Abstract
Quantitative phosphorylation analysis is essential to understanding cellular signal transductions. Here we present a novel technology for the highly efficient assay of protein phosphorylation in high throughput format without the use of phospho-specific antibodies. The technique is based on a water-soluble, nanosize polymer, termed pIMAGO that is multi-functionalized with titanium (IV)ions for specific binding to phosphoproteins, and with biotin groups that allow for enzyme-linked spectrometric detection. The sensitivity, specificity and quantitative nature of pIMAGO for phosphorylation assays were examined with standard phosphoproteins and with purified phosphoproteins from whole cell extracts. As low as 100 pg of phosphoprotein can be measured quantitatively with the pIMAGO chemiluminescence assay. The pIMAGO assay was applied to an in vitro kinase assay, kinase inhibitor screening, and measurement of endogenous phosphorylation events. The technique provides a universal, quantitative method for global phosphorylation analysis with high sensitivity and specificity.
Introduction
Protein phosphorylation is an essential post-translational modification that regulates numerous cellular functions, including cell cycle progression, proliferation, differentiation, signal transduction and apoptosis.1, 2 Changes in phosphorylation dynamics within the cell have been linked to the onset and development of numerous diseases, most notably cancer.3 Accordingly, phosphorylation analysis, in particular the quantitative measurement of changes in phosphorylation, is vital to understand how signaling networks interact and function, and how they are mis-regulated in disease states.
Current methods for phosphorylation analyses include the use of phospho-specific antibodies, 32P radioactive labeling, and mass spectrometry. The method of choice may vary depending on many factors, including the specific question being asked and availability of specialized equipment or reagents. Mass spectrometry is a powerful tool that allows for the identification of novel phosphorylated proteins and sites of phosphorylation.4–6 Mass spectrometry, however, is biased toward certain phosphorylated sites and it is in general unquantitative. Therefore isotope dilution and MRM/SRM (multiple/single reaction monitoring) mass spectrometry are typically used for quantitative measurement with relatively high sensitivity.7, 8 Furthermore, many research groups do not have access to the required instrumentation, and therefore routine analysis of phosphorylation using mass spectrometry is often impractical. Similarly, inductively coupled plasma (ICP) mass spectrometry can be used for accurate detection of absolute amounts of phosphorus in the sample but the method is unlike for daily analysis.9 Many researchers would benefit greatly from a simpler technique that allows detection of phosphorylation in the average research lab. The most commonly used methods in this category include the utilization of phospho-specific antibodies and 32P labeling in a Western blot or Enzyme-Linked-Immunosorbent Assay (ELISA)formats. A classical approach to directly measure protein phosphorylation involves the incubation of whole cells with radiolabeled 32P-orthophosphate, the generation of cellular extracts, separation of proteins by SDS-PAGE, and exposure on film. This labor-intensive method requires many multi-hour incubations and the use of large doses of radioisotopes, which are toxic to the cells.10 Therefore, 32P radioisotope labeling has more frequently been used in in vitro kinase assays where kinase activity within a biological sample is measured in vitro by incubating the immunoprecipitated kinase with an exogenous substrate in the presence of ATP-γ-32P. Measurement of phosphorylated substrates can be assessed by autoradiography or scintillation counting. The use of radioisotopes is more efficient with this method, but it is still a serious safety concern. In light of these radioisotope related issues, the development of phosphorylation-dependent antibodies was a welcomed event for researchers. The main caveat in utilizing phospho-specific antibodies, however, is that successful detection is dependent on the specificity, availability, and affinity of the antibody for the phosphoprotein of interest. With increasing discovery of new phosphorylation events, there is an urgent need for simple and general technology for assaying protein phosphorylation.
We introduce here a novel strategy based on a multifunctionalized soluble nanopolymer for the detection of protein phosphorylation in a 96-well plate. The procedure is similar to ELISA that has long been used for successful identification and quantitation of biological molecules and their activities. Though quantitation of protein amounts is probably the most common application for ELISA, recently, phosphorylation assay by ELISA has been available as kits for individual phosphorylation events.11–13 Typically, a synthetic peptide substrate and a phosphospecific antibody are employed for the assay.14 Alternatively, a general antibody for the protein of interest is immobilized on a solid-phase support and then incubated with a sample mixture containing the antigen.15 After antigen binding and washing, a phospho-antibody is added that is specific to the antigen’s site of phosphorylation. The technique allows for quantitative measurement of phosphorylation (sometimes absolute) in comparison to total amount of protein present, provided that a phosphorylated standard is available. Despite frequent usage of the currently available phospho-ELISA methods, there are multiple shortcomings that limit their applications. First, in order to use the assay, the site of phosphorylation of interest has to be known beforehand, thus limiting the analysis to only well-characterized phosphorylation events. Second, an effective phosphosite-specific antibody has to be made for every single phosphosite and phosphoprotein, making the assays very costly and difficult if the antibody is unavailable. Though good generic phosphotyrosine antibodies are accessible for use, the generation of a universal phosphoserine or phosphothreonine antibody has had only limited success. Third and finally, while much progress has been made in the field of antibody generation, development of phosphosite-specific antibodies faces the challenges of poor selectivity, overall reduced quality, and higher cost compared to regular antibodies.
There have been several non-antibody assay formats based on the chelation of trivalent metal ions, such as Fe(III), with phosphate groups, including Immobilized Metal Ion Affinity-Based Fluorescence Polarization (IMAP),16 IQ,17 QTL Lightspeed18 and so on.19 While these assays show great promise in terms of versatility, they are limited by low specificity of binding between trivalent metal ions and phosphate groups. The detection is typically achieved through measuring fluorescence polarization or quenching, and therefore it requires fluorescent labeling on proteins or peptides.19 Additionally, homogeneous technologies like IMAP suffer from interference from high concentrations of ATP and other negative ions, as well as from the problem of the recombinant kinase preparations being insufficiently active for the fluorescence polarization or transfer formats.20 The applications of these non-antibody-based assays have been limited to mainly measure kinase activity using labeled peptides as artificial substrates.
To alleviate the afore-mentioned problems, the new non-antibody based technique we present here is capable of detecting phosphoproteins using a commonly available ELISA format. The core of the technique is a water-soluble nanopolymer (e.g., dendrimer), termed pIMAGO (phospho-image), that has been functionalized with Ti ions for selective binding to phosphorylated residues21–23 and with biotin molecules for versatile detection. The technique was characterized with quantitative measurements of standard phosphoproteins and phosphoproteins isolated directly from whole cell extracts. Applications of the technique were demonstrated with in vitro kinase assays, kinase inhibitor screening and the measurement of endogenous cell cycle-dependent phosphorylation.
Experimental Section
Experimental details in materials, synthesis of pIMAGO reagent, purification of Acm1 and Cdk, in vitro kinase assay, inhibition assay, and Cdh1 immunoprecipitation and detection were included in the Supporting Information.
pIMAGO-based detection of phosphoproteins in a 96-well plate
Individual proteins, i.e., the standard proteins(α-casein, β-casein, BSA, and α-lactalbumin), purifiedAcm1, kinase substrates band3 and Cdh1, were adsorbed onto a polystyrene 96-well plate by incubating each well with the designated amount in 100 uL of carbonate buffer, pH 9.6, overnight at 4°C. The protein solution was then removed and the wells were blocked for 30 minutes with 200 uL of SuperBlock T20 blocking buffer supplemented with 1% BSA. Where specified, wells were incubated for 1 hour with 2 uL of Calf Intestine Alkaline Phosphatase (CIAP) in 1X CIAP buffer at 30°C in order to dephosphorylate the proteins. Successively, each well was incubated with 500 nL of the prepared pIMAGO reagent in 100 uL of 500 mM glycolic acid/1% trifluoroacetic acid solution for 1 hour. The wells were washed 4 times with the 500 mM glycolic acid/1% trifluoroacetic acid solution, twice with the SuperBlock T20 blocking buffer, and blocked again for 30 minutes with the SuperBlock T20 blocking buffer containing 1% BSA. Finally, the wells were incubated for 1 hour with 100 uL of the 1:2,000 dilution of avidin-HRP in SuperBlock T20 blocking buffer containing 1% BSA. After washing with the SuperBlock T20 blocking buffer, 100 uL of the HRP substrate was added (either for colorimetric or chemiluminescent detection) and the plate was incubated for 2–3 minutes to allow the signal to develop. For colorimetric assays, further signal development was stopped by addition of 150 uL of 2% oxalic acid. Finally, the plate was subjected to either absorbance detection at 415 nm wavelength or luminescence detection using the Biotek plate reader. All of the phosphoprotein signals were normalized by subtracting any background signal produced by nonphosphorylated proteins, which were used at higher concentration to eliminate any potential false positives.
Results and Discussion
Validation of pIMAGO strategy for phosphorylation assay
The ability to confidently detect whether a given protein is phosphorylated using an easy and straightforward method would be extremely useful. Recently, we introduced a novel reagent, which we named PolyMAC(polymer-based metal affinity capture), based on a soluble nanopolymer functionalized with Ti ions for the isolation of phosphopeptides with exceptionally high selectivity (>95% selectivity from whole cell extracts).23 PolyMAC reagent is a soluble polyamidoamine synthetic nanopolymer (e.g. dendrimer) with a hyperbranched surface that can be functionalized with desired chemical groups. The advantages of using a dendrimer include outstanding solubility, high structural and chemical homogeneity, compact spherical shape, and controlled surface functionalities.24 The homogeneous and hyper-branched nature of the reagent exhibited superior specificity for phosphophopeptides, unparalleled yield, and fast binding kinetics.
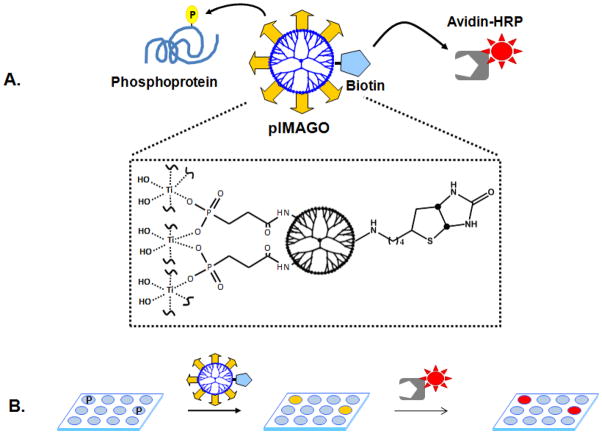
We applied the same concept with a few modifications for specific detection of phosphorylated proteins bound to a 96-well plate in ELISA format. As a result, a novel reagent, pIMAGO (Figure 1A), was prepared by multi functionalization with titanium metal ion groups, enabling strong and selective binding to phosphoproteins, and with multiple biotin molecules (the major difference between pIMAGO and PolyMAC which is functionalized with aldehyde groups)that can bind to enzyme-linked avidin for superior detection sensitivity. Multiple titanium and biotin groups on each dendrimer improve binding efficacy and enhance the signal, allowing for efficient detection of low abundant phosphoproteins. The procedure for the detection of phosphoproteins by pIMAGO in a 96-well format is outlined in Figure 1B. Briefly, after the immobilization of proteins/phosphoproteins on a well plate, the well is blocked to prevent nonspecific binding and then incubated with the pIMAGO reagent under highly acidic conditions to prevent non-specific binding. After washing away the unbound reagent, horseradish peroxidase-linked avidin (avidin-HRP) is added to the well, coupling the biotin groups on the dendrimer to a detection system. Final detection is achieved by adding substrates of HRP for colorimetry-or chemiluminescence-based measurement. The measurements are carried using a plate reader at a 415 nm OD setting for colorimetry assays or chemiluminescence detection setting for chemiluminescent assays.
Figure 1.
A) A schematic representation of the pIMAGO reagent. A soluble nanopolymer dendrimer is functionalized with multiple titanium ions for selective binding to the phosphoproteins, and with biotin groups for sequential detection by avidin-linked HRP.B) Experimental workflow for pIMAGO-based phosphoprotein detection in a 96-well plate format. After phosphoproteins of interest are bound to the wells and desired manipulations are performed, the wells are briefly incubated with the pIMAGO reagent which selectively binds to phosphosites. HRP-linked avidin is then added into the wells, exclusively attaching to the biotin groups of pIMAGO. Finally, a colorimetric or chemiluminescent HRP substrate is added to the wells for the detection.
We first demonstrated the practicality and selectivity of the pIMAGO assay method on a number of standard phosphoproteins bound to the plate. When compared to unphosphorylated bovine serum albumin (BSA) and α-lactoglobulin, the colorimetry-based signals detected from equal amounts(100 ng)of the phosphorylated α-casein and β-casein were approximately 20- and 10-fold stronger, respectively (Figure S1). To illustrate that the observed differences in signal were due to protein phosphorylation, we treated all proteins with Calf Intestine Alkaline Phosphatase (CIAP), a general phosphatase capable of dephosphorylating most sites and motifs. After treatment, the pIMAGO-avidin-based signal was only slightly above the background of the nonphosphoproteins. We further validated that the signal was indeed due to the specific binding of the pIMAGO technology by including a number of controls that ommited one or more step s in the protocol or different components in the pIMAGO reagent. As shown in Figure S2, when the pIMAGO or avidin-HRP reagents were not included in the procedure or pIMAGO had missing functional groups, no signal was detected from either of the proteins. Consequently, after the addition of both reagents, the signal detected for β-casein was greatly increased above the background. Because a tiny signal increase was also observed in the control wells containing no protein or BSA(a very common occurance during standard ELISA analyses), we subsequently used the signal from a control nonphosphoprotein as the backround for the remainder of experiments.
Quantitative capabilities of pIMAGO-based detection
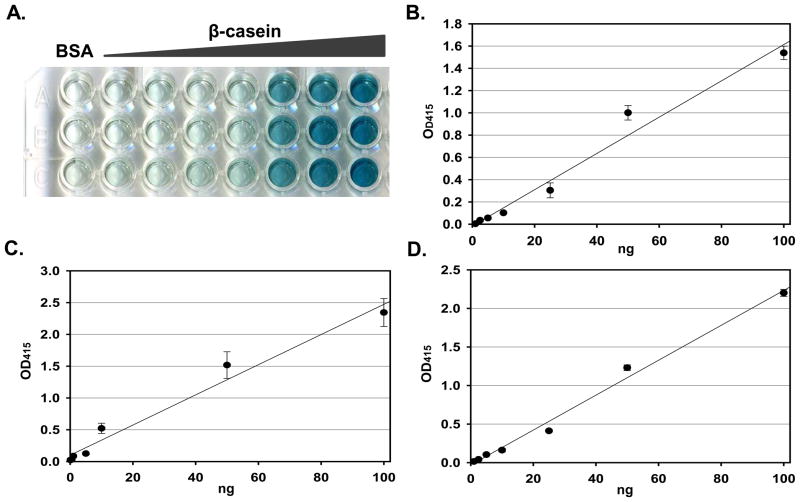
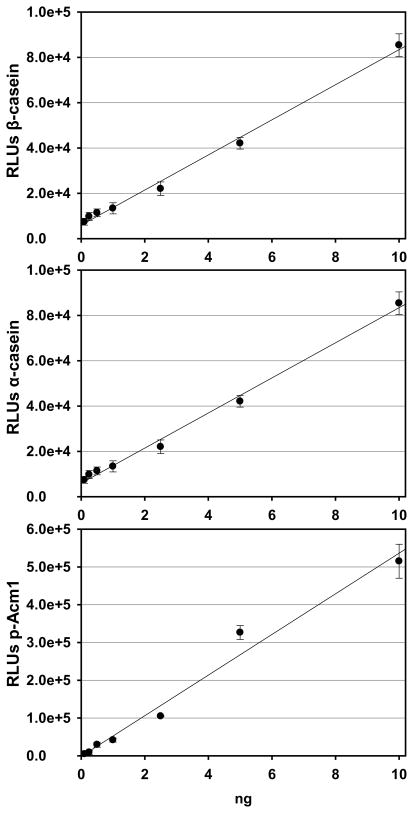
In the next step, the quantitative capability of pIMAGO-based phosphorylation assay was investigated. Two standard phosphoproteins, α-casein and β-casein, and a purified phosphorylated yeast protein, phospho-Acm1, were employed for quantitative analyses. Nonphosphorylated proteins(100 ng of BSA or unphosphorylated Acm1) were used as the background controls (see Figure 2A for a visual signal from the β-casein sample detection, and Figure 2B–D for graphical signal representation against BSA or Acm1 background). The experiments based on colorimetry demonstrated outstanding quantitative capabilities of the approach with the detection limit of 1 ng of the phosphoproteins. Furthermore, we contemplated that the approach could be more sensitive if we replace d the colorimetric substrate with a chemiluminescent substrate.25 The pIMAGO method can detect the signal from as little as 1 pg of α-casein (Figure S3). However, the nonphosphoprotein BSA also yielded detectable signal and quantitation was not inaccurate at such low range of concentations. On the other hand, the signal resulting from 100 pg of α-casein was much stronger than that of 100 pg of BSA, indicating that the lowest limit of pIMAGO for selective detection of phosphoproteins is around 100 pg, based on the model system. As shown in Figure 3, the chemiluminescence procedure allowed for the detection of a range of phosphoprotein concentrations, starting at just 100 pg, while still retaining quantitative capabilities. The experiments revealed linear dynamic range of the assay in the range of 100 pg-10 ng (expanded graphs of the lower ranges of concentrations are shown in Figure S4). These results demonstrate that pIMAGO can be successfully applied for the highly sensitive and quantitative measurement of phosphorylation content in proteins.
Figure 2.
Quantitation of colorimetric signals from pIMAGO-based detection of purified phosphoproteins. A–B. Different amounts of β-casein were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 100 ng of BSA background; B is the graphical representation of A). C. Different amounts of α-casein were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 100 ng of BSA background). D. Different amounts of purified phosphorylated Acm1 were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 100 ng of nonphosphorylated Acm1 background). Error bars represent standard deviations from the average of 3 experiments.
Figure 3.
Quantitation of chemiluminescence signal from pIMAGO-based detection of standard phosphoproteins. Top: Different amounts of β-casein were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 10 ng of BSA background). Middle: Different amounts of α-casein were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 10 ng of BSA background). Bottom: Different amounts of purified phosphorylated Acm1 were adsorbed onto the plate and measured by pIMAGO-based assays (the signal was finalized by subtracting the 10 ng of unphosphorylated Acm1 background). Error bars represent standard deviations from the average of 3 experiments.
Utility of pIMAGO for in vitro kinase assays and inhibitor screening
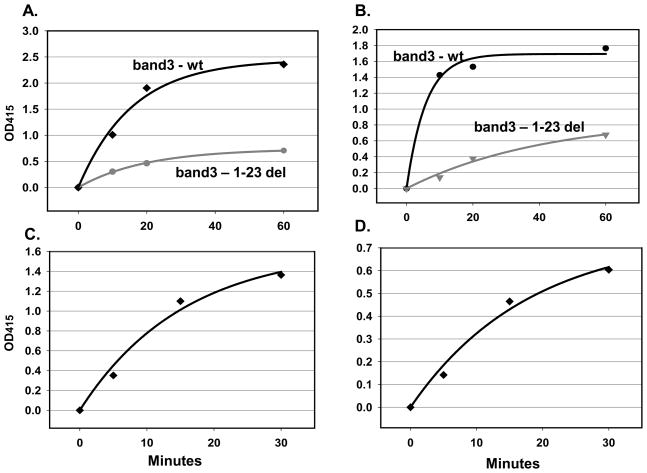
Since pIMAGO reagent is capable of detecting general phosphorylation, it can be utilized as a universal detection reagent for any phosphorylation assay. Therefore, it can be used to not only detect and quantify the phosphorylation level of a protein, but also to examine kinase or phosphatase activities (in vitro or in vivo). To demonstrate this, we immobilized full-length and 1–23 deletion mutant of band3 protein on a 96-well plate. Band3 was used as an effective substrate for the analysis of Spleen tyrosine kinase (Syk) activity in vitro26, 27. As demonstrated in Figure 4A, pIMAGO was capable of effectively detecting the increase in phosphorylation of band3 substrate over time, exhibiting typical enzymatic kinetics. As a comparison, we used BSA instead of band3 as a control to show the specificity of the reaction, which produced no change in signal (data not shown). Furthermore, the signal was greatly decreased when the band3 1–23 deletion mutant was utilized as a substrate because this mutant is missing 2 major Syk-dependant tyrosine phosphorylation sites, again demonstrating the specificity and sensitivity of pIMAGO. We were able to corroborate these results using a general anti-phosphotyrosine (pTyr) antibody instead of pIMAGO (Figure 4B).
Figure 4.
In vitro tyrosine kinase assay based on pIMAGO. Syk activity with full-length or 1–23 deletion mutant of band3 substrate detected by A) pIMAGO and B) anti-phosphotyrosine antibody. In vitro serine/threonine kinase assay of Cdk activity with C) Acm1 as the substrate, or D) Cdh1 as the substrate, detected by pIMAGO. The signal was recorded by subtracting the background of the kinase assay lacking the kinases.
Phosphorylation assay based on pIMAGO would be particularly useful to analyze serine/threonine substrate phosphorylation, for which an effective general antibody is lacking. Currently, detection of Ser/Thr phosphorylation usually requires specific antibodies against individual phosphorylation sites in phospho-ELISA assays, thus requiring the knowledge of phosphorylation site a priori. We demonstrated the general utility of pIMAGO for serine/threonine kinase assays with two purified substrates (Cdh128 and Acm129) of cyclin-dependent kinase 1 (Cdk1) from budding yeast. After the two substrates were individually immobilized on a plate, a 30-minute kinase reaction was carried out with purified Cdk1 (Figure 4C, D). Both in vitro kinase assays demonstrated the ability of pIMAGO to successfully detect changes in general phosphorylation level of the substrates. Again, BSA was also included as a control, producing no change in signal(data not shown). These results indicate that pIMAGO can be successfully utilized for selective and sensitive kinase assay, overcoming the limitation of expensive or unavailable antibodies. This approach can be particularly useful when the sites of phosphorylation are not known or when there are multiple putative sites that would require the use of multiple phosphosite-specific antibodies.
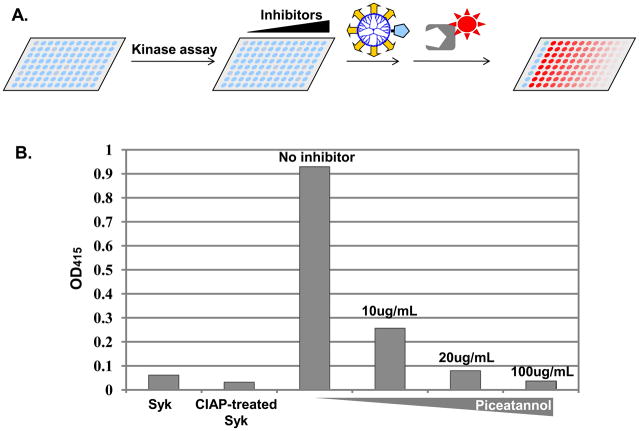
The pIMAGO-based kinase assay was further expanded to test the applicability of pIMAGO in kinase inhibitor screening. Since the great success of Gleevac® (Imatinib),30 tyrosine kinases have become major drug targets, and numerous new inhibitors have been screened every year. Typically, such screenings involve the construction of ELISA plates immobilized with known substrates and the generation of phosphosite-specific antibodies that accomodate each known site of phosphorylation. Needless to say, such an approach is expensive for high throughput screening and the assay may be biased if only one substrate is screened. The current procedure is typically able to detect the phosphorylation level of only the well-known sites, completely ignoring the less-known or currently unknown sites, which might be just as relevant. To address this challenge, we propose to employ the pIMAGO reagent for the determination of inhibitor effects on kinase activity regardless of the site of phosphorylation, with a workflow for pIMAGO-based inhibitor screening illustrated in Figure 5A. We utilized Syk kinase as the kinase of interest and examined its autophosphorylation level with or without piceatannol, a known Syk inhibitor.31 Because Syk is typically phosphorylated, we dephosphorylated purified Syk in a number of wells and then introduced ATP and MnCl2 into the wells to promote its autophosphorylation. In the specified cases, piceatannol was added along with the ATP and MnCl2 at different concentrations (10, 20 or 100 μg/mL). The signal was recorded by subtracting the background of BSA that was also incubated with ATP and MnCl2 to eliminate any false positive signal due to nonspecifically bound ATP. In the experiments, we did not see any increase in BSA signal after incubation with ATP(data not shown). As shown in Figure 5B, the signal was reduced after dephosphorylation to approximately the background level, and then greatly increased after the in vitro kinase assay. The phosphorylation signal increase was compromised when piceatannol was introduced. This is particularly true when100 ug/mL of piceatannol was applied, where the signal remained at the level of the dephosphorylated Syk. Therefore, the approach has demonstrated the capability of pIMAGO for determination of changes in kinase activity and it can be applied as a general and cost-effective method for kinase inhibitor screening.
Figure 5.
A) An illustration of experimental workflow for pIMAGO-based screening of kinase inhibitors. B) Measurement of Syk activity inhibition by piceatannol via pIMAGO assay. See text for experimental details.
Use of pIMAGO for the detection of endogenous phosphorylation
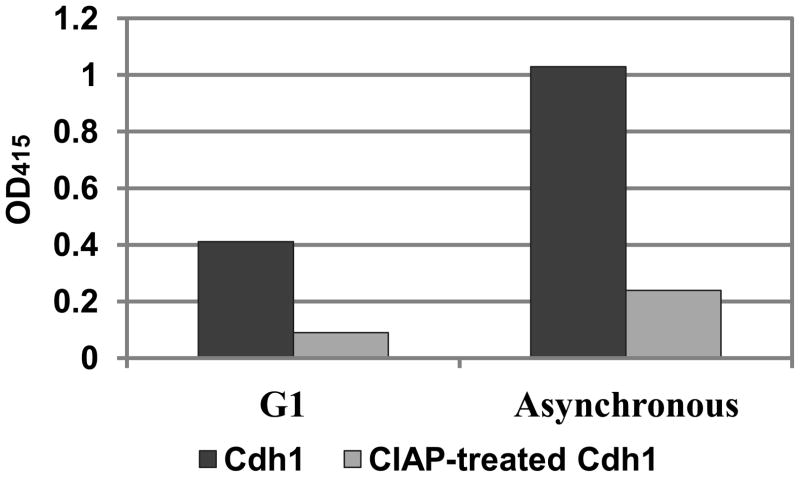
Finally, we applied the pIMAGO assay to detect the differences in physiological phosphorylation in situ. It has remained a great challenge to identify and quantify changes resulting from in vivo phosphorylation because good phosphospecific antibodies are required for each site of interest. Moreover, only a limited number of options are available when the modified sites are not known (e.g. mass spectrometry). To test whether pIMAGO is capable of detecting the changes in physiological phosphorylation, we used the immunoprecipitation approach followed by pIMAGO detection of the Cdh1 yeast protein purified at two different stages of cell cycle. Cdh1 has an important role in cell cycle regulation by establishing a stable G1 state until cells receive the appropriate signal to divide. In G1, Cdh1 is in a dephosphorylated and active state, and it is able to target specific substrates to the anaphase-promoting complex (APC) for ubiquitination and degradation. Once cells commit to division in late G1, Cdh1 is phosphorylated by Cdk, rendering it unable to recruit its substrates to the APC, and allowing accumulation of proteins that promote the subsequent S and M phase.28, 32, 33 In late M, Cdh1 is dephosphorylated by the phosphatase Cdc14, promoting mitotic exit and initiating another cycle.28 To examine the cell cycle-dependent phosphorylation status of Cdh1, we collected asynchronous growing cells, which contain mostly S, G2 and M phase cells with phosphorylated Cdh1, and cells arrested in the G1 phase, which contain primarily unphosphorylated Cdh1. FLAG-tagged Cdh1 was immunoprecipitated from cell extracts derived from the two cell populations using anti-FLAG antibody-coated resin. The amount of total protein was normalized and the level of phosphorylation was detected with pIMAGO. As expected, the phosphorylation state ofCdh1 purified from asynchronous cells was much higher than that from the G1-arrested cells(Figure 6), consistent with previous results.28 We further confirmed that the signal detected was due to phosphorylation by treating the bound Cdh1 with CIAP, which resulted in significant decrease in signal. The results reveal that the pIMAGO assay is particularly appealing to measure new phosphorylation events on endogenous proteins under different conditions without a priori knowledge of sites of phosphorylation.
Figure 6.
Detection of Cdh1 phosphorylation at different stages of cell cycle with pIMAGO. The samples were also dephosphorylated by CIAP to examine signal specificity. The signal was recorded by subtracting the 100 ng of BSA background.
Conclusion
A novel, universal technique for the detection of phosphorylation in a 96-well format was presented. The pIMAGO-based ELISA-like procedure is simple and efficient for qualitative and quantitative assessment of protein phosphorylation without the use of phosphospecific antibodies. The technique can detect any global changes in phosphorylation originally from an increase in phosphorylation of a particular site or from phosphorylation of any additional sites. When combined with other affinity enrichments on the plate, it is conceivable that the technology can be applied to directly analyze a complex sample such as a cell lysate. The pIMAGO approach offers a high-throughput, sensitive method for a variety of assays, ranging from quantitation of phosphorylation and in vitro kinase assays, to kinase inhibitor screening. The new technique has the potential to become a major analytical tool for phosphorylation assays.
Supplementary Material
Acknowledgments
This project has been funded in part by an NSF CAREER award, a 3M general fund, and by National Institutes of Health grant 1R01GM088317. We thank Dr. Phillip Low for general contribution of purified band3 protein.
References
- 1.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Pawson T. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 3.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, Ong SE, Gronborg M, Steen H, Jensen ON, Pandey A. Trends Biotechnol. 2002;20:261–268. doi: 10.1016/s0167-7799(02)01944-3. [DOI] [PubMed] [Google Scholar]
- 5.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Tao WA, Wollscheid B, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 7.Iliuk A, Galan J, Tao WA. Anal Bioanal Chem. 2009;393:503–513. doi: 10.1007/s00216-008-2386-0. [DOI] [PubMed] [Google Scholar]
- 8.Tao WA, Aebersold R. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 9.Kruger R, Zinn N, Lehmann WD. Methods in molecular biology (Clifton, NJ) 2009;527:201–218. ix. doi: 10.1007/978-1-60327-834-8_15. [DOI] [PubMed] [Google Scholar]
- 10.Cooper PC, Burgess AW. Analytical biochemistry. 1985;144:329–335. doi: 10.1016/0003-2697(85)90125-3. [DOI] [PubMed] [Google Scholar]
- 11.Bianco C, Giovannetti E, Ciardiello F, Mey V, Nannizzi S, Tortora G, Troiani T, Pasqualetti F, Eckhardt G, de Liguoro M, Ricciardi S, Del Tacca M, Raben D, Cionini L, Danesi R. Clin Cancer Res. 2006;12:7099–7107. doi: 10.1158/1078-0432.CCR-06-0833. [DOI] [PubMed] [Google Scholar]
- 12.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, 3rd, Edeen PT, Floyd M, Ford JM, Grotzfeld RM, Herrgard S, Insko DE, Mehta SA, Patel HK, Pao W, Sawyers CL, Varmus H, Zarrinkar PP, Lockhart DJ. Proc Natl Acad Sci U S A. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rani CS, Qiang M, Ticku MK. Molecular pharmacology. 2005;67:2126–2136. doi: 10.1124/mol.104.007872. [DOI] [PubMed] [Google Scholar]
- 14.Yu JS, Chang SH, Chan WH, Chen HC. J Biochem. 2001;129:243–251. doi: 10.1093/oxfordjournals.jbchem.a002851. [DOI] [PubMed] [Google Scholar]
- 15.Pattoli MA, MacMaster JF, Gregor KR, Burke JR. J Pharmacol Exp Ther. 2005;315:382–388. doi: 10.1124/jpet.105.087569. [DOI] [PubMed] [Google Scholar]
- 16.Sportsman JR, Daijo J, Gaudet EA. Combinatorial chemistry & high throughput screening. 2003;6:195–200. doi: 10.2174/138620703106298374. [DOI] [PubMed] [Google Scholar]
- 17.Morgan AG, McCauley TJ, Stanaitis ML, Mathrubutham M, Millis SZ. Assay and drug development technologies. 2004;2:171–181. doi: 10.1089/154065804323056512. [DOI] [PubMed] [Google Scholar]
- 18.Xia W, Rininsland F, Wittenburg SK, Shi X, Achyuthan KE, McBranch DW, Whitten DG. Assay and drug development technologies. 2004;2:183–192. doi: 10.1089/154065804323056521. [DOI] [PubMed] [Google Scholar]
- 19.Olive DM. Expert Rev Proteomics. 2004;1:327–341. doi: 10.1586/14789450.1.3.327. [DOI] [PubMed] [Google Scholar]
- 20.Sharlow ER, Leimgruber S, Yellow-Duke A, Barrett R, Wang QJ, Lazo JS. Nat Protoc. 2008;3:1350–1363. doi: 10.1038/nprot.2008.111. [DOI] [PubMed] [Google Scholar]
- 21.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Anal Chem. 2004;76:3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 22.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Iliuk AB, Martin VA, Alicie BM, Geahlen RL, Tao WA. Mol Cell Proteomics. 2010;9:2162–2172. doi: 10.1074/mcp.M110.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boas U, Heegaard PM. Chem Soc Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- 25.Lehel C, Daniel-Issakani S, Brasseur M, Strulovici B. Analytical biochemistry. 1997;244:340–346. doi: 10.1006/abio.1996.9894. [DOI] [PubMed] [Google Scholar]
- 26.Harrison ML, Isaacson CC, Burg DL, Geahlen RL, Low PS. J Biol Chem. 1994;269:955–959. [PubMed] [Google Scholar]
- 27.Brunati AM, Bordin L, Clari G, James P, Quadroni M, Baritono E, Pinna LA, Donella-Deana A. Blood. 2000;96:1550–1557. [PubMed] [Google Scholar]
- 28.Jaspersen SL, Charles JF, Morgan DO. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 29.Hall MC, Jeong DE, Henderson JT, Choi E, Bremmer SC, Iliuk AB, Charbonneau H. J Biol Chem. 2008;283:10396–10407. doi: 10.1074/jbc.M710011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, Lydon NB. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 31.Oliver JM, Burg DL, Wilson BS, McLaughlin JL, Geahlen RL. J Biol Chem. 1994;269:29697–29703. [PubMed] [Google Scholar]
- 32.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.