Abstract
The reliable and sensitive detection of cancer-specific biomarkers is important for the diagnosis and treatment of cancer. Hence, detection of these biomarkers has to be reliably and rapidly performed in diverse settings. A limitation of the conventional biomarker-screening method of ELISA is the employment of labile components, such as hydrogen peroxide and horseradish peroxidase. Previously, we reported that nanoceria is able to oxidize various colorimertic dyes at acidic pH, such as TMB and AzBTS, and an assay was designed for screening the folate receptor. Herein, we show that the ability of nanoceria to oxidize a substrate can be tuned by modulating the pH. Results showed that nanoceria can oxidize the non-fluorescent substrate ampliflu, either to the very stable fluorescent product resorufin at pH 7.0 or to the non-fluorescent resazurin at pH 4.0. Based on these findings, we conjugated Protein G to immobilize antibodies on the surface of nanoceria, in order to detect the expression of prototypic cancer biomarkers at pH 7.0, such as the folate receptor and EpCAM. We found that within 3 h, nanoceria identified the expression of the folate receptor and EpCAM on lung carcinoma and breast adenocarcinoma cells respectively. Traditional ELISA had a readout time of 15 h and a higher detection threshold, while requiring multiple washing steps. Considering these results and nanoceria’s ability to oxidize ampliflu to its stable fluorescent product at neutral pH, the use of antibody-carrying nanoceria in the lab and point-of-care molecular diagnostics is anticipated.
Keywords: nanoceria, cerium oxide, oxidase, ampliflu, resorufin, resazurin, ELISA
In recent years, the unique and tunable properties of nanomaterials have been utilized to create sensitive, fast and robust diagnostic assays.1–11 Recently, a colorimetric assay to detect the expression of cell surface receptors on cancer cells, using TMB (3,3’,5,5’-tetramethylbenzydine) as a chromogenic substrate and cerium oxide (nanoceria) as an oxidase-mimetic agent has been developed. One major benefit of using nanoceria in enzyme-linked immunoassay (ELISA) applications is the fact that no hydrogen peroxide is needed to facilitate the oxidation of the dye.12 However, one of the drawbacks of this colorimetric nanoceria-based ELISA method is that oxidation of the dye (TMB) by nanoceria is optimal in acidic conditions, limiting the use of antibodies and other pH-labile biomolecules as targeting ligands. Therefore, detection at neutral pH would be ideal to advance the wide application of this method, particularly when dyes that can be easily oxidized by nanoceria at normal pH are utilized. Although various colorimetric dyes are widely used in ELISA, they often have low sensitivity. In contrast, fluorescent dyes are more sensitive and their use in nanoceria-based ELISA could advance the application of this technique. Therefore, a chromophore that develops a stable fluorescence upon oxidation by nanoceria, particularly at neutral pH, would be ideal for developing a more robust nanoceria-based ELISA. Among the most widely used fluorigenic substrate is 10-Acetyl-3,7-dihydroxyphenoxazine (ampliflu red). In the presence of hydrogen peroxide, ampliflu can be enzymatically oxidized by horseradish peroxidase (HRP) to the fluorescent product resorufin (Scheme 1a).13 However, in the presence of H2O2 resorufin is quickly oxidized to the non-fluorescent product resazurin (Scheme 1a).13,14, limiting the application of ampliflu, as its fluorescent readout quicky diminishes and has to be recorded within minutes upon initiating the HRP/H2O2 reaction.
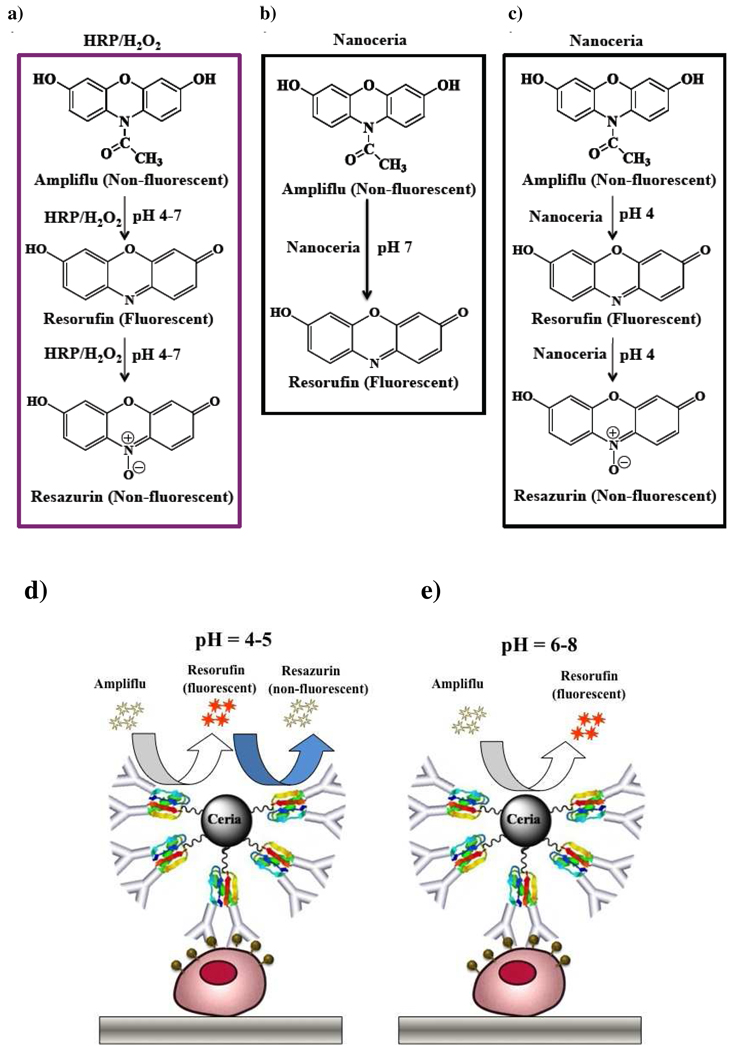
Scheme 1.
Schematic showing the HRP/H2O2 and nanoceria mediated oxidation of ampliflu. In the pH range 4–7, HRP/H2O2 oxidizes Ampliflu to a non-fluorescent final product (Resazurin). (a) In contrast, nanoceria oxidizes Ampliflu to the intermediate oxidation fluorescent product (Resorufin) at pH 7 (b), while at or below pH 5.0, nanoceria yields the terminal oxidized non-fluorescent product resazurin. (c). The ability of nanoceria to oxidize ampliflu to a stable fluorescent product in the pH range 6–8, will facilitate its use in ELISA. (d, e) without the use of H2O2.
Considering these limitations, there is a need to develop reaction conditions for the mild oxidation of ampliflu, yielding the intermediate fluorescent product (resorufin) without further oxidation to the non-fluorescent product (resazurin). Since nanoceria possess unique oxidase activity that can be tuned by changing the solution’s pH, behaving as a strong oxidant at acidic pH and weak oxidant at neutral pH,12 we hypothesized whether the oxidation of ampliflu to the fluorescent resorufin can be achieved by tuning the oxidase-like activity of nanoceria through the pH (Scheme 1b and 1c). We reasoned that at neutral pH, the weak oxidase-like activity of nanoceria could facilitate the partial oxidation of ampliflu to the fluorescent resorufin (Scheme 1b). Meanwhile, the strong oxidase-like activity of nanoceria at acidic pH would trigger the complete oxidation of ampliflu to the non-fluorescent resazurin (Scheme 1d), similar to the HRP/H2O2 system. Following this approach, a fluorigenic nanoceria-based ELISA system using ampliflu as a substrate at neutral pH could be developed (Scheme 1e)
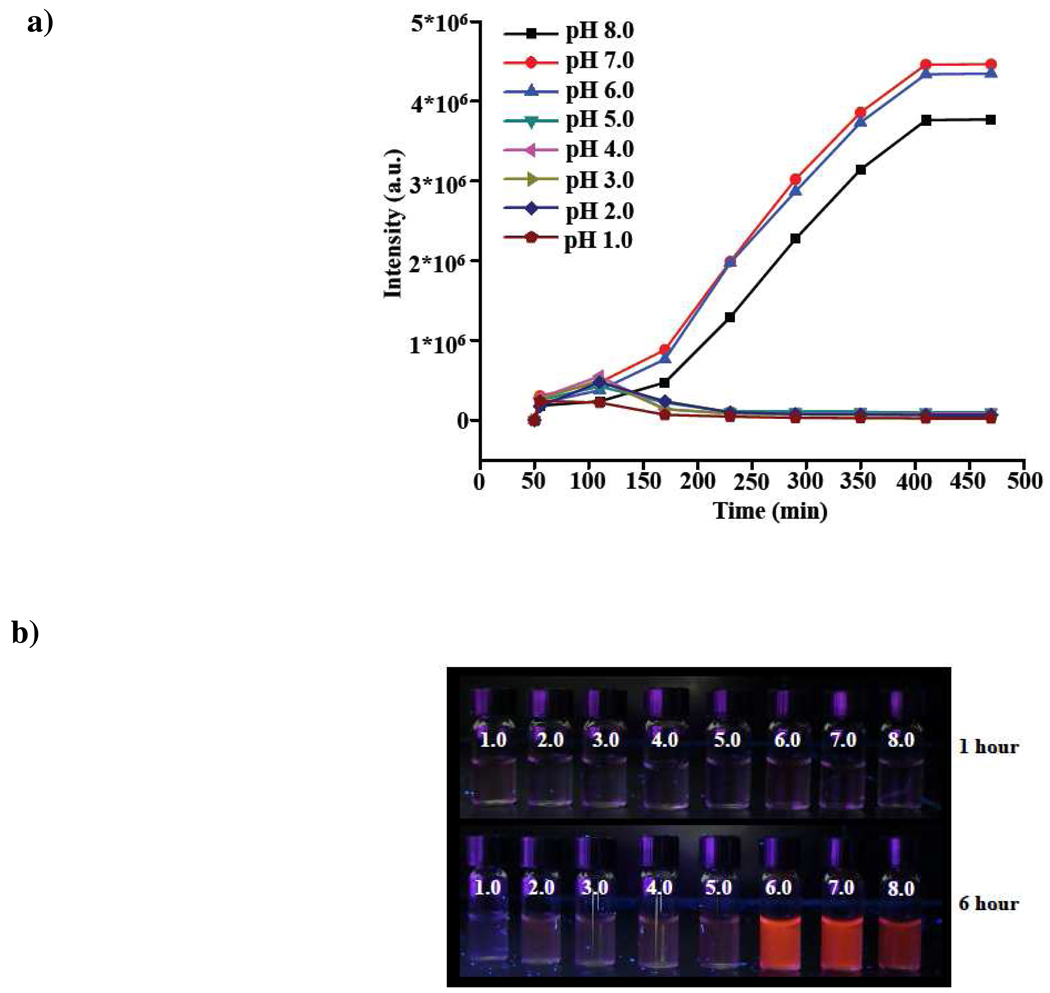
To test our hypothesis, we used polyacrylic acid-coated nanoceria (PNC) (1.0 mM) suspended in buffers ranging from pH 1.0 to 8.0. Ampliflu (1.25 µg) was then added and the fluorescence emission of the suspension was recorded at different time intervals. Results showed that nanoceria was able to oxidize ampliflu in a pH-dependent manner. Nanoceria quickly oxidized ampliflu to a stable pink-colored fluorescent product within the pH range of 6.0 to 8.0 (Figure 1a and 1b). The fluorescence emission maximum of this product was at 585 nm, which corresponds to resorufin.13,14 This fluorescence emission intensity increased until reaching a plateau after 8 h. In contrast, when nanoceria oxidized ampliflu in the pH range from 1.0 to 5.0, a weakly fluorescent product was obtained within an hour upon incubation of ampliflu with the nanoparticles (Figure 1a and 1b). The fluorescent intensity of this oxidized product quickly decreased, eventually becoming non-fluorescent after 2 h. These results indicate that nanoceria in the pH range from 6.0 to 8.0 oxidized ampliflu to the fluorescent resorufin with emission at 585 nm. In contrast, from pH 1.0 to 5.0 nanoceria transiently formed a fluorescent product that eventually was converted to non-fluorescent resazurin (Figure 1a).
Figure 1.
Fluorimetric analysis of the nanoceria-mediated oxidation of ampliflu at different pH. a) In the pH range 6–8, the fluorescence intensity at 585 nm increases with time, reaching a plateau after 6 hours (360 mins).In the pH range 1–5, fluorescence starts to develop within the first hours, but decreases right after 100 minutes, reaching zero after 3 hours (180 minutes). b) Photograph taken under UV illumination of vials containing nanoceria within the pH-range 1–8 after 1 or 6 hours of incubation at the corresponding pH.
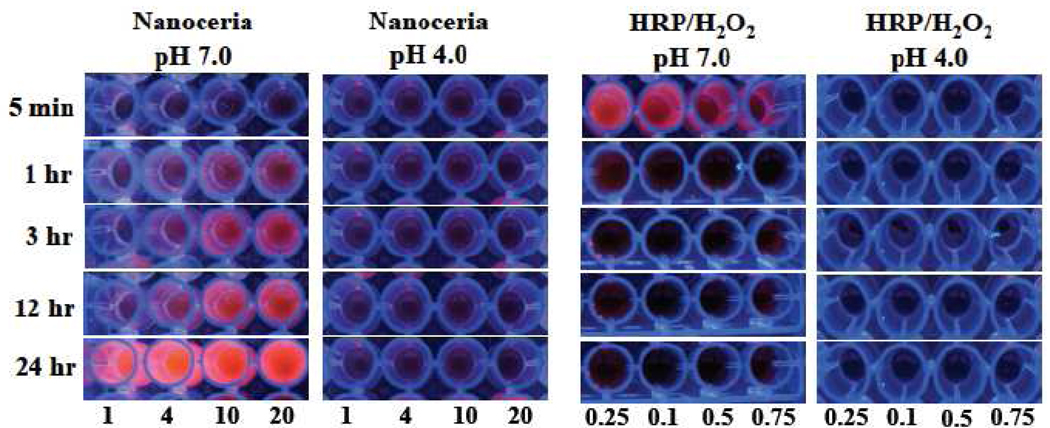
It has been previously reported that the use of high levels of hydrogen peroxide in ELISA results in inactivation of HRP and decrease in the fluorescence of the oxidized substrate.14–16 In addition, pH also plays an important role in the oxidation of ampliflu.13 Fluorescence imaging studies under UV illumination of 96-well plates containing ampliflu (1.25 µg) with increasing concentrations of nanoceria (PNC) or HRP/H2O2 revealed that the HRP/H2O2 system quickly oxidized ampliflu, developing a fluorescent product within minutes (Figure 2) that quickly decreased after 5 minutes, becoming practically undetectable. These results were expected, as it has been reported that HRP/H2O2 rapidly oxidizes ampliflu to the fluorescent product resorufin at pH 7.0 but subsequently oxidizes resorufin to the non-fluorescent resazurin.13 In contrast, when ampliflu was oxidized with nanoceria at pH 7.0, fluorescent emission started to develop within an hour and continued increasing until reaching its maximum fluorescence within 24 hours. The observed increase in fluorescence dependented on the concentration of nanoceria, as the higher the amount of nanoceria the higher fluorescence intensity was observed (Figure 2 and Figure S1 Supporting Information). This indicates that the mild oxidase activity of nanoceria at pH 7 was able to facilitate the oxidation of ampliflu to the fluorescent intermediate resorufin without further oxidazing the substrate to the non fluorescent product resazurin. Interestingly, nanoceria at pH 4.0 yielded no fluorescence, suggesting that nanoceria at pH 4.0 may primarily oxidize ampliflu to the non-fluorescent resazurin (Figure 2). As expected the HRP/H2O2 system at pH 4.0 was unable to generate a fluorescent signal, due to the instability of horseradish peroxidase at this pH (Figure 2). Taking together, these results demonstrate that nanoceria is an ideal system for long ELISA readouts at pH 7.0, as opposed to the commonly used HRP/H2O2 system.
Figure 2.
Comparison of the oxidation of ampliflu by nanoceria (PNC) and HRP/H2O2. a) Nanoceria yields a stable fluorescent product at pH 7.0, while at pH 4.0 a weakly fluorescent product is obtained. HRP/H2O2 rapidly oxidizes ampliflu to a fluorescent product at pH 7.0, which is then converted to a non-fluorescent product. At pH 4.0, HRP/H2O2 forms a non-fluorescent product. b) Photographs of nanoceria-mediated and HRP/H2O2-mediated oxidation of ampliflu at pH 7.0 and pH 4.0 under UV illumination. (numbers shown at lower bottom of photographs are various concentrations of nanoceria or HRP in uM.)
Based on these findings, we hypothesized that nanoceria at pH 7.0 could provide better detection than the HRP/H2O2 system in cancer biomarker screening assays. Considering that at pH 7.0 nanoceria is able to oxidize ampliflu to the stable fluorescent product resorufin, we reasoned that a more robust and stable fluorigenic detection system could be developed. Additionally, since no acidic conditions are required to achieve detection, the possibility of protein denaturation is reduced.
Furthermore, as nanoceria does not utilize hydrogen peroxide for a substrate’s oxidation due to the nanoparticles’ intrinsic oxidase-like activity, terminal non-fluorescent oxidation of ampliflu is also minimal. Towards these objectives, we conjugated Protein G to the polyacrylic acid-coated nanoceria (PNC) by EDC/NHS carbodiimide chemistry (Scheme 1 Supporting Information), resulting in Protein G-polyacrylic-acid-coated nanoceria (Protein G-PNC). Protein G introduces versatility to our nanoceria-based detection system, as it can immobilize the desirable antibodies for the detection of specific molecular targets simply by changing the targeting antibody. Successful conjugation of Protein G to PNC was confirmed through the BCA protein assay (0.33 µg of total protein per µL of the nanoceria preparation) and by zeta potential measurements, as upon conjugation of Protein G to nanoceria a shift in the zeta potential from −75.3 (PNC) to −35.1 mV (Protein G-PNC) (Figure S2 Supporting Information) was observed. In addition, further confirmation of Protein G binding was assessed by the ability of the resulting Protein G-PNC to immobilize antibodies for the folate receptor and EpCAM, which are important cancer biomarkers.
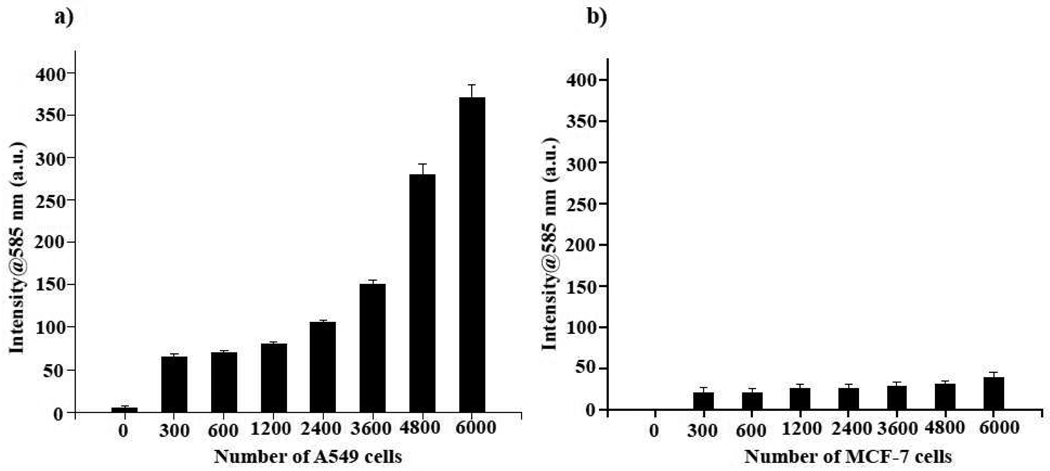
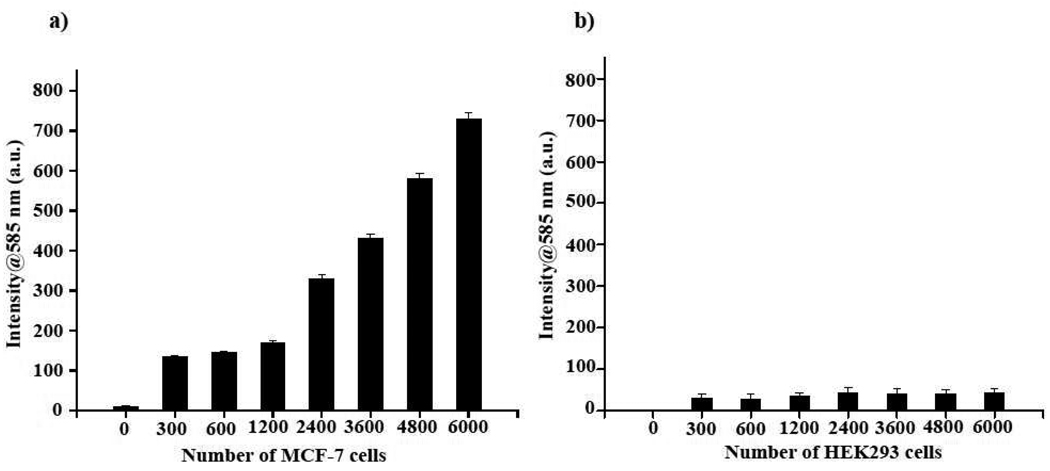
Typically in a traditional immunoassay, a horseradish peroxidase (HRP)-labeled secondary antibody is utilized to assess the binding of a specific primary antibody to a particular target or surface receptor. This binding event is assessed by the ability of HRP/H2O2 to oxidize a substrate, such as ampliflu. However, the instability of HRP and H2O2, in addition to the H2O2-induced quenching of fluorigenic substrates, could result in erroneous results. Therefore, we reasoned that an antibody-Protein G-PNC conjugate would be more robust than current HRP-based assays, since neither HRP, H2O2 nor a secondary antibody would be needed for detection. First, we immobilized an anti-folate-receptor antibody (Santa Cruz Biotech) on the Protein G-PNC conjugate. This antibody recognizes the folate receptor, which is overexpressed in many tumors.17,18 Experiments were performed using the lung cancer cell line A-549, which has been shown to overexpress this receptor.19,20 In control experiments, we used a breast carcinoma cell line (MCF-7) that does not express the folate receptor.21 Although both cell lines were grown in media having high amounts of folate (2.3 µM), subsequent treatments with the anti-folate-receptor antibody–Protein G-PNC conjugate were carried out in 1X PBS having no folate. This ruled out the possibility of any false positive results or interference from the folate in the medium. In our initial experiments, both the lung carcinoma (A-549) and breast carcinoma (MCF-7) cells were plated at cell densities, ranging from 0 to 6000 cells per well, and incubated with the anti-folate-receptor antibody–Protein G-PNC conjugate. The cells were then washed with 1X PBS and incubated with ampliflu (25 µL of 0.5 mg/mL) for 3 h before fluorescence emission was recorded at 585 nm, using a microtiter plate reader. As expected, a target-concentration-dependent signal intensity was observed in the lung carcinoma cell line (Figure 3a), as opposed to the breast carcinoma cells that lacks the folate receptor (Figure 3b). The obtained fluorescent signal was stable even after 24 hours, being able to detect as little as 1200 A-549 cells with the nanoceria conjugate.
Figure 3.
Fluorigenic determination of the expression of the folate receptor in cancer cells using an anti-folate-receptor antibody–Protein G-PNC conjugate at pH 7.0. a) Lung carcinoma cells (A-549) that overexpress the folate receptor on their surface effectively oxidized ampliflu, due to the interaction between the anti-folate-receptor antibody–Protein G-PNC conjugate and the folate receptor. b) MCF-7 breast carcinoma cells, lacking the folate receptor, showed minimal fluorescence due to the absence of any interaction with the nanoceria conjugate
In subsequent studies, we immobilized an anti-EpCAM antibody (Santa Cruz Biotech) on the Protein G PNC-conjugate and screened MCF-7 cells for the expression of this protein. EpCAM (37–40 kDa) is overexpressed on the plasma membrane of most human epithelial cancers, including the MCF-7 breast cancer cell line, which makes this biomarker a promising tumor-associated antigen for detection and therapy.22–24 Results showed an increasing trend in fluorescence intensity as the number of MCF-7 cells increased (Figure 4a). Moreover, the signal from each well was stable even after 24 h. As negative control, we used human embryonic kidney cells (HEK293), as these cells lack the EpCAM receptor. As expected, a nominal signal was observed with these cells (Figure 4b). In addition, saturation experiments adding excess anti-EpCAM antibody to the MCF-7 cells, before addition of the EpCAM antibody nanoceria conjugate, abograted the signal (data not shown). These results clearly indicate that the anti-EpCAM antibody conjugated to nanoceria (PNC) via Protein G was able to selectively assess the presence of the EpCAM receptor in MCF-7 cancer cells.
Figure 4.
Nanoceria-facilitated fluorigenic determination of the expression of EpCAM, an important cancer biomarker, on breast cancer cells. a) MCF-7 cells overexpressing EpCAM on their surface were successfully quantified using an EpCAM antibody-Protein G-PNC conjugate. b) Non-EpCAM expressing HEK293 cells (negative control) did not show any significant signal when incubated with the EpCAM antibody-Protein G-PNC conjugate.
In our previously published report, we used folate-conjugated nanoceria to monitor the expression of folate receptor utilizing TMB as the chromogenic substrate.13 However, TMB, being a colorimetric substrate, may not provide good quantification and sensitive detection as opposed to a fluorigenic substrate like ampliflu. In addition, the low pH required for detection using nanoceria and TMB could limit that assay, particularly when proteins including antibodies and other labile biomacromolecules (HRP) are used.
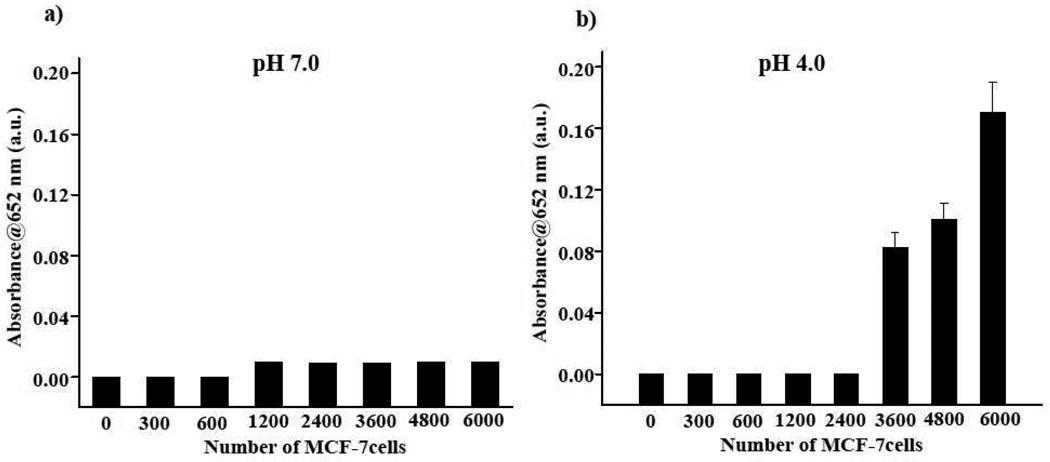
Finally, we compared our results with those obtained with the colorimetric TMB detection of EpCAM in MCF-7 cells at pH 7.0 and 4.0 using the EpCAM antibody-Protein G-PNC conjugate. Results show that when detection was performed with TMB (0.04 mM) at pH 7.0, quantification of the MCF-7 cells was not possible, as low absorbance at 652 nm was obtained (Figure 5a). However, in experiments performed at pH 4.0, TMB identified EpCAM in a higher number of cells (Figure 5b), in contrast with ampliflu at pH 7.0 (Figure 4) that had a lower detection limit. Overall, these results indicate that the nanoceria-mediated oxidation of a fluorigenic substrate at neutral pH provides a more sensitive immunoassay with impoved detection limit for cellular ELISA applications. The fluorigenic substrate ampliflu works efficiently at pH 7.0, eliminating any extreme conditions, such as the acidic pH and oxidizing agents (H2O2).
Figure 5.
Colorimetric determination of EpCAM expression using EpCAM antibody–Protein G-PNC conjugate. TMB was used as the substrate. (a) At pH 7.0, absence of substrate oxidation was observed, as TMB is a poor substrate at this pH.25 (b) Expression of EpCAM was determined, due to the oxidation of TMB by nanoceria at pH 4.0, with a detection limit of 3600 cells.
In summary, we demonstrated that the oxidase-like activity of nanoceria can be tuned by changing the pH of the solution, facilitating the mild oxidation of a substrate to yield a product with enhanced fluorescent properties. Specifically, we showed that ampliflu can be oxidized at pH 7 by nanoceria to the fluorescent product resorufin, without further oxidizing the substrate to the non fluorescent product resazurin. This unique capability of nanoceria is due to its mild oxidizing activity at neutral pH that prevents further oxidation of the substarte to resazurin in the absence of hydrogen peroxide. The selective nanoceria-mediated oxidation of ampliflu can be used to develop sensitive cell-based ELISA, offering various advantages over the HRP/H2O2 ELISA. The complexities associated with the HRP/H2O2 system, such as instability, quenching/bleaching of the fluorigenic substrate and other labile components in immunoassays, can be potentially avoided when a nanoceria-antibody conjugate system is employed. In addition, nanoceria-mediated oxidation of a fluorigenic substrate is independent of hydrogen peroxide, introduces stability in the assay, increases sensitivity, and is an ideal candidate for long readouts, overall rendering it a more reliable setup for target detection. Finally, when compared to gold nanoparticle based ELISA methods that use quartz crystal microbalances (QCM) or surface plasmon resonsnce (SPR) detectors that require complex instrumentation26, the fluorescent signal generated by the nanoceria based ELISA method can be easy read visually under UV-ilumination or by a fluorimeter. Furthermore, when compared to iron oxide based ELISA methods that take advantage of the intrinsic peroxidase activity of these nanoparticles in the presence of hydrogen peroxide25, our method has the unique advantage that no hydrogen peroxide is needed to oxidize the flurigenic substrate, facilitating detection.. Taken together, the use of antibody-immobilized nanoceria based ELISA is expected in the clinic and field as a robust nanoprobe for efficient and sensitive cellular assays.
Synthesis of PAA-coated nanoceria (PNC)
Polyacrylic acid-coated nanoceria was synthesized as described by Asati et al.8
Nanoceria mediated oxidation of amplilfu at various pH (1–8)
Polyacrylic acid-coated nanoceria (PNC) (1.0 mM) suspended in PBS buffer at different pH ranging from pH 1.0 to pH 8.0. Then ampliflu (1.25 µg) was added and the fluorescence emission of the suspension was recorded at different time point intervals spectrum (UV-visible and fluorescence emission).
Concentration-dependent oxidation of ampliflu via nanoceria and HRP/H2O2
Various concentrations of nanoceria (1.0 µM, 4.0 µM, 10 µM and 20 µM) and HRP (0.25 µM, 0.1 µM, 0.5 µM and 0.75 µM) were used, while keeping the hydrogen peroxide concentration (20.0 µL of 0.03%) constant. Amount of amplilfu used for the experiment was 1.25 µg (Stock 0.5 mg/mL solution in DMSO/PBS). Reaction was carried out in a 96-well plate and pictures were taken at different time points.
Fluorescence intensity measurement of concentration dependent oxidation of ampliflu using nanoceria
In a 96-well plate, 1.25 µg of amplilfu (Stock 0.5 mg/mL solution in DMSO/PBS) was added into different wells and treated with various concentration of nanoceria (1.0 µM, 4.0 µM, 10 µM and 20 µM) and intensity was recorded at 3 hours using a Synergy HT plate reader (BIOTEK) with an excitation filter at 525/25 nm and a 590/25 nm emission filter.
Conjugation of Protein G to polyacrylic acid-coated nanoceria (PNC)
Carbodiimide chemistry
To 1.0 mL suspension of PAA-coated nanoceria (50.0 mg Ce/mL) in MES buffer (1.0 mL, pH = 6.0), a solution of EDC (40.0 mg) and NHS (26.0 mg) in MES buffer (0.5 mL) was added and incubated for 3 minutes. To the resulting reaction mixture, Protein G (3.0 mg) in 1X PBS (0.25 mL, pH = 7.4) was added drop-wise, incubated for 2 h at room temperature and then incubated at 4 °C for 3 h. The resulting solution was then dialyzed against DI water to remove unbound Protein G and other reagents, using a cellulose membrane (MWCO 50K). The concentration of Protein G conjugated on the nanoparticle was determined through the BCA assay. The results indicate that the protein concentration of the Protein-G-PNC was 0.33 µg/µL. The final preparation was stored at 4 °C until further use.
Immbolization of antibody on Protein-G-conjugated nanoceria
To immobilize antibodies on nanoceria, 7.0 µL of the antibody (anti-folate and anti-EpCAM, SantaCruz Biotech) was added to 1.0 mL solution of Protein-G-PNC and allowed to incubate overnight at 4 °C. Final concentration of antibody on nanoceria was 1.4 µg/mL.
Cellular ELISA using Amplilfu as a substrate
Lung cancer (A-549) and breast carcinoma (MCF-7) were obtained from ATCC, USA. Lung cancer cells were grown in Kaighn’s modification of Ham’s F12 medium (F12K) supplemented with 5% fetal bovine serum, L-glutamine, streptomycin, amphotericin B, and sodium bicarbonate. All cell lines were maintained at 37 °C, 5% CO2 in a humidified incubator. Cells were plated in a 96-well plate at densities of 0 to 6000 cells per well. After 24 h growth, they were treated with 25.0 µL of antibody-Protein G-PNC conjugate. For the lung carcinoma cell line, anti-folate-receptor antibody–Protein G-PNC conjugate was used, and for the breast carcinoma cells and human embryonic kidney 293 cells (HEK293) the EpCAM antibody-Protein G-PNC conjugate was utilized. Nanoparticle conjugates were allowed to incubate with the cells overnight. Afterwards, the cells were washed with 1X PBS and then incubated with 50.0 µL of 0.5 mg/mL of ampliflu in PBS for 3 h to allow development of the fluorescent product. The product of the reaction was quantified fluorimetrically, using a Synergy HT plate reader (BIOTEK) with an excitation filter at 525/25 nm and 590/25 nm emission filter.
Cellular ELISA using saturation with free anti-EpCAM antibody
MCF-7 cells were preincubated with free anti-EpCAM antibody (0.05 µg/mL) for 3 hour. Rest of the procedure is same as above.
Cellular ELISA using TMB as substrate
TMB was purchased from Sigma. Similar experiments were performed as described in previous section with the breast carcinoma cell line. After incubation with nanoceria conjugate and washing, 100 µL of (0.04 mM) TMB was added and incubated for 3 h. The product of the reaction was quantified spectrophotometrically at 652 nm using a Synergy HT plate reader (BIOTEK).
Supplementary Material
ACKNOWLEDGMENT
The authors aknowledge funding from the National Institutes of Health (CA10178) and a UCF-NSTC Start Up Fund, all to J. M. P.
Footnotes
SUPPORTING INFORMATION AVAILABLE: This is material is available free of chage via the Internet at http://pubs.acs.org.
REFERENCE
- 1.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JE, Medley CD, Tang ZW, Shangguan D, Lofton C, Tan WH. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 3.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 4.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 5.Alivisatos P. Nat. Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 6.Ambrosi A, Airo F, Merkoci A. Anal. Chem. 2010;82:1151–1156. doi: 10.1021/ac902492c. [DOI] [PubMed] [Google Scholar]
- 7.Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Anal. Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaittanis C, Santra S, Perez JM. Adv. Drug Deliv. Rev. 2010;62:408–423. doi: 10.1016/j.addr.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 10.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 11.Teeparuksapun K, Hedstorm M, Wong EY, Tang S, Hewlett IK, Mattiasson B. Anal. Chem. 2010;82:8406–8411. doi: 10.1021/ac102144a. [DOI] [PubMed] [Google Scholar]
- 12.Asati A, Santra S, Kaittanis C, Nath S, Perez JM. Angew. Chem. Int. Ed. 2009;48:2308–2312. doi: 10.1002/anie.200805279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towne V, Will M, Oswald B, Zhao QJ. Anal. Biochem. 2004;334:290–296. doi: 10.1016/j.ab.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin D, Grover T, Aust S. Biochemistry. 1997;36:139–147. doi: 10.1021/bi961465y. [DOI] [PubMed] [Google Scholar]
- 15.Arno MB, Acosta M, Rio JA, Varon R, Garciacanovas F. Biochim. Biophys. Acta. 1990;1041:43–47. doi: 10.1016/0167-4838(90)90120-5. [DOI] [PubMed] [Google Scholar]
- 16.Arno MB, Acosta M, Rio JA, Varon R, Garciacanovas F. Biochim. Biophys. Acta. 1990;1038:85–89. doi: 10.1016/0167-4838(90)90014-7. [DOI] [PubMed] [Google Scholar]
- 17.Nelson ME, Loktionova NA, Pegg AE, Moschel RC. J. Med. Chem. 2004;47:3887–3891. doi: 10.1021/jm049758+. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XG, Miao J, Dai YQ, Du YZ, Yuan H, Hu FQ. Int. J. Pharm. 2008;361:239–244. doi: 10.1016/j.ijpharm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Nelson ME, Loktionova NA, Pegg AE, Moschel RC. J. Med. Chem. 2004;47:3887–3891. doi: 10.1021/jm049758+. [DOI] [PubMed] [Google Scholar]
- 20.Yuan H, Miao J, Du YZ, You J, Hu FQ, Zeng S. Int. J. Pharm. 2008;348:137–145. doi: 10.1016/j.ijpharm.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Chung KN, Saikawa Y, Paik TH, Dixon KH, Mulligan T, Cowan KH, Elwood PC. J. Clin. Invest. 1993;91:1289–1294. doi: 10.1172/JCI116327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong D, Steurer M, Obrist P, Barbieri V, Margreiter R, Amberger A, Laimer K, Gastl G, Tzankov A, Spizzo G. J. Clin. Pathol. 2008;61:31–35. doi: 10.1136/jcp.2006.037333. [DOI] [PubMed] [Google Scholar]
- 23.Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, Hannun YA, Cole DJ, Gillanders WE. Cancer. Res. 2004;64:5818–5824. doi: 10.1158/0008-5472.CAN-04-0754. [DOI] [PubMed] [Google Scholar]
- 24.Winkler J, Martin-Killias P, Pluckthun A, Zangemeister-Wittke U. Mol. Cancer Ther. 2009;8:2674–2683. doi: 10.1158/1535-7163.MCT-09-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao LZ, Zhuang J, Nie L, Zhang JB, Zhang Y, Gu N, Wang TH, Feng J, Yang DL, Perrett S, Yan X. Nat. Nanotechnol. 2007;2:577–583. doi: 10.1038/nnano.2007.260. [DOI] [PubMed] [Google Scholar]
- 26.Cliffel DV, Turner BN, Huffman B. Adv. Rev. 2009;1:47–59. doi: 10.1002/wnan.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.