM EDTA and 2.0 M NaCl] and then incubated with 50 pmoles of biotinylated dsDNA/mg DYNAL beads for 30 min at room temperature in incubation buffer [5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA and 1.0 M NaCl]. Strepatividin beads bound to biotinylated DNA were washed twice with incubation buffer and stored at 4°C until further use. The beads were washed twice with reaction buffer [20 mM HEPES, pH 7.2, 80 mM KCl, 10% glycerol, 0.1 mM EDTA, 8 ng/µl of poly (dI-dC)] and incubated with whole cell extracts (∼100 mg total protein at 20 mg/ml, which was extracted from ∼250 g of Arabidopsis tissue) for 30 min at 4°C with gentle shaking. Beads were then washed twice with reaction buffer containing 0.5 mg/ml of salmon sperm DNA (ssDNA), followed by three washes with 100 mM ammonium bicarbonate and one wash with 50 mM ammonium bicarbonate solution. The beads were stored at 4°C in 50 mM ammonium bicarbonate solution until further use. Protein bound to the beads was digested by addition of 10 µl of diluted sequencing-grade Promega trypsin (13 ng/ul) (Promega, Madison, WI) and incubation at 37°C for 6 hr. Supernatant containing the digested peptides were removed and acidified with trifluoroacetic acid to a final concentration of 0.1%. The samples were stored at −80°C and were analyzed by LC-MS/MS.
LC-MS/MS analysis
Nano LC-MS/MS analysis was performed on a LTQ linear ion trap mass spectrometer (Thermo-Scientific, San Jose, CA), equipped with a Picoview nanospray source (New Objective, Woburn, MA), and an Eksigent nano 2d HPLC and autosampler (Eksigent, Dublin, CA). The tryptic peptide mixture was separated on a 75 µm ID PicoFrit column packed in-house with Magic C18AQ (Michrom BioResources, Auburn, CA) to a length of 15 cm with a 100% MeOH slurry of C18 reversed-phase material (100 Å pore size, 3 µm particle size) using a high-pressure cell pressurized with helium. The column was pre-equilibrated for 10 min at 2% solvent B [0.1% (v/v) formic acid in acetonitrile] and 98% solvent A [0.1% (v/v) formic acid in water] at a flow rate of 300 nL/min. Separation was achieved using a linear gradient from 2 to 60% solvent B in 45 min at a flow rate of 300 nL/min. The LTQ mass spectrometer was operated in the data dependent acquisition mode using a standard TOP10 method: 1 full-scan MS acquired was followed by 10 MS/MS scans.
Database searching and criteria for protein identification
Tandem mass spectra were extracted by Bioworks version 3.2. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version TORNADO (2010.01.01.4)). X! Tandem was set up to search the Uniprot Knowledge Base Arabidopsis thaliana complete proteome set database (Release 2010_08 July 12 2010, 31975 entries) and the cRAP common laboratory artifacts database (release 1.0, 112 entries) plus an equal number of reverse decoy sequences assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 1.8 Da. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulphone of methionine, tryptophan oxidation to formylkynurenin of tryptophan and acetylation of the n-terminus were specified in X! Tandem as variable modifications. Next, Scaffold (version Scaffold_3_00_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability as specified by the Peptide Prophet algorithm [74]. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm [75]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Using the above criteria the false discovery rate (FDR, Decoy/Target) was calculated as 0.1% on the peptide level and 2.4% on the protein level using a target-decoy (reverse) search strategy [76].
Plant materials
T-DNA insertion mutant rve8-1 (SALK_053482) was obtained from ABRC; PCR with RP 5′-AGTTTGCTGCTGATTTCTGAG-3′ and LP 5′-TTCAGCAAAATCAGGAACACC-3′ generates an approximately 1.1 kb band in Col but not in the mutant. RP with LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′ gives a band of 700 bp in the mutant. Binary vector containing CCR2:: LUC+ [77] was transformed into rve8-1 and wild type (Col) by Agrobacterium mediated transformation [78]. Primary transformants were screened on MS medium containing 6.5 mg/ml gentamycin (EMD Chemicals) to select transformants. TOC1::LUC reporter [77] was introgressed into rve8-1 by crossing. For overexpression and rescue experiments, binary vectors containing either CaMV35S::HA-RVE8 or RVE8:: RVE8-HA were transformed by vacuum infiltration [78] into Col reporter lines also containing CCR2::LUC+. Binary vector containing PRR5::LUC2+ reporter was also transformed into Col and rve8-1 by Agrobacterium mediated transformation [78]. Primary transformants were selected on MS medium containing 150 µM basta (Chem Services, West Chester, PA).
Vectors
To generate the overexpression binary vector containing CaMV35S::HA-RVE8, a 0.9 Kb RVE8 cDNA was PCR-amplified with Pfu ultra high fidelity enzyme (Stratagene) using primers F: 5′- CACCAGCTCGTCGCCGTCAAGAAATCC -3′ and R: 5′-TTGTTATGCTGATTTGTCGCTTGTTGAG -3′ and the plasmid U19901 obtained from Arabidopsis Biological Resource Center (ABRC) as a template. The PCR amplified product was cloned into pENTR/D-TOPO (Invitrogen) followed by LR recombination with pEarleyGate201. To generate the RVE8 rescue construct, a 2.5Kb genomic fragment containing ∼0.7Kb RVE8 upstream sequence was amplified from Arabidopsis genomic DNA using primers F: 5′- CACCTGTTTCGTAAGATTTGAATACAAAACCG -3′ and R: 5′- TGCTGATTTGTCGCTTGTTGAGTTC -3′ and Pfu ultra high fidelity enzyme (Stratagene) according to the manufacturer's protocol. The PCR amplified product was cloned into pENTR/D-TOPO (Invitrogen) followed by LR recombination with pEarleyGate301 to generate a binary vector containing RVE8::RVE8-HA. The 3kb region upstream of PRR5 was PCR-amplified using primers F 5′-CACCAGATTTTGTCACGCATCATTTTT-3′and R 5′-CAGCAAAATACTGTATACGAGACAAA-3′and using Col DNA as template. This fragment was cloned into pENTR/D-TOPO followed by LR recombination with pEARLYGATE 301-LUC2 [28] to generate a binary vector containing PRR5::LUC2. All clones were sequenced for any PCR-generated errors before being moved into Agrobacterium strain GV3101.
Yeast one-hybrid
The full length cDNA of RVE8 was moved from the pENTR/D-TOPO vector to pDEST22 containing GAL4AD by LR recombination. Yeast one-hybrid assays were performed as previously described [28].
Gel shift assay
The His-RVE8 vector was generated by LR recombination of full length RVE8 cDNA in pENTR/D-TOPO with pDEST 15 (Invitrogen). Gel shift assays were performed as previously described [28].
Luciferase imaging
Luciferase imaging was performed as previously described [28]. Seeds were entrained in 12 hour white light (50 mol m−2 sec−1, provided by cool white fluorescent bulbs)/12 hour dark cycles at 22°C for 6 days before being released into the indicated conditions for luciferase activity analysis, using either an ORCA II ER (Hamamatsu) or a DU434-BV (Andor Technology) CCD camera. Illumination was provided by red and/or blue LED Snap-Lites (Quantum Devices). Neutral density filters (RoscoLux no. 98 and no. 398) were used to obtain specific fluence levels for the fluence rate response curves. Images were analyzed using MetaMorph (Molecular Devices) and rhythms were estimated by Fourier Fast Transform-Non-Linear Least Squares [79].
Hypocotyl length and flowering time
For hypocotyl measurements, seeds were stratified at 4°C in the dark for 48 hours and sown on MS medium (0.8% agar and 3% sucrose) in petri plates. The plates were treated with white light (55 µmol m-2s-1) for 6 hours, and then either kept either in darkness or grown either under constant white light or in 8 hour light/16 hours dark cycles using neutral density filters (RoscoLux no. 98and no. 398) to obtain the indicated fluence rates. After 6 days of growth, seedlings were transferred to transparencies and scanned. Individual hypocotyl was measured using the application ImageJ (http://rsb.info.nih.gov/ij). For flowering time analysis, seeds were soaked in 0.1% agar in the dark at 4°C for 3 days and then sown in soil. Plants were grown either in short day (8 hours light/16 hours dark) or long day (16 hrs light/8 hours dark) conditions at 22°C, and monitored daily for bolting.
qRT–PCR
Plants were grown, RNA isolated, and qRT-PCR performed as previously described [80], with the following modifications. Seedlings were entrained in 12 hour white light (50 mol m−2 sec−1, provided by cool white fluorescent bulbs)/12 hour dark cycles at 22°C for 6 days before being released into either constant white light (50 mol m−2 sec−1) provided by cool white fluorescent bulbs or constant red light (35 mol m−2 sec−1) provided by red LED Snap-Lites (Quantum Devices). Samples were run in triplicate using iQ5 (Bio-Rad), and starting quantity was estimated from critical thresholds using the standard curve method. Data for each sample were normalized to the respective PROTEIN PHOSPHATASE 2a (PP2a) expression level. Primer sets used for amplification of CCA1, LHY, TOC1, PRR5, PRR7, and PP2a have been described [80], [81]. RVE8 mRNA was amplified using primers F 5′- GGGAAGCTCAAGCCGAACAGTATC-3′ and R 5′- GGCCTCTCGTTTCAGGATCAAAGA-3′, which flank the T-DNA insertion site in rve8-1.
Immunoblot analysis
For each time point, approximately forty RVE8::RVE8-HA or 35S::RVE8-HA seedlings were collected, frozen in liquid nitrogen and stored at −80°C until analysis. Plant tissue was ground in homogenization buffer (25 mM MOPS (pH 7.8), 0.25 M sucrose, 0.1 mM MgCl2, Complete EDTA-free protease-inhibitor cocktail (Roche) at 4°C. Protein concentrations of total cell extracts were then determined by Bradford assay (Bio-rad). 50 µg of each sample was analyzed by immunoblotting, using anti-HA-antibody conjugated to peroxidase (Roche, 3F10), or anti-UGPase antibody (AS05086, AgriSera, Vännäs, Sweden) followed by a secondary antibody, goat anti-rabbit IgG-HRP (1858415, Pierce). ECL Plus reagent (GE Healthcare) was used to generate chemiluminescence which was then detected with BioMax Light Film (Kodak). Data was quantified using ImageQuant software (GE Healthcare).
Chromatin immunoprecipitation (ChIP)
ChIP on wild type or plants expressing 35S::HA-RVE8 or RVE8::RVE8-HA was carried out as previously described [82]. Plants were entrained in 12 hour white light (50 mol m−2 sec−1, provided by cool white fluorescent bulbs)/12 hour dark cycles at 22°C for 6 days before being released into constant red light (35 mol m−2 sec−1) provided by red LED Snap-Lites (Quantum Devices). Seedlings were harvested at ZT48 (subjective dawn) or ZT32 (subjective afternoon). Immunoprecipitation was carried out using either an anti-HA antibody (Sigma, catalog #SAB4300603) or an anti-GST antibody (Santa Cruz Biotechnology, catalog #sc-459) as a negative control. Primers used to amplify the region -468/-345 basepairs upstream of the predicted PRR5 translational start site (containing one EE) were: (F) 5′-TGCAAACCTATGTACCAAACAGA-3′ and (R) 5′-TCCCACTCGTGACTTT-3′. Primers used to amplify the region -881/-701 basepairs upstream of the predicted TOC1 translational start site (containing one EE) were: (F): 5′-TGGTTTGGTCTGATCTGGTCAT-3′ and (R): 5′-AGGCCACGTCATCTTGGAGAAA-3′. Primers used to amplify the region -915/-765 basepairs upstream of the predicted PRR7 translational start site (containing three CCA1 binding sites) were: (F): 5′-CACGTGTAATGGTGGGTAAGG-3′ and (R): 5′-TGGGTTAAAATCTTTTTGAATGG-3′. The primer set used to amplify the UBQ10 locus as a negative control has been previously described [14].
Accession numbers
The mass spectrometry data associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hash:
4fTHRVBPxyFD+GzvduEyt/sCUPO+bIIc4aNGUl1EUNGIaTr0jgZdcpdX5Ivu19clTLIQiHcaUDNICymEkeEGuhyPP+YAAAAAAAAL8A = =
The hash may be used to prove exactly what files were published as part of this manuscript's dataset, and the hash may also be used to check that the data has not changed since publication.
Supporting Information
Isolation of plants overexpressing or deficient for RVE8. (A, B) RVE8 expression was determined in Col and RVE8-OX (A) and rve8-1 (SALK_053282) (B) plants using qRT-PCR. Values are expressed relative to PP2a. Error bars represent ± SE. (C) Locations of the T-DNA insertion site in rve8-1 and the primers used for qRT-PCR analysis. The boxes represent exons; open boxes signify untranslated regions while filled boxes signify coding regions.
(0.05 MB PDF)
All peptides derived from RVE family proteins identified in LC-MS/MS experiments. In three independent experiments, plant extracts were incubated with either wild-type or mutant versions of the EE bound to beads. Purified proteins were digested with trypsin and peptides were identified after LC-MS/MS as described in the Materials and Methods section. All peptides that could have been derived from a RVE family protein are shown; the percentages indicate the probability that the indicated peptide spectrum match (PSM) is correct according to the Scaffold 3 proteomics software (Proteome Software, Portland OR). Note that some peptides could have been derived from multiple RVE proteins.
(0.01 MB XLS)
All peptides identified in LC-MS/MS experiments. In three independent experiments, plant extracts were incubated with either wild-type or mutant versions of the EE bound to beads. Purified proteins were digested with trypsin and peptides were identified after LC-MS/MS as described in the Materials and Methods section. The peptide numbers reported are based on scaffold grouping. Complete protein identification data and statistics may be downloaded from the ProteomeCommons.org Tranche network using the following hash: 4fTHRVBPxyFD+GzvduEyt/sCUPO+bIIc4aNGUl1EUNGIaTr0jgZdcpdX5Ivu19clTLIQiHcaUDNICymEkeEGuhyPP+YAAAAAAAAL8A = =
(0.10 MB XLS)
Acknowledgments
We would like to thank Julie Pelletier for technical advice on ChIP experiments and J. N. Maloof and members of the Harmer laboratory for helpful discussion and comments on the manuscript. The rve8-1 mutant was generated by the Ecker laboratory [38] and obtained from the Arabidopsis Biological Resource Center.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the National Institutes of Health grant GM069418 and National Science Foundation grant 0616179 (SLH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 2.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 2003;132:629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 6.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 7.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap JC, Loros JJ, Colot HV, Mehra A, Belden WJ, et al. A circadian clock in Neurospora: how genes and proteins cooperate to produce a sustained, entrainable, and compensated biological oscillator with a period of about a day. Cold Spring Harb Symp Quant Biol. 2007;72:57–68. doi: 10.1101/sqb.2007.72.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehra A, Baker CL, Loros JJ, Dunlap JC. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 14.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. Plant Cell Physiol; 2005. Pseudo-Response Regulators, PRR9, PRR7, and PRR5, Play Together Essential Roles Close to the Circadian Clock of Arabidopsis thaliana. [DOI] [PubMed] [Google Scholar]
- 16.Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 17.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 20.Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara S, Wang L, Han L, Suh SS, Salome PA, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo- response regulator proteins. J Biol Chem. 2008 doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 22.Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 25.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003;133:1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci U S A. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuno N, Moller SG, Shinomura T, Xu X, Chua NH, et al. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell. 2003;15:2476–2488. doi: 10.1105/tpc.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Chen Y, Wang ZY, Chen Z, Gu H, et al. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 2007;51:512–525. doi: 10.1111/j.1365-313X.2007.03156.x. [DOI] [PubMed] [Google Scholar]
- 31.Gong W, Hea K, Covington MF, Dinesh-Kumar SP, Snyder M, et al. The development of protein microarrays and their applications in DNA–protein and protein–protein interaction analyses of Arabidopsis transcription factors. Molecular Plant. 2008;1:27–41. doi: 10.1093/mp/ssm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170:387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 34.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 35.Nozue K, Maloof JN. Diurnal regulation of plant growth. Plant Cell Environ. 2006;29:396–408. doi: 10.1111/j.1365-3040.2005.01489.x. [DOI] [PubMed] [Google Scholar]
- 36.Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr Opin Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 38.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 39.Kim JY, Song HR, Taylor BL, Carre IA. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. Embo J. 2003;22:935–944. doi: 10.1093/emboj/cdg075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farre EM, Kay SA. PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 2007;52:548–560. doi: 10.1111/j.1365-313X.2007.03258.x. [DOI] [PubMed] [Google Scholar]
- 41.Ito S, Nakamichi N, Kiba T, Yamashino T, Mizuno T. Rhythmic and light-inducible appearance of clock-associated pseudo-response regulator protein PRR9 through programmed degradation in the dark in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:1644–1651. doi: 10.1093/pcp/pcm122. [DOI] [PubMed] [Google Scholar]
- 42.Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 44.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci U S A. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- 47.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 48.McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, et al. ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 2007;144:391–401. doi: 10.1104/pp.107.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, et al. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell. 1997;9:491–507. doi: 10.1105/tpc.9.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Craigon DJ, James N, Okyere J, Higgins J, Jotham J, et al. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 2004;32:D575–577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oecking C, Jaspert N. Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr Opin Plant Biol. 2009;12:760–765. doi: 10.1016/j.pbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, et al. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tosini G, Menaker M. The tau mutation affects temperature compensation of hamster retinal circadian oscillators. Neuroreport. 1998;9:1001–1005. doi: 10.1097/00001756-199804200-00009. [DOI] [PubMed] [Google Scholar]
- 55.Konopka RJ, Pittendrigh C, Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J Neurogenet. 1989;6:1–10. doi: 10.3109/01677068909107096. [DOI] [PubMed] [Google Scholar]
- 56.Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18:1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salome PA, Weigel D, McClung CR. The Role of the Arabidopsis Morning Loop Components CCA1, LHY, PRR7, and PRR9 in Temperature Compensation. Plant Cell. 2010;22:3650–3661. doi: 10.1105/tpc.110.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dowson-Day MJ, Millar AJ. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999;17:63–71. doi: 10.1046/j.1365-313x.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 59.Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kevei E, Gyula P, Hall A, Kozma-Bognar L, Kim WY, et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 2006;140:933–945. doi: 10.1104/pp.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito S, Nakamichi N, Nakamura Y, Niwa Y, Kato T, et al. Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:971–983. doi: 10.1093/pcp/pcm063. [DOI] [PubMed] [Google Scholar]
- 62.Kaczorowski KA, Quail PH. Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell. 2003;15:2654–2665. doi: 10.1105/tpc.015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanovsky MJ, Whitelam GC, Casal JJ. fhy3-1 retains inductive responses of phytochrome A. Plant Physiol. 2000;123:235–242. doi: 10.1104/pp.123.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, et al. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 65.Jones MA, Covington MF, Ditacchio L, Vollmers C, Panda S, et al. Proc Natl Acad Sci U S A; 2010. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ. Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta. 2003;218:159–162. doi: 10.1007/s00425-003-1106-4. [DOI] [PubMed] [Google Scholar]
- 67.Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell. 2009;21:2563–2577. doi: 10.1105/tpc.109.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oh E, Kang H, Yamaguchi S, Park J, Lee D, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernard F, Krejci A, Housden B, Adryan B, Bray SJ. Specificity of Notch pathway activation: twist controls the transcriptional output in adult muscle progenitors. Development. 2010;137:2633–2642. doi: 10.1242/dev.053181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ito S, Matsushika A, Yamada H, Sato S, Kato T, et al. Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:1237–1245. doi: 10.1093/pcp/pcg136. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto Y, Sato E, Shimizu T, Nakamich N, Sato S, et al. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 2003;44:1119–1130. doi: 10.1093/pcp/pcg148. [DOI] [PubMed] [Google Scholar]
- 72.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 73.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 74.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 75.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 76.Tabb DL. What's driving false discovery rates? J Proteome Res. 2008;7:45–46. doi: 10.1021/pr700728t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 78.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 79.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 80.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007;143:473–486. doi: 10.1104/pp.106.088757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci U S A. 2004;101:12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, et al. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc Natl Acad Sci U S A. 2008;105:3151–3156. doi: 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci U S A. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolation of plants overexpressing or deficient for RVE8. (A, B) RVE8 expression was determined in Col and RVE8-OX (A) and rve8-1 (SALK_053282) (B) plants using qRT-PCR. Values are expressed relative to PP2a. Error bars represent ± SE. (C) Locations of the T-DNA insertion site in rve8-1 and the primers used for qRT-PCR analysis. The boxes represent exons; open boxes signify untranslated regions while filled boxes signify coding regions.
(0.05 MB PDF)
All peptides derived from RVE family proteins identified in LC-MS/MS experiments. In three independent experiments, plant extracts were incubated with either wild-type or mutant versions of the EE bound to beads. Purified proteins were digested with trypsin and peptides were identified after LC-MS/MS as described in the Materials and Methods section. All peptides that could have been derived from a RVE family protein are shown; the percentages indicate the probability that the indicated peptide spectrum match (PSM) is correct according to the Scaffold 3 proteomics software (Proteome Software, Portland OR). Note that some peptides could have been derived from multiple RVE proteins.
(0.01 MB XLS)
All peptides identified in LC-MS/MS experiments. In three independent experiments, plant extracts were incubated with either wild-type or mutant versions of the EE bound to beads. Purified proteins were digested with trypsin and peptides were identified after LC-MS/MS as described in the Materials and Methods section. The peptide numbers reported are based on scaffold grouping. Complete protein identification data and statistics may be downloaded from the ProteomeCommons.org Tranche network using the following hash: 4fTHRVBPxyFD+GzvduEyt/sCUPO+bIIc4aNGUl1EUNGIaTr0jgZdcpdX5Ivu19clTLIQiHcaUDNICymEkeEGuhyPP+YAAAAAAAAL8A = =
(0.10 MB XLS)