Abstract
Phytochemical investigation of the whole plant of Lepisorus contortus (Christ) Ching led to the isolation of five new phenylethanoid glycosides (1–5), each containing a caffeoyl group, a new flavonoid glycoside (10), as well as 14 known compounds (6–9 and 11–15, syringic acid, vanillic acid, phloretic acid, diplopterol, and β-sitosterol). This is the first report of phenylethanoid glycosides from the family Polypodiaceae. Compounds 1–15 were evaluated for their cancer chemopreventive potential based on their ability to inhibit tumor necrosis factor alpha (TNF-α)-induced NF-κB activity, nitric oxide (NO) production, aromatase, quinone reductase 2 (QR-2), and COX-1/-2 activities. Quercetin-3-O-β-D-glucoside (15) demonstrated inhibition against QR2 with an IC50 value of 6.7 µM, which confirmed kaempferol/quercetin glycosides as the active compounds to inhibit QR2. The compound also demonstrated NF-κB activity with an IC50 value of 33.6 µM. In addition, compounds 1, 2, 4 and 6 showed aromatase activity with IC50 values of 30.7, 32.3, 26.8, and 35.3 µM, respectively.
Lepisorus contortus, belonging to the family Polypodiaceae, is a fern that is widely distributed in China and India. It grows on rocks and tree trunks under moist conditions at 1450–2600 m above sea level, and has been used in a folk medicine for trauma, burns, and scald injuries.1,2 Although the genus Lepisorus contains approximately 70 species, only two have been phytochemically investigated previously, which led to the identification of five flavonoids and three steroids from L. ussuriensis,3,4 and two phenylpropanoids and one flavonoid from L. thunbergianus.5 In our search for new cancer chemoprentive agents from medicinal plants of Yunnan, China, we isolated 20 natural compounds including five new phenylethanoid glycosides (1–5) and a new flavone glycoside (10) from the fern L. Contortus. This is also the first report of phenylethanoid glycosides from the family Polypodiaceae. The compounds 1–15 have been evaluated for their cancer chemopreventive potential in the assays for TNF-α-induced NF-κB, NO production, aromatase, QR-2, and COX-1/-2 activities. Some of these compounds (1, 2, 4, and 6) showed inhibitory activities against TNF-α-induced NF-κB and aromatase. Our study further revealed that the kaempferol/quercetin glycosides contained in this plant are QR-2 inhibitors. The current paper reports the isolation and structure identification of these compounds as well as the evaluation of their cancer chemopreventive potentials, based on their ability to inhibit TNF-α-induced NF-κB activity, nitric oxide (NO) production, aromatase, quinone reductase 2 (QR-2), and COX-1/-2 activities.
Results and Discussion
The 95% EtOH extract of L. contortus was sequentially partitioned with petroleum ether and EtOAc to yield two extracts, which were combined and subjected to a series of column chromatographic separation including silica gel and Sephadex LH-20 gel permeation chromatography to afford 20 compounds (1–15, and syringic acid, vanillic acid, phloretic acid, diplopterol, and β-sitosterol).
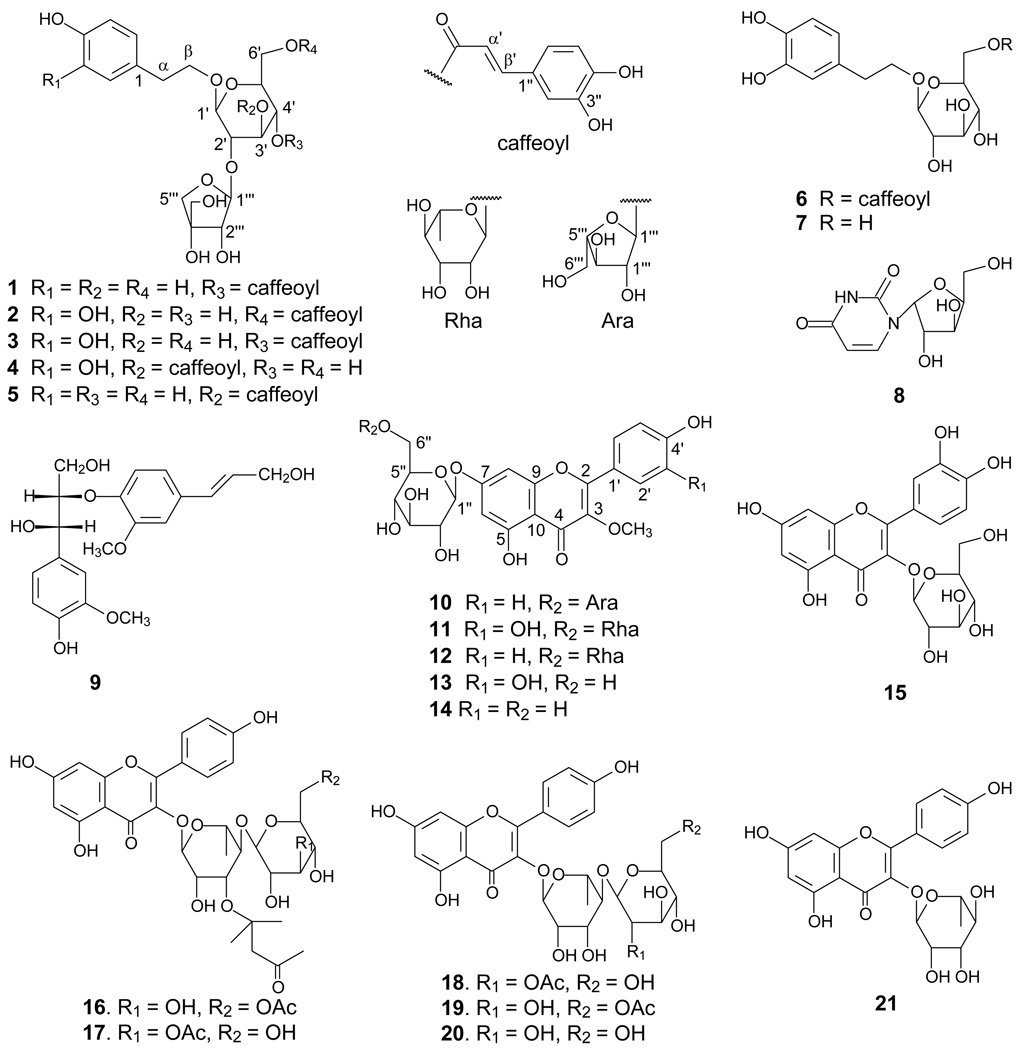
Compound 1, a brown syrup, was determined to have a molecular formula of C28H34O14 by a negative FABMS deprotonated molecule at m/z 593 [M-H]− and a negative HRESIMS deprotonated molecule at m/z 593.1864 ([M-H]−, calcd for 593.1870), which was supported by 13C NMR and DEPT data. The 1H and 13C NMR spectra (Tables 1 and 2) revealed the presence of a para-substituted benzene ring due to the proton and carbon signals at δH 7.10 (2H, d, J = 8.6 Hz, H-2 and H-6), 6.73 (2H, d, J = 8.6 Hz, H-3 and H-5), and δC 131.7 (s, C-1), 131.4 (d, C-2 and C-6), 116.8 (d, C-3 and C-5) and 157.2 (s, C-4), and a caffeoyl moiety due to the aromatic and olefinic proton and carbon signals at δH 7.63 (1H, d, J = 16.0 Hz, H-β′), 6.32 (1H, d, J = 16.0 Hz, H-α′), 6.82 (1H, d, J = 8.1 Hz, H-5″), 6.96 (1H, dd, J = 8.1, 1.3 Hz, H-6″), 7.08 (1H, d, J = 1.3 Hz, H-2″), and δC 168.6 (s, ester carbonyl carbon), 115.2 (d, C-α′), 148.1 (d, C-β′), 128.2 (s, C-1″), 115.7 (d, C-2″), 147.3 (s, C-3″), 150.2 (s, C-4″), 117.0 (d, C-5″) and 123.5 (d, C-6″).6,7 The 1H, 13C NMR, and DEPT spectra also showed signals of an oxygenated methylene group at δH 3.70 and 4.10 (each 1H, m, H2-β) and δC at 72.3 (t, C-β), a methylene group at δH 2.86 (2H, t, J = 7.5 Hz, H2-α) and δC 36.9 (t, C-α), a glucopyranosyl at δH 4.81 (1H, d, J = 8.5 Hz, H-1′) and δC 100.9 (d, C-1′) and an apiofuranosyl moiety at δH 5.15 (1H, d, J = 1.2 Hz, H-1‴) and δC 107.3 (d, C-1‴), indicating 1 as a diglycoside. The presence of correlations between the two methylenes in the 1H-1H COSY and HMBC spectra indicated that the two groups were linked together as an ethylene group, which was further determined to be connected to the para-substituted benzene moiety to form a 4-hydroxyphenylethoxy group evidenced by the presence of the cross peaks between the signals of the methylenes and the 4-hydroxybenzene ring in the HMBC spectrum (Figure 1).
Table 1.
1H NMR Data (500 MHz, methanol-d4) [δH, (J in Hz)] for 1–5
| position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 2 | 7.10, d (8.6) | 6.70, d (1.9) | 6.70, d (1.5) | 6.69, d (1.5) | 7.09, d (8.6) |
| 3 | 6.73, d (8.6) | 6.72, d (8.6) | |||
| 5 | 6.73, d (8.6) | 6.66, d (8.1) | 6.73, d (8.0) | 6.71, d (8.0) | 6.72, d (8.6) |
| 6 | 7.10, d (8.6) | 6.56, dd (8.1, 1.9) | 6.58, dd (8.0, 1.5) | 6.58, dd (8.0, 1.5) | 7.09, d (8.6) |
| α | 2.86 t (7.5) | 2.78 t (7.2) | 2.79 t (7.5) | 2.79 t (7.5) | 2.79 t (7.4) |
| βa | 4.10, m | 3.99, m | 4.09, m | 4.10, m | 4.09, m |
| βb | 3.70, m | 3.68, m | 3.71, m | 3.70, m | 3.70, m |
| Glc-1′ | 4.81, d (8.5) | 4.82, d (8.0) | 4.82, d (8.2) | 4.74, d (8.0) | 4.75, d (8.0) |
| 2′ | 3.59, m | 3.32, m | 3.60, m | 3.82, m | 3.82, m |
| 3′ | 4.50, m | 4.28, m | 4.52, m | 5.75, m | 5.75, m |
| 4′ | 4.82, m | 3.57, m | 4.83, m | 3.76, m | 3.76, m |
| 5′ | 4.05, m | 4.00, m | 4.07, m | 3.67, m | 3.67, m |
| 6′ | 3.76, m | 4.50, m | 3.75, m | 3.90, m | 3.89, m |
| 3.63, m | 4.33, m | 3.64, m | 3.72, m | 3.72, m | |
| 2″ | 7.08, d (1.3) | 7.06, d (1.9) | 7.08, d (1.3) | 7.09, d (1.3) | 7.08, d (1.2) |
| 5″ | 6.82, d (8.1) | 6.79, d (8.1) | 6.82, d (8.1) | 6.87, d (8.1) | 6.83, d (8.0) |
| 6″ | 6.96, dd (8.1, 1.3) | 6.92, dd (8.1, 1.9) | 6.96, dd (8.1, 1.3) | 7.00, dd (8.1, 1.3) | 7.00, dd (8.0, 1.2) |
| α′ | 6.32, d (16.0) | 6.31, d (16.0) | 6.32, d (16.0) | 6.37, d (16.0) | 6.38, d (16.0) |
| β′ | 7.63, d (16.0) | 7.58, d (16.0) | 7.63, d (16.0) | 7.62, d (16.0) | 7.62, d (16.0) |
| Api-1‴ | 5.15, d (1.2) | 5.17, d (1.0) | 5.19, d (1.5) | 5.18, d (1.2) | 5.22, d (1.2) |
| 2‴ | 3.97, d (1.2) | 4.00, d (1.0) | 3.98, d (1.5) | 3.78, d (1.2) | 3.78, d (1.2) |
| 4‴a | 3.96, d (9.9) | 4.00, d (9.8) | 3.96, d (9.8) | 3.89, d (9.8) | 3.89, d (9.8) |
| 4‴b | 3.79, d (9.9) | 3.77, d (9.8) | 3.79, d (9.8) | 3.74, d (9.8) | 3.74, d (9.8) |
| 5‴ | 3.65, s | 3.63, s | 3.63, s | 3.58, s | 3.58, s |
Table 2.
13C NMR Data (125 MHz, methanol-d4) (δC, mult.) for 1–5
| position | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 131.7, C | 132.1, C | 132.2, C | 132.1, C | 132.2, C |
| 2 | 131.4, CH | 117.6, CH | 117.7, CH | 117.6, CH | 131.4, CH |
| 3 | 116.8, CH | 146.5, C | 146.1, C | 146.5, C | 116.9, CH |
| 4 | 157.2, C | 145.0, C | 145.0, C | 145.0, C | 157.1, C |
| 5 | 116.8, CH | 117.0, CH | 117.1, CH | 117.0, CH | 116.9, CH |
| 6 | 131.4, CH | 121.8, CH | 121.7, CH | 121.8, CH | 131.4, CH |
| α | 36.9, CH2 | 37.2, CH2 | 37.1, CH2 | 37.2, CH2 | 37.2, CH2 |
| β | 72.3, CH2 | 72.4, CH2 | 72.3, CH2 | 72.2, CH2 | 72.2, CH2 |
| Glc-1′ | 100.9, CH | 101.0, CH | 100.9, CH | 101.3, CH | 101.3, CH |
| 2′ | 75.3, CH | 75.6, CH | 75.2, CH | 76.5, CH | 76.5, CH |
| 3′ | 67.4, CH | 69.4, CH | 67.4, CH | 71.8, CH | 71.9, CH |
| 4′ | 71.0, CH | 69.7, CH | 71.0, CH | 67.8, CH | 67.8, CH |
| 5′ | 73.7, CH | 73.4, CH | 73.7, CH | 73.7, CH | 73.8, CH |
| 6′ | 62.9, CH2 | 65.6, CH2 | 62.9, CH2 | 62.2, CH2 | 63.2, CH2 |
| 1″ | 128.2, C | 128.2, C | 128.2, C | 128.3, C | 128.3, C |
| 2″ | 115.7, CH | 115.6, CH | 115.8, CH | 115.7, CH | 116.5, CH |
| 3″ | 147.3, C | 147.2, C | 146.5, C | 146.5, C | 147.2, C |
| 4″ | 150.2, C | 150.0, C | 150.1, C | 150.1, C | 150.0, C |
| 5″ | 117.0, CH | 116.9, CH | 116.9, CH | 116.8, CH | 117.0, CH |
| 6″ | 123.5, CH | 123.6, CH | 123.7, CH | 123.5, CH | 123.5, CH |
| α′ | 115.2, CH | 115.4, CH | 115.2, CH | 115.7, CH | 115.8, CH |
| β′ | 148.1, CH | 147.6, CH | 148.2, CH | 147.6, CH | 147.7, CH |
| CO | 168.6, C | 169.7, C | 168.6, C | 169.4, C | 168.4, C |
| Api-1‴ | 107.3, CH | 107.2, CH | 107.3, CH | 107.4, CH | 107.4, CH |
| 2‴ | 78.5, CH | 78.5, CH | 78.5, CH | 78.2, CH | 78.2, CH |
| 3‴ | 81.2, C | 81.2, C | 81.3, C | 81.2, C | 81.3, C |
| 4‴ | 75.9, CH2 | 75.9, CH2 | 75.9, CH2 | 76.1, CH2 | 76.1, CH2 |
| 5‴ | 66.6, CH2 | 66.5, CH2 | 66.6, CH2 | 66.9, CH2 | 66.9, CH2 |
Figure 1.
Key HMBC correlations for 1–5 and 10.
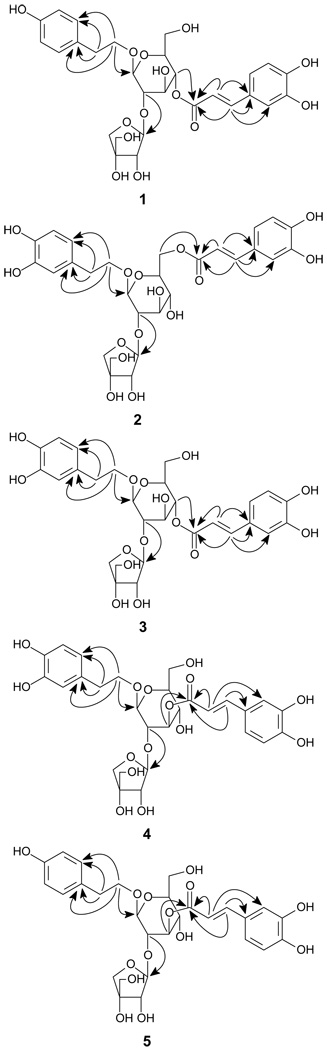
The glucose moiety was determined to have a β-configuration at C-1 due to a large coupling constant for the anomeric proton of the sugar unit at δH 4.81 (1H, d, J = 8.5 Hz), and the apiose unit was also determined to have a β-configuration at C-1 due to the chemical shift of its anomeric carbon signal in the 13C-NMR at δC 107.3.8,9 The presence of a three bond correlation between the glucose C-2′ signal at δC 75.3 and the apiose anomeric proton signal at δH 5.15 in the HMBC spectrum suggested that the interglycosyl linkage is apiosyl-(1→2)-glucose. The location of the caffeoyloxy group in 1 was fixed at C-4′ of the glucose residue by the presence of the HMBC correlation from the acyl carbonyl carbon signal at δC 168.6 to the glucosyl C-4′ proton signal at δH 4.82. The glycosidation position was unambiguously determined by the presence of a three bond correlation between the glucosyl anomeric proton H-1′ at δH 4.81 and the oxygenated methylene group at δC 72.3 (C-β) in the HMBC spectrum. Thus, compound 1 was identified as β-(4-hydroxyphenyl)ethyl-4-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside.
Compound 2 was also obtained as a brown syrup. The molecular formula was determined as C28H34O15 from a negative FABMS ion at m/z 609 [M-H]− and a negative HRESIMS ion at m/z 609.1804 ([M-H]−, calcd for 609.1819), suggesting the presence of an additional hydroxy group in comparison with 1. Its 1H, 13C NMR, and DEPT spectra (Tables 1 and 2) were similar to those of 1, indicating that 2 was also a phenylethanoid diglycoside with a caffeoyl group. Analysis of the 1H, 13C NMR, and DEPT data showed that the aglycone of 2 was a 3, 4-dihydroxyphenylethanol group due to the signals at δH 6.70 (1H, d, J = 1.9 Hz, H-2), 6.66 (1H, d, J = 8.1 Hz, H-5), 6.56 (1H, dd, J = 8.1, 1.9 Hz, H-6), δC 132.1 (s, C-1), 117.6 (d, C-2), 146.5 (s, C-3), 145.0 (s, C-4), 117.0 (d, C-5), and 121.8 (d, C-6). The presence of the correlation between an acyl carbonyl carbon signal at δC 169.7 and the glucosyl C-6′ proton signals at δH 4.50 and 4.33 (each 1H, m, H2-6′) in the HMBC spectrum (Figure 1) assigned the caffeoyloxy group at C-6′ of the glucose residue. Thus, compound 2 was identified as β-(3, 4-dihydroxyphenyl)ethyl-6-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside.
Compound 3 was obtained as a brown syrup. It was determined to have the same molecular formula (C28H34O15) as that of 2 by a negative FABMS ion at m/z 609 [M − H]− and a negative HR-ESI-MS ion at m/z 609.1807 ([M − H]−, calcd for 609.1819). It showed similar 1H and 13C NMR data to those of 1 and 2 (Tables 1 and 2). Compound 3 differs from 1 by having a 3, 4-dihydroxyphenylethanol group instead of a 4-hydroxyphenylethanol in 1, and from 2 only by the location of the caffeoyl group. The caffeoyloxy group in 3 was determined to be located at C-4′ of the glucose residue by the presence of the HMBC correlation (Figure 1) from the acyl carbonyl resonance at δC 168.6 to the glucosyl C-4′ proton signal at δH 4.83. Thus, compound 3 was identified as β-(3, 4-dihydroxyphenyl)ethyl-4-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside.
Compound 4 was obtained as a brown syrup. The molecular formula was determined to be the same as that of 3 (C28H34O15) from a negative FABMS ion at m/z 609 [M − H]− and a negative HRESIMS ion at m/z 609.1809 ([M − H]−, calcd for 609.1819). The 1H, 13C NMR, and DEPT spectra (Tables 1 and 2) of 4 were similar to those of 3, differing only by the location of the caffeoyl group. The caffeoyloxy group in 3 was determined at C-3′ of the glucose residue by the presence of the HMBC correlation (Figure 1) from the acyl carbonyl resonance at δC 169.4 to the glucosyl C-3′ proton signal at δH 5.75. Accordingly, compound 4 was identified as β-(3, 4-dihydroxyphenyl)ethyl-3-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside.
Compound 5, a brown syrup, was deduced to have the same molecular formula as that of 1 (C28H34O14) by the negative FABMS (m/z 593 [M − H]−) and the negative HR-ESI-MS ([M − H]− m/z 593.1864, calcd for 593.1870). The 1H, 13C NMR, and DEPT data (Tables 1 and 2) were similar to those of 1. The difference between 5 and 1 is similar to that between 4 and 3 in that the caffeoyl group is at a different location. Analysis of the HMBC data (Figure 1) determined the caffeoyloxy group in 5 at C-3′. Thus, compound 5 was identified as β-(4-hydroxyphenyl)ethyl-3-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside.
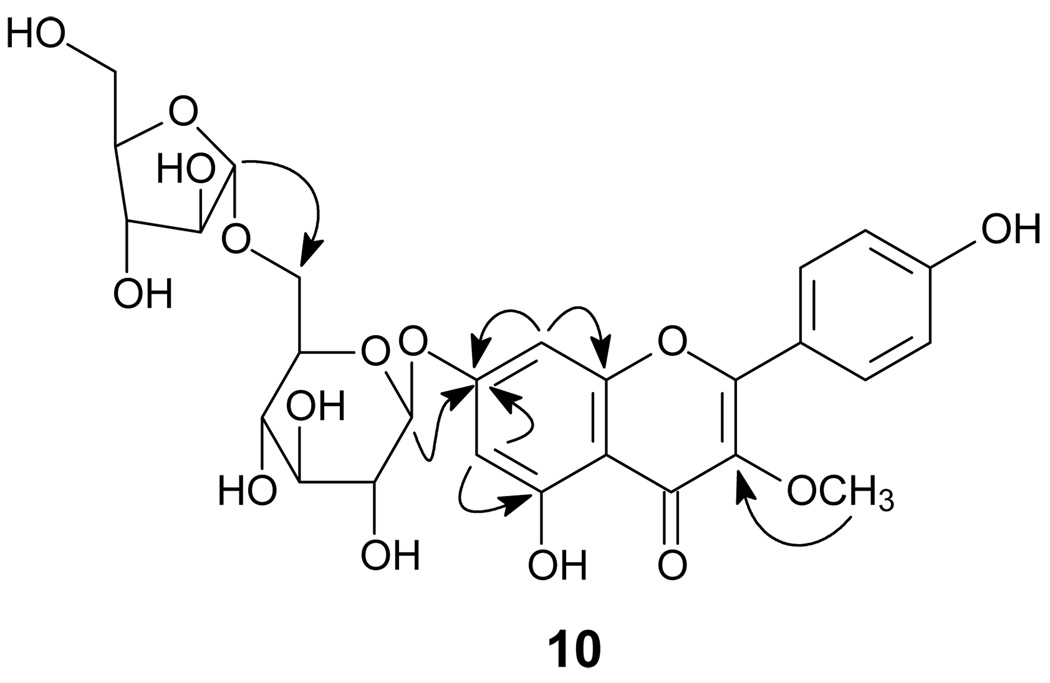
Compound 10 was obtained as a yellow amorphous powder, and its molecular formula was deduced as C27H30O15 by the negative HRESIMS (found 593.1493, calcd for 593.1506). The 1H and 13C NMR spectra (Table 3) showed typical signals of a flavonoid [δH 8.05 (2H, d, J = 8.5 Hz, H-2′ and H-6′), 6.96 (2H, d, J = 8.5 Hz, H-3′ and H-5′), 6.53 (1H, brs, H-6), 6.78 (1H, brs, H-8) and 15 sp2 hybrid carbon signals including a conjugated carbonyl group at δC 180.6 (C-4, s)].10,11 The presence of the two doublets corresponding to the AA′BB′ spin system at δH 8.05 and 6.96 suggested a p-substituted ring B. The proton signals of the two broad singlets at 6.53 and 6.78 were assigned, respectively, to meta-coupled H-6 and H-8 of ring A in 10, which was confirmed by the presence of the 1H-1H COSY correlation between the two protons and the presence of the HMBC correlations from H-6 to C-8 (δC 96.4, d) and from H-8 to C-6 (δC 101.3, d). Additionally, the 1H NMR spectrum showed the resonance of a methoxy singlet at δH 3.82 (3H, s), which was assigned at C-3 (δC 140.2, s) due to the presence of the correlation between the methoxy protons and C-3 in the HMBC experiment (Figure 1).
Table 3.
1H (500 MHz) [δH, (J, Hz)] and 13C NMR (125 MHz) (δC) Data of 10 in methanol-d4
| 10 | ||
|---|---|---|
| position | 1H | 13C |
| 2 | 159.2, C | |
| 3 | 140.2, C | |
| 4 | 180.6, C | |
| 5 | 163.2, C | |
| 6 | 6.53, brs | 101.3, CH |
| 7 | 165.1, C | |
| 8 | 6.78, brs | 96.4, CH |
| 9 | 158.4, C | |
| 10 | 108.2, C | |
| 1′ | 122.9, C | |
| 2′ | 8.05, d (8.5) | 132.1, CH |
| 3′ | 6.96, d (8.5) | 117.1, CH |
| 4′ | 162.3, C | |
| 5′ | 6.96, d (8.5) | 117.1, CH |
| 6′ | 8.05, d (8.5) | 132.1, CH |
| Glc-1″ | 5.08, d (7.2) | 102.0, CH |
| 2″ | 3.52, m | 75.2, CH |
| 3″ | 3.53, m | 78.2, CH |
| 4″ | 3.40, m | 72.1, CH |
| 5″ | 3.65, m | 77.5, CH |
| 6″a | 4.11, m | 68.5, CH2 |
| 6″b | 3.73, m | |
| Ara-1‴ | 4.95, d (2.0) | 110.5, CH |
| 2‴ | 4.08, m | 83.5, CH |
| 3‴ | 3.85, m | 79.3, CH |
| 4‴ | 4.01, m | 86.5, CH |
| 5‴a | 3.73, m | 63.5, CH2 |
| 5‴b | 3.63, m | |
| MeO-3 | 3.82, s | 61.0 CH3 |
The 1H and 13C NMR spectra also showed two anomeric protons at δH 5.08 (1H, d, J = 7.2 Hz, H-1″) and 4.95 (1H, d, J = 2.0 Hz, H-1‴) and 11 oxygenated carbons, suggesting 10 as a flavonol diglycoside possessing a pentosyl and a hexosyl moiety. The anomeric configurations of the glucose and the arabinose units were determined as β and α, respectively, based on the coupling constants of 7.2 Hz for H-1″ and 2.0 Hz for H-1‴.12,13 The glucose C-6″ signal appeared at δC 68.5, suggesting that the interglycosyl linkage is arabinosyl-(1→6)-glucose, which was confirmed by the presence of the HMBC correlation between H-1‴ and C-6″. The glycosidation position was further determined by the presence of the three-bond HMBC correlation between the glucosyl anomeric proton H-1″ and C-7 of ring A. The structure of 10 was identified as 4′, 5, 7-trihydroxy-3-methoxyflavone-7-O-α-L-arabinofuranosyl (1→6)-β-D-glucopyranoside.
Acid hydrolysis of 1–5 with 5% H2SO4 in EtOH yielded respectively two sugars, which were separated by column chromatography. The two sugars were identified as glucose and apiose, respectively, by comparison of their 1H NMR and optical rotation data with literature reports and authentic samples. Acid hydrolysis of 10 with 5% H2SO4 in EtOH afforded D-glucose and L-arabinose, which were separated by column chromatography and identified by comparison of their 1H NMR and optical rotation data with literature reports and authentic samples. The total assignment of the 1H- and 13C-NMR spectroscopic data of compound 1–5 and 10 was carried out by a combination of 1H-1H COSY, HSQC, HMBC, and ROESY experiments.
The 14 known isolates were identified as calceolarioside B (6),14 β-(3, 4-dihydroxylphenyl) β-D-glucopyranoside (7),15 arauridine (8),16,17 threo-1-(4-hydroxy-3-methoxyphenyl)-2-{4-[(E)-3-hydroxy-1-propenyl]-2-methoxyphenoxy}-1, 3-propane-diol (9),18 3′, 4′, 5, 7-tetrahydroxy-3-methoxyflavone-7-O-α-L-rhamnopyranosyl-(1→6)-β-D-glucopyranoside (11),19 4′, 5, 7-trihydroxy-3-methoxyflavone-7-O-rutinoside (12), transilin (13), 4′, 5, 7-trihydroxy-3-methoxyflavone-7-O-β-D-glucopyranoside (14),20 quercetin-3-O-β-D-glucoside (15),21 syringic acid,22 vanillic acid,23 phloretic acid,24 diplopterol,25,26 and β-sitosterol by comparison of their physical data with reported data.
Although flavonoids are found abundantly in ferns, phenylethanoids are rare. This is the first report of phenylethanoid glycosides from the family Polypodiaceae. Compounds 1–15 were evaluated for their cancer chemopreventive potential based on their ability to inhibit TNF-α-induced NF-κB activity, nitric oxide (NO) production, aromatase, quinone reductase 2 (QR-2), and COX-1/-2 activities (Table 4).
The nuclear transcription factor (NF-κB), the signaling molecule (NO) and the enzymes (aromatase, QR-2, and COX-1/-2) all play different roles in cells, respectively.27,28 NF-κB is a ubiquitous transcription factor associated with cell apoptosis, differentiation, and migration, and may promote cell proliferation and prevent cell death through anti-apoptotic factors upon being activated.29 Aromatase catalyzes the conversion of androgen to estrogen, the female sex hormone associated with proliferation of breast cancer cells.30,31 Although NO production has a beneficial role for protection of an organ such as the liver from ischemic damage, excessive and sustained levels of NO expression may damage tissues which may result in vascular collapse, inflammation, or even carcinomas. Studies have shown that NO impacts many physiological and pathological processes associated with the development of cancers in the early stages.32 QR2 may transform certain quinone substrates into more highly reactive species, is abundantly expressed in some cancer cells,33–39 and is also associated with various neurological disorders including Parkinson’s disease40,41 and schizophrenia.42 Although the precise function of QR2 remains to be determined, some chemopreventive agents such as resveratrol and melatonin were found to have potent binding activities with QR2,37,38 suggesting QR2 as a potential new target for the development of chemopreventive agents. COX-1/-2 are associated with neoplastic transformation.43 Thus, inhibition against each of NF-κB signaling, NO production, and the enzyme activities of aromatase, QR2 and COX-1/-2 may have beneficial effects for the treatment or prevention of cancer.
Among the evaluated isolates, only quercetin-3-O-β-D-glucoside (15) demonstrated inhibition of TNF-α-induced NF-κB activity with an IC50 value of 33.6 µM; and compounds 1, 2, 4 and 6 showed aromatase activity with IC50 values of 30.7, 32.3, 26.8, and 35.3 µM, respectively. Compounds 1–15 were also evaluated for their inhibition against NO production and COX-1/-2 enzymes, but none of them showed significant NO activity at a concentration of 20 µg/mL (in a range of 27 to 82 µM, depending on compounds), and none of them showed COX activity at a concentration of 10 µg/mL (in a range of 13 to 41 µM, depending on compounds).
Compounds 1–15 were further evaluated for their ability to interact with QR2 using an LC-MS ultrafiltration binding assay. Compounds 10 and 14 were shown to mediate a positive response (Table 4). Since neither of the two compounds showed more than 50 % inhibition against QR2, it may be suggested they interact with a binding site of the enzyme rather than the catalytic site. A similar phenomenon was also observed in our previous study for compounds 16–21 (Table 4).28 Our studies showed that only the kaempferols/quercetin with a glycoside unit at C-3 (15, 17, 19, and 21) were able to inhibit QR2. Further, the kaempferols/quercetin with a single sugar unit at C-3 (15 and 21) showed stronger binding activity than other kaempferol/quercetin derivatives. To confirm the findings, we further evaluated QR2 inhibition activity for the commercially purchased compound 21 at different concentrations (Figure 2). The compound was found to have strong inhibition activity against QR2 with an IC50 value of 3.84 µM.
Figure 2.
Titration of 21: the compound was shown to inhibit QR2 with an IC50 value of 3.84 µM.
To determine cytotoxicity, compounds 1–15 were evaluated against Hepa1c1c7 and MCF7 cells. None of them showed any growth inhibitory effects against the two cell lines at a concentration of 20 µg/mL (in a range of 27 to 82 µM, depending on compounds). The absence of general toxicity is considered beneficial in view of cancer chemoprevention since a high therapeutic index is required for disease prevention. Further studies to elucidate the modulation of unique chemopreventive targets of these compounds, especially the kaempferol/quercetin glycosides, may provide a rationale for modification of the structures to produce more potent chemopreventive molecules.
Experimental Section
General Experimental Procedures
Optical rotations were measured on a Perkin-Elmer 241 polarimeter. MS were determined on a Finnigan MAT 90 instrument and a VG Auto Spec-3000 spectrometer. NMR spectra were measured on a Bruker DRX-500 spectrometer. Silica gel (200–300 mesh, Qingdao Marine Chemical Co., China), MCI GEL® (Mitsubishi Chemical Corp.), and Sephadex LH-20 (25–100 µm, Pharmacia Fine Chemical Co. Ltd.) were used for column chromatography (CC), and silica gel GF254 was used for TLC (Qingdao Marine Chemical Co., China). Solvents were of industrial purity and distilled prior to use.
Plant Material
The whole plant of L. contortus was collected from Gongshan County, Yunnan, China in October, 2005 and identified by Prof. Shugang Lu, School of Life Science, Yunnan University, where a voucher specimen (No.0510017) is deposited.
Extraction and Isolation
Air-dried powder of the whole plant (1.1 kg) was extracted with 95% EtOH (×5) at room temperature. The EtOH extract (108 g) was partitioned sequentially with petroleum ether and EtOAc to yield petroleum ether and EtOAc extracts, respectively. The two extracts were combined (17.5 g) and fractionated by CC over silica gel with a gradient from petroleum ether to EtOAc to give six fractions, fractions I-VI. Fraction II (0.1 g) was separated by silica gel CC eluted with petroleum ether-EtOAc (15:1) to afford diplopterol (21 mg) and β-sitosterol (110 mg). Fraction IV (2.5 g) was separated using a MCI GEL® ion exchange column eluted with a gradient of 50–90% MeOH in H2O to afford four subfractions (IV-1- IV-4). Fraction IV-1 (50 mg), IV-2 (1.2 g), IV-3 (163 mg) and IV-4 (163 mg) were further subjected to a Sephadex LH-20 column, respectively, eluting with MeOH to yield 9 (4 mg), syringic acid (5 mg), vanillic acid (8 mg), phloretic acid (6 mg), 7 (240 mg), 12 (163 mg), and 15 (13 mg). Fraction V (3.1 g) was subjected to CC separation over MCI GEL® ion exchange eluted with 50–90% MeOH in H2O to yield fractions V-1-V-5. Fractions V-1 (800 mg), V-2 (600 mg), V-3 (300 mg) and V-4 (56 mg) were further chromatographed on a Sephadex LH-20 column, respectively, eluting with MeOH to afford 13 (250 mg), 14 (25 mg), 6 (4 mg), 8 (24 mg), 2 (14.5 mg), 3 (52 mg), 4 (10.5 mg), 1 (7 mg), and 5 (20 mg). Fraction VI (2.7 g) was chromatographed on a silica gel RP-18 column, eluting with MeOH-H2O (6: 4 →1: 0, v/v) in a stepwise system to get five subfractions (VI-1-VI-5). Fractions VI-2 (20 mg) and VI-3 (590 mg) were purified by a Sephadex LH-20 column, respectively, eluting with MeOH to yield compounds 10 (12 mg) and 11 (210 mg).
β-(4-Hydroxyphenyl)ethyl-4-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside (1)
brown syrup; [α]25D −73.2 (c 1.5, MeOH); UV (MeOH) λmax (Abs.) 324 (0.30), 203.5 (0.58) nm; IR (KBr) νmax 3425, 2927, 1692, 1607, 1516, 1164 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; negative FABMS (glycerol matrix) m/z 593 [M-H]−; negative HRESIMS m/z 593.1864 [M-H]− (calcd for C28H33O14, 593.1870).
β-(3, 4-Dihydroxyphenyl)ethyl-6-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside (2)
brown syrup; [α]25D −63.4 (c 5.4, MeOH); UV (MeOH) λmax (Abs.) 330 (0.20), 290 (0.18), 204 (0.57) nm; IR (KBr) νmax 3430, 2927, 1694, 1607, 1519, 1447, 1283, 1092 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; negative FABMS (glycerol matrix) m/z 609 [M-H]−; negative HRESIMS m/z 609.1804 [M-H]− (calcd for C28H33O15, 609.1819).
β-(3, 4-Dihydroxyphenyl)ethyl-4-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside (3)
brown syrup; [α]25D −71.0 (c 6.4, MeOH); UV (MeOH) λmax (Abs.) 331 (0.26), 291.0 (0.21), 203.5 (0.60) nm; IR (KBr) νmax3423, 2934, 1691, 1605, 1520, 1446, 1360, 1283,1162 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; negative FABMS (glycerol matrix) m/z 609 [M-H]−; negative HRESIMS m/z 609.1807 [M-H]− (calcd for C28H33O15, 609.1819).
β-(3, 4-Dihydroxyphenyl)ethyl-3-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside (4)
brown syrup; [α]25D −86.4 (c 2.9, MeOH); UV (MeOH) λmax (Abs.) 328.5 (0.20), 290.0 (0.18), 203.5 (0.55) nm; IR (KBr) νmax 3439, 2928, 1694, 1607, 1519, 1447, 1281, 1159 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; negative FABMS (glycerol matrix) m/z 609 [M-H]−; negative HRESIMS m/z 609.1809 [M-H]− (calcd for C28H33O15, 609.1819).
β-(4-Hydroxyphenyl)ethyl-3-O-E-caffeoyl-O-[β-D-apiofuranosyl-(1→2)]-β-D-glucopyranoside (5)
brown syrup; [α]25D −65.1 (c 4.6, MeOH); UV (MeOH) λmax (Abs.) 318.0 (0.22), 291.5 (0.21), 203.5 (0.52) nm; IR (KBr) νmax 3424, 2927, 1694, 1606, 1516, 1446, 1268, 1159 cm−1; 1H NMR and 13C NMR data, see Tables 1 and 2; negative FABMS (glycerol matrix) m/z 593 [M-H]−; negative HRESIMS m/z 593.1864 [M-H]− (calcd for C28H33O14, 593.1870).
4′, 5, 7-Trihydroxy-3-methoxyflavone-7-O-α-L-arabinofuranosyl (1→6)-β-D-glucopyranoside (10)
yellow amorphous powder; [α]25D = −41.6 (c 1.6, MeOH); UV (MeOH) λmax (Abs.) 332.5 (0.22), 268.5 (0.25), 205 (0.45) nm; IR (KBr) νmax 3415, 2921, 1652, 1609, 1341, 1180 cm−1; 1H NMR and 13C NMR data, see Table 3; negative HRESIMS m/z 593.1493 [M-H]− (calcd for C27H29O15, 593.1506).
Acid Hydrolysis of Compounds 1–5 and 10
Each compound (5 mg) was individually refluxed in 5 % sulfuric acid in EtOH (5.0 mL) on a water bath for 4 hr. After cooling, the reaction mixture was neutralized with 8 % NaOH and concentrated under reduced pressure. The residue was separated on a silica gel column, eluting with MeCN-H2O (8:1) to yield D-glucose (0.7 mg), [α]D25 +43.3 (c 0.7, H2O) and D-apiose (0.5 mg), [α]D25 +5.6 (c 0.5, H2O) for 1, D-glucose (0.6 mg), [α]D25 +40.6 (c 0.6, H2O) and D-apiose (0.4 mg), [α]D25 +3.4 (c 0.5, H2O) for 2, D-glucose (0.5 mg), [α]D25 +41.6 (c 0.5, H2O) and D-apiose (0.5 mg), [α]D25 +4.4 (c 0.5, H2O) for 3, D-glucose (0.4 mg), [α]D25 +40.2 (c 0.4, H2O) and D-apiose (0.5 mg), [α]D25 +3.8 (c 0.5, H2O) for 4, D-glucose (0.5 mg), [α]D25 +41.3 (c 0.5, H2O) and D-apiose (0.5 mg), [α]D25 +3.6 (c 0.5, H2O) for 5, and D-glucose (0.5 mg), [α]D25 +37.5 (c 0.5, H2O) and L-arabinose (0.4 mg), [α]D25 +93.5 (c 0.4, H2O) for 10, respectively.
Evaluation of Biological Activity
NF-κB luciferase, aromatase, quinone reductase 2 (QR2), cyclooxygenase-1 (COX-1), and cyclooxygenase-2 (COX-2), ultrafiltration LC-MS for QR2 and cytotoxicity assays were conducted as previously described. Below is brief description of each assay.
NF-κB Luciferase Assay
Panomic (Fremont, CA) has established a number of stably-transfected NF-κB reporter cell lines. We have employed human embryonic kidney cells 293 for monitoring any changes occurring along NF-κB pathway. Cells were seeded into sterile 96-well plates at a density of 20 × 103 cells per well. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen Co., Carlsbad, CA), supplemented with 10% FBS, 100 units/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine. After a 48 hr incubation, medium was replaced and various concentrations of test compounds were added (dissolved in PBS). Human recombinant TNF-α (2 ng/mL; 0.14 nM) (Calbiochem, Gibbstown, NJ) was used as activator. The plate was incubated for 6 hr. Spent media were discarded and the cells were washed once with PBS. Then, the cells were lysed by adding 50 µL/well of Reporter Lysis Buffer (diluted 5-fold with water) (Promega, Madison, WI) and incubating for 5 min on a shaker. At this point, plates can be stored at −80°C for subsequent analysis. The luciferase assay was performed using the Luc assay system from Promega. The gene product, luciferase enzyme, reacts with luciferase substrate, emitting light which was detected using a luminometer (LUMIstar Galaxy BMG). Dose-response curves were constructed and data were expressed as IC50 values (i.e., concentration of tested sample required to inhibit TNF-α–induced NF-κB activity by 50%). Na-tosyl-L-phenylalanine chloromethyl ketone (TPCK) (IC50 = 3.76 µM) was used as a positive control.44
Aromatase Assay
Test compounds (3.5 µL) were preincubated with 30 µL of a NADPH-regenerating system (2.6 mM NADP+, 7.6 mM glucose 6-phosphate, 0.8 U/mL glucose 6-phosphate dehydrogenase, 13.9 mM MgCl2, and 1 mg/mL albumin in 50 mM potassium phosphate buffer, pH 7.4) for 10 min at 37 °C. The enzyme and substrate mixture [33 µL of 1 µM CYP19 enzyme (BD Biosciences), 0.4 µM dibenzylfluorescein, 4 mg/mL albumin, in 50 mM potassium phosphate, pH 7.4] was added, and the plate was incubated for 30 min at 37 °C before quenching with 25 µL of 2 N NaOH. After termination of the reaction and shaking for 5 min, the plate was further incubated for 2 hr at 37 °C. This enhances the ratio of signal to background. Fluorescence was measured at 485 nm (excitation) and 530 nm (emission). IC50 values and dose-response curves were based on three independent experiments performed in duplicate using five concentrations of test substance. Naringenin (IC50 = 0.23 µM) was used as a positive control.45
Nitric Oxide (NO) Assay
The blocked production of NO is a potential mechanism for chemoprevention. RAW 264.7 cells were incubated in a 96-well culture plate for 24 hr. The cells were treated with various concentrations of compounds dissolved in phenol red- free DMEM for 30 min, followed by 1 µg/mL of LPS treatment for 24 hr. NO was oxidized to the stable end product, nitrite, by the addition of Griess reagent [1:1 mixture (v/v) of 1% sulfanilamide and 0.1% N-(1-naphthyl) ethylenediamine in 2.5% H3PO4], and absorbance was measured at 540 nm. A standard curve was created by using known concentrations of NaNO2. Na-L monomethyl arginine (L-NMMA) (IC50 = 19.7 µM) was used as a positive control.46
QR2 Assay
The activity of QR2 under steady-state conditions was evaluated on a Molecular Devices SpectraMax Plus 384 UV–visible spectrophotometer by monitoring the decrease in absorbance of the enzyme co-substrate NMeH (N-methyldihydronicotinamide) at 360 nm at 25 °C. Reactions were carried out in a 96-well plate and were initiated by the addition of QR2 to the assay buffer (50 mM Tris/HCl, pH 8.0, 100 mM NaCl and 0.1% Triton X-100) containing various concentrations of menadione (5–75 µM), and various concentrations of NMeH (10–140 µM). Stock QR2 enzyme concentrations were determined using the Bio-Rad protein assay. The final enzyme concentration was 5 nM in a reaction volume of 200 µL. The plate was shaken vigorously for 5 s to mix reagents, and the loss in absorbance upon oxidation of NMeH was monitored until the reaction reached completion. Reaction rates were converted into specific activity using ε360=7060 M−1·cm−1 for NMeH, with a well pathlength of 0.445 cm. The specific activity of QR2 is expressed in µM of NMeH oxidized per mL per min per mg of QR2 added (units/mg). One unit of activity is defined as 1 µL of NMeH oxidized per min. Data were expressed as percentage of inhibition or IC50 values (concentration required to inhibit QR2 activity by 50 %). Resveratrol was used as a positive control, which showed 50 % inhibition against QR2 at a concentration of 0.96 µM.38
COX-1 and COX-2 Assays
COX-2 (0.2 µg) or COX-1 (0.2 µg) was activated by adding 146 µL Tris-HCl buffer (pH 8.0), 2 µL hematin (1 µM final), 10 µL L-epinephrine (2 mM final) at room temperature for 2 min on ice. Then, 2 µL of each test solution was added and preincubated for 10 min in a water bath at 37°C for 10 min. Negative control incubations were identical except that 2 µL of Tris-HCl buffer was added instead of the test solution. Celecoxib and indomethacin were used as positive controls in the COX-2 and COX-1 inhibition assays, respectively. The reactions were initiated by adding 20 µL arachidonic acid (5 µM, final concentration) and terminated after 2 min by adding 10 µL 2.0 M HCl. 20 µL d4-[PGE2] at 50 ng/mL was then added as internal standard. Both PGE2 and [d4]-PGE2 were extracted from incubates using 800 µL H2O saturated with EtOAc. The EtOAc phase was then collected, evaporated to dryness, and reconstituted in 100 µL MeOH/H2O (50:50, v/v). The formation of the COX product prostaglandin E2 was measured using the LC-MS-MS method as described previously (Cao et al., 2008). An Applied Biosystems (Foster City, CA) API 4000 triple quadrupole mass spectrometer with negative ion electrospray and a collision energy of 22 eV equipped with a Shimadzu (Columbia, MD) Prominence UFLC system with a Waters (Milford, MA) XTerra MS C18 (2.1 × 50 mm, 3.5 µm) analytical column was used for PGE2 measurement. Indomethacin was used as a positive control for the COX-1 inhibition assay. At10 µM, indomethacin produced 85% inhibition, and 125 nM produced 65% inhibition of ovine COX-1. For assays of COX-2 inhibition, celecoxib was used as a positive control at two different concentrations. At 33 µM, celecoxib inhibited human COX-2 93 %, and at 46 nM, celecoxib (approximately the IC50 value) produced 49% inhibition. 47
Ultrafiltration LC-MS Binding Assay for QR2
Test compounds were incubated with ovine QR2 for 1 hr at 37 °C. The mixture was then filtered through a 30,000 Da molecular weight cut-off ultrafiltration membrane. After washing each sample 3 times with buffer, the ligands were dissociated from QR2 using methanol. The ligand ultrafiltrates were dried under nitrogen and reconstituted in 50 % aqueous methanol prior to LC-MS analysis.48
Cytotoxicity Assay
Hepa1c1c7 cells were maintained in α-MEM (Alpha Minimum Essential Medium) medium. MCF-7 cells were maintained in MEME (Eagle’s Minimum Essential Medium) medium containing 10 mg/L of insulin. In each case, PSF (100 units/mL penicillin G, 100 µg/mL streptomycin sulfate, 250 ng/mL amphotericin B) was added. All media were supplemented with 10 % heat-inactivated FBS. The 190 µL cell suspension (3×104 cells in 1 mL media) was incubated with 10 µL sample solutions, in triplicate, in 96-well tissue culture plate at 37°c in a humidified atmosphere of 5% CO2 in air for 72 hours. 10% Aqueous DMSO (10 µL) was used as control group. Then the cells were fixed to plastic substratum by the addition of 100 µL cold 20% aqueous trichloroacetic acid and washing with H2O after incubation at 4°c for 30 min. After staining cells with 100 µL of 0.4% sulforhodamine B in 1% aqueous HOAc for 30 min, unbound dye was removed by rinsing with 1% aqueous HOAc. The bound dye was solubilized with 200 µL 10 mM unbuffered Tris base, pH 10, and the optical density was measured at 515 nm using an ELISA plate reader. The average data were expressed as a percentage, relative to the negative control. Paclitaxel and vinblastine were used as positive controls. At 23.4 µM, paclitaxel produced 64% and 57 % inhibition of MCF-7 and Hepa1c1c7, respectively. At 49.4 µM, vinblastine produced 74% and 90 % inhibition of MCF-7 and Hepa1c1c7, respectively.49
Supplementary Material
Acknowledgment
This investigation was supported in part by a program project P01 CA48112 awarded by the National Cancer Institute USA, a grant (2005DFA30670) for international collaborative research from the Chinese Ministry of Science and Technology, a grant (30560178) from the Natural Science Foundation of China, a Program for New Century Excellent Talents in University (NCET-05-0824), and the West Light Program from the Chinese Academy of Sciences.
Footnotes
Supporting Information Available. Spectroscopic data (1D and 2D NMR spectra) of compounds 1–5 and 10, are available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Flora Reipublicae Popularis Sinicae. 2. Vol. 6. Beijing: Science Press; 2000. Delectis Florae Reipublicae Popularis Sinicae Agendae, Academiae Sinicae Edita; p. 63. [Google Scholar]
- 2.Wu ZY. XinHuaBenCaoGangYao. Vol. 3. Shanghai: Shanghai Press of Science and Technology; 1990. p. 710. [Google Scholar]
- 3.Choi YH, Lim YH, Yeo H, Kim J. Phytochemistry. 1996;43:1111–1113. doi: 10.1016/s0031-9422(96)00417-7. [DOI] [PubMed] [Google Scholar]
- 4.Choi YH, Kim JW, Choi YH. Phytochemistry. 1999;51:453–456. [Google Scholar]
- 5.Lee MW. Saengyak Hakhoechi. 1998;29:142–145. [Google Scholar]
- 6.Kostadinova EP, Alipieva KI, Kokubun T, Taskova RM, Handjieva NV. Phytochemistry. 2007;68:1321–1326. doi: 10.1016/j.phytochem.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Jensen SR. Phytochemistry. 1996;43:777–783. [Google Scholar]
- 8.Mathias L, Vieira IJC, Braz R, Rodrigues E. J. Nat. Prod. 1998;61:1158–1161. doi: 10.1021/np9800598. [DOI] [PubMed] [Google Scholar]
- 9.Kitagawa I, Hori K, Sakagami M, Hashiuchi F, Yoshikawa M, Ren J. Chem. Pharm. Bull. 1993;41:1350–1357. doi: 10.1248/cpb.41.1350. [DOI] [PubMed] [Google Scholar]
- 10.Bao KH, Jin WY, Thuong PT, Min BS, Na MK, Lee YM, Kang SS. Fitoterapia. 2007;78:409–413. doi: 10.1016/j.fitote.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Kotsos MP, Aligiannis N, Mitakou S. Biochem. Syst. Ecol. 2007;35:381–385. [Google Scholar]
- 12.Agrawal PK. Phytochemistry. 1992;31:3307–3330. doi: 10.1016/0031-9422(92)83678-r. [DOI] [PubMed] [Google Scholar]
- 13.Tian JM, Hao XJ, He HP. Helv. Chim. Acta. 2006;89:291–298. [Google Scholar]
- 14.Damtoft S, Jensen SR. Phytochemistry. 1994;37:441–443. doi: 10.1016/0031-9422(94)85075-5. [DOI] [PubMed] [Google Scholar]
- 15.Toyota M, Saito T, Asakawa Y. Phytochemistry. 1996;43:1087–1088. [Google Scholar]
- 16.Hruska FE, Blonski, Wayne JP. Can. J. Chem. 1982;60:3026–3032. [Google Scholar]
- 17.Akhrem AA, Mikhailopulo IA, Abramov AF. Org. Magn. Reson. 1979;12:247–253. [Google Scholar]
- 18.Li S, Lundquist K, Wallis FA. Phytochemistry. 1998;49:2125–2128. [Google Scholar]
- 19.Mcghie TK, Markham KR. Phytochem. Anal. 1994;5:121–126. [Google Scholar]
- 20.Breton F, Jose L, Gonzalez A, Rodriguez RM. Anal. Quim. 1969;65:297–301. [Google Scholar]
- 21.Li C, Liu Y, Gao Y, Zhang CZ. J. Chin. Med. Mat. 2005;28:101–102. [PubMed] [Google Scholar]
- 22.Cong Y, Wang JH, Guo HR, Li X. Chin. J. Med. Chem. 2003;13:349–352. [Google Scholar]
- 23.Chen Q, Wu LJ, Ruan LJ. J. Shenyang Pharma. Univ. 2002;19:257–259. [Google Scholar]
- 24.Xiao YP, Chen JJ, Zhang YH, Shao ZY, Xu DQ. Chin. J. Marine Drugs. 2004;23:11–13. [Google Scholar]
- 25.Wu TS, Furukawa H. J. Nat. Prod. 1982;45:721–724. doi: 10.1021/np50024a013. [DOI] [PubMed] [Google Scholar]
- 26.Grammes C, Burkhardt G, Becker H. Phytochemistry. 1994;35:1293–1296. [Google Scholar]
- 27.Jutiviboonsuk A, Zhang HJ, Kondratyuk TP, Herunsalee A, Chaukul W, Pezzuto JM, Fong HHS, Bunyapraphatsara N. Pharm. Biol. 2007;45:185–194. [Google Scholar]
- 28.Yang JH, Kondratyuk TP, Marler LE, Qiu X, Choi YS, Cao HM, Sturdy M, Pegan S, Liu Y, Wang LQ, Mesecar AD, van Breemen RB, Pezzuto JM, Fong HHS, Chen YG, Zhang HJ. Phytochemistry. 2010;71:641–647. doi: 10.1016/j.phytochem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldwin AS. J. Clin. Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubet RA, Steele VE, Casebolt TL, Eto I, Kelloff GJ, Grubbs CJ. Carcinogenesis. 1994;15:2775–2780. doi: 10.1093/carcin/15.12.2775. [DOI] [PubMed] [Google Scholar]
- 31.Gunson DE, Steele RE, Chau RY. Br. J. Cancer. 1995;72:72–75. doi: 10.1038/bjc.1995.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowell JA, Steele VE, Sigman CC, Fay JR. Mol. Cancer Ther. 2003;2:815–823. [PubMed] [Google Scholar]
- 33.Chen S, Wu K, Knox R. Free Radic. Biol. Med. 2000;29:276–284. doi: 10.1016/s0891-5849(00)00308-7. [DOI] [PubMed] [Google Scholar]
- 34.Long DJ, 2nd, Gaikwad A, Multani A, Pathak S, Montgomery CA, Gonzalez FJ, Jaiswal AK. Cancer Res. 2002;62:3030–3036. [PubMed] [Google Scholar]
- 35.Long DJ, 2nd, Iskander K, Gaikwad A, Arin M, Roop DR, Knox R, Barrios R, Jaiswal AK. J. Biol. Chem. 2002;277:46131–46139. doi: 10.1074/jbc.M208675200. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Hsieh TC, Zhang Z, Ma Y, Wu JM. Biochem. Biophys. Res. Commun. 2004;323:743–749. doi: 10.1016/j.bbrc.2004.08.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buryanovskyy L, Fu Y, Boyd M, Ma YL, Hsieh TC, Wu JM, Zhang ZT. Biochemistry. 2004;43:11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calamini B, Santarsiero BD, Boutin JA, Mesecar AD. Biochem. J. 2008;413:81–91. doi: 10.1042/BJ20071373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maiti A, Reddy PVN, Sturdy M, Marler L, Pegan SD, Mesecar AD, Pezzuto JM, Cushman M. J. Med. Chem. 2009;52:1873–1884. doi: 10.1021/jm801335z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Le WD, Pan TH, Stringer JL, Jaiswal AK. J. Gerontology: Biol. Sci. 2008;63A:127–134. doi: 10.1093/gerona/63.2.127. [DOI] [PubMed] [Google Scholar]
- 41.Fu Y, Buryanovskyy L, Zhang ZT. J. Biol. Chem. 2008;283:23829–23835. doi: 10.1074/jbc.M801371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harada S, Tachikawa H, Kawanishi Y. Psychiatr. Genet. 2003;13:205–209. doi: 10.1097/00041444-200312000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Cuendet M, Pezzuto JM. Drug Metab. Drug Interac. 2000;17:109–157. doi: 10.1515/dmdi.2000.17.1-4.109. [DOI] [PubMed] [Google Scholar]
- 44.Homhual S, Zhang HJ, Bunyapraphatsara N, Kondratyuk TP, Santasiero BD, Mesecar AD, Herunsalee A, Chaukul W, Pezzuto JM, Fong HHS. Planta Med. 2006;72:255. doi: 10.1055/s-2005-873171. [DOI] [PubMed] [Google Scholar]
- 45.Lee D, Bhat KPL, Fong HHS, Farnsworth NR, Pezzuto JM, Kinghorn AD. J. Nat. Prod. 2001;64:1286–1293. doi: 10.1021/np010288l. [DOI] [PubMed] [Google Scholar]
- 46.Schupp P, Kohlert-Schupp C, Whitefield S, Engemann A, Hemscheidt T, Pezzuto JM, Kondratyuk TP, Park EJ, Marler L, Rostama B, Wright AD. Nat. Prod. Commun. 2009;65:1717–1728. [PMC free article] [PubMed] [Google Scholar]
- 47.Cao H, Xiao L, Park G, Wang X, Azim AC, Christman JW, van Breemen RB. Anal. Biochem. 2008;372:41–51. doi: 10.1016/j.ab.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Breemen RB, Huang CR, Nikolic D, Woodbury CP, Zhao YZ, Venton DL. Anal. Chem. 1997;69:2159–2164. doi: 10.1021/ac970132j. [DOI] [PubMed] [Google Scholar]
- 49.Mi QW, Lantvit D, Reyes-Lim E, Chai HB, Zhao WM, Lee IS, Peraza-Sanchez S, Ngassapa O, Kardono LBS, Riswan S, Hollingshead MG, Mayo JG, Farnsworth NR, Cordell GA, Kinghorn AD, Pezzuto JM. J. Nat. Prod. 2002;65:842–850. doi: 10.1021/np010322w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.