Abstract
H2O2 is commonly generated in biological habitats by environmental chemistry and by cellular immune responses. H2O2 penetrates cells, disrupts metabolism, and blocks growth; it therefore is of interest to identify the major cellular molecules that H2O2 damages and the strategies by which cells protect themselves from it. We used a strain of Escherichia coli that lacks catalases and peroxidases to impose protracted low-grade H2O2 stress. Physiological analysis indicated that the pentose–phosphate pathway, in particular, was poisoned by submicromolar intracellular H2O2. Assays determined that ribulose-5-phosphate 3-epimerase (Rpe) was specifically inactivated. In vitro studies demonstrated that Rpe employs a ferrous iron atom as a solvent-exposed cofactor and that H2O2 rapidly oxidizes this metal in a Fenton reaction. The oxidized iron is released immediately, causing a loss of activity. Most Rpe proteins could be reactivated by remetallation; however, a small fraction of proteins were irreversibly damaged by each oxidation cycle, and so repeated cycles of oxidation and remetallation progressively led to permanent inactivation of the entire Rpe pool. Manganese import and iron sequestration are key elements of the H2O2 stress response, and we found that manganese can activate Rpe in vitro in place of iron, converting the enzyme to a form that is unaffected by H2O2. Indeed, the provision of manganese to H2O2–stressed cells protected Rpe and enabled the pentose–phosphate pathway to retain function. These data indicate that mononuclear iron enzymes can be primary targets of H2O2 stress and that cells adapt by shifting from iron- to manganese-centered metabolism.
Keywords: hydroxyl radical, MntH, oxidative damage, OxyR
Catalases and peroxidases were among the first enzymes discovered, and their ubiquity among the biota prompted biologists to speculate that H2O2 must be a common by-product of the aerobic metabolism and that, if not scavenged, it must critically harm cells (1). Since then, workers have recognized that H2O2 also is generated by extracellular processes, including photochemical processes in natural waters and the autoxidation of S and metal species at interfaces between anoxic and oxic sediments. Perhaps more critically, H2O2 is generated by the antimicrobial responses of both plant and mammalian higher organisms as a strategy to stave off infection (2, 3). Local H2O2 concentrations at plant wound sites and in macrophages rise to micromolar levels, which are sufficient to suppress microbial growth.
Virtually all microbes engage specialized stress responses to fend off exogenous H2O2. Among the bacteria, the OxyR and PerR systems are widespread (4, 5). OxyR, which has been closely studied in enterics, is a transcription factor that is activated when H2O2 oxidizes a key cysteine residue; the subsequent formation of a disulfide bond locks the protein into an active form that stimulates the transcription of ca. two dozen genes (6, 7). Mutants that lack OxyR grow poorly or not at all in environments that contain H2O2.
Still, fundamental aspects of H2O2 stress remain poorly understood. Perhaps most importantly, the primary intracellular targets of H2O2 have not been identified fully. A technical difficulty has hindered work on this problem: When laboratory cultures are stressed with micromolar H2O2, growth stops briefly, but the H2O2 is scavenged quickly, and growth resumes. Under these circumstances it has been difficult to identify the specific cellular processes and biomolecules that are affected. One way around this problem has been the use of strains that lack scavenging enzymes (8). Escherichia coli catalase/peroxidase mutants (katG katE ahpCF, denoted Hpx−) grow at wild-type rates in anaerobic cultures. When inoculated into aerobic medium, H2O2 accumulates to ca. 1 micromolar, which equilibrates between the cytoplasm and the extracellular environment. This concentration exceeds the threshold that activates OxyR, and indeed Hpx− ΔoxyR mutants die quickly in aerobic medium (9). Thus, the Hpx− strains offer the opportunity to identify the primary biomolecules that H2O2 damages as well as the key defensive strategies that OxyR triggers.
Thus far it has become clear that H2O2 damages DNA through standard Fenton chemistry:
The Fe that is involved belongs to the cellular pool of unincorporated Fe, some of which presumably associates with nucleic acids (10). A ferryl radical (FeO2+) is the immediate product, but ultimately a hydroxyl radical is released that can attack directly both the sugar and base moieties of DNA (11). Intracellular H2O2 also disables a family of dehydratases that contain [4Fe-4S] clusters; the chemistry is similar, in that H2O2 directly oxidizes a catalytic Fe atom that is exposed to solvent (12). The oxidized cluster is unstable, the key Fe atom dissociates, and enzyme activity is lost. These enzymes were first shown to be oxidative targets in studies of superoxide (13–17). Exacerbating this situation, H2O2 also inhibits the cellular Fe–S cluster assembly machinery, probably by oxidizing nascent clusters as they are assembled on the IscU scaffold protein (18). Finally, H2O2 has been shown to disrupt the Fur Fe-sensing system (19). In this case the proximate cause has not been demonstrated, but it seems likely that H2O2 oxidizes the Fe2+ atom that normally binds to the Fur transcription factor.
The OxyR regulon stimulates the synthesis of several proteins whose roles are well-suited to addressing these effects. The HPI catalase and AhpCF peroxidase are induced to drive back down the intracellular H2O2 level (7). The Dps Fe-storage protein sequesters unincorporated Fe and thereby diminishes the amount of DNA damage (9, 20–22). An alternative Fe–S assembly machine, the Suf system, is resistant to H2O2 and is induced to compensate for the failure of the Isc system (18). The Fur regulator is induced to higher levels as a hedge against its lower efficiency (19, 23).
Recently we found that the induction of an Mn import system also is critical, because Hpx− ΔmntH mutants stop growing when they are aerated (24). E. coli normally does not import a significant amount of Mn. We speculated that E. coli might use ferrous Fe as a cofactor in nonredox enzymes, which would place such enzymes at risk during periods of H2O2 stress. Induction of the MntH transporter might enable Mn to replace Fe in these enzymes; because Mn does not react with H2O2, the enzymes might be protected.
In the present study we sought to identify cellular processes that are inhibited by micromolar H2O2. We identified the pentose–phosphate pathway as one such target and determined that the bottleneck arises because of damage to a mononuclear enzyme that contains ferrous Fe. When Mn is provided to these cells, the enzyme is protected, and the pathway function is restored.
Results
Ribulose-5-Phosphate 3-Epimerase Is Vulnerable to H2O2 Damage in Vivo.
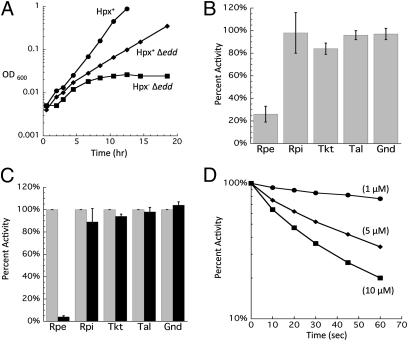
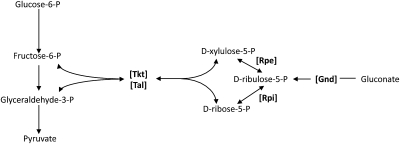
Strains of E. coli that cannot scavenge H2O2 gradually accumulate ca. 1 micromolar H2O2 (8). This level moderately exceeds the dose that activates the OxyR response, and so it is likely that these strains confront protracted H2O2 stress of a magnitude similar to that found in some natural habitats. Growth studies of this strain have shown that most catabolic and biosynthetic pathways continue to function, as is consistent with the observation that this level of H2O2 is innocuous to the vast majority of enzymes. However, in the process of testing the H2O2 sensitivity of central metabolic pathways, we noted that an Hpx− Δedd mutant is unable to grow in aerobic gluconate medium (Fig. 1A). This strain lacks 6-phosphogluconate dehydratase and therefore requires gluconate flux through the pentose–phosphate pathway (Fig. 2). The cells began to lag when the H2O2 concentration had reached ∼0.8 μM (Fig. S1), indicating that very low levels of H2O2 can inactivate this pathway.
Fig. 1.
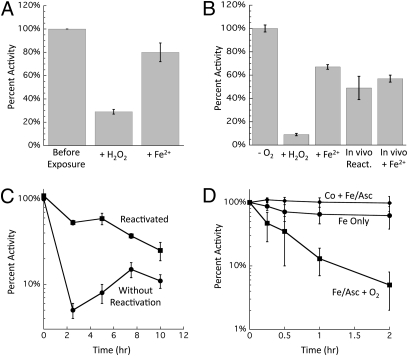
(A) Hpx− Δedd cells cannot grow on gluconate because of damage to Rpe. After anaerobic preculture, cells were diluted into aerobic minimal gluconate medium at time 0. Data presented are representative of multiple experiments. (B) Activities measured in extracts prepared from Hpx− Δedd cells grown aerobically. Values were normalized to those of anaerobic cells. Gnd, 6-phosphogluconate dehydrogenase; Rpi, ribose-5-phosphate isomerase; Tal, transaldolase; Tkt, transketolase. (C) Activities measured before (gray bars) and after (black bars) 10 μM H2O2 was added to cell extracts prepared from anaerobically grown Hpx− cells. Error bars represent SD from the mean of three independent experiments. (D) Time course of the inactivation at 0 °C of Rpe in cell extracts treated with the indicated concentrations of H2O2. Extracts were prepared from anaerobically grown Hpx− cells.
Fig. 2.
The pentose–phosphate pathway. Relevant intermediates are listed, with key enzymes denoted in brackets. Arrows indicate direction of each reaction.
The fact that Hpx− edd+ cells can grow on gluconate via the alternative Entner–Doudoroff pathway implied that the block must occur downstream of gluconate import. The five pentose–phosphate pathway enzymes were assayed in extracts that had been prepared from aerobic cultures of wild-type and Hpx− strains. No differences were found in the levels of 6-phosphogluconate dehydrogenase, ribose-5-phosphate isomerase, transketolase, and transaldolase (Fig. 1B). However, ribulose 5-phosphate 3-epimerase (Rpe) showed low activity. To determine whether H2O2 can inactivate Rpe directly, Hpx− cells were grown anaerobically, cell extracts were prepared, and the extracts then were briefly challenged with H2O2 in vitro. Rpe rapidly lost activity (Fig. 1C). Even at 0 °C, 5 μM H2O2 inactivated most of the enzyme within 1 min (Fig. 1D); the measured rate constant for inactivation (4,000 M−1 s−1) approximated those of H2O2-sensitive Fe/S dehydratases (12).
Rpe Can Be Metallated by Various Divalent Metals That Result in Variable Properties.
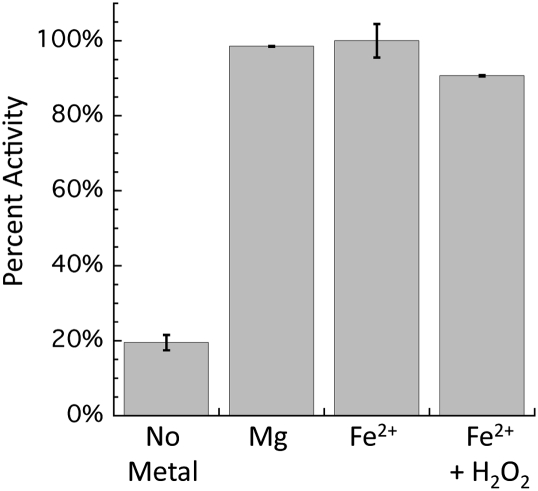
To study Rpe in vitro, Rpe from E. coli was purified using a His tag, which was removed after purification. Rpe is known to require a divalent metal for activity, but the identity of the metal used in vivo remained uncertain (25). A previous study of Rpe from Streptococcus pyogenes reported that it copurified with substantial Zn2+, leading the authors to speculate that this is its physiological metal. Akana et al. (25) reported that Mn2+ and Co2+ also would activate Rpe in vitro, but Mg2+ and Fe2+ would not. However, these experiments were conducted under aerobic conditions, and the Fe2+ probably oxidized to Fe3+. We used chelators to remove any associated metals from the purified Rpe from E. coli, and we then tested various metals for their ability to activate the enzyme in vitro under anaerobic conditions. Of the nine metals used, Ca2+, Cd2+, Cu2+, Mg2+, and Ni2+ did not activate Rpe above background levels. However, Co2+, Fe2+, Mn2+, and Zn2+ were able to activate Rpe in vitro.
The kinetic constants of the four metalloforms of Rpe were determined (Table 1). Strikingly, the Zn2+ metalloform had a much lower kcat and higher KM than the other metalloforms; thus the catalytic efficiency (kcat/KM) of the Zn2+ metalloform was much worse than those of the Mn2+, Fe2+, and Co2+ forms, which were roughly comparable. The dissociation rates for each of the activating metals were measured also (Table 1). Despite providing the most catalytically efficient metalloform, Mn2+ rapidly dissociated from Rpe, but the Fe2+ metalloform was moderately more stable. Interestingly, Zn2+ bound tightly to the enzyme.
Table 1.
Properties of various Rpe metalloforms
| Metal | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) | t1/2 of metal dissociation |
| Fe 2+ | 67,000 ± 7,700 | 1.6 ± 0.2 | 4.2 × 107 | 50 min |
| Mn 2+ | 30,400 ± 1,700 | 1.8 ± 0.1 | 1.7 × 107 | 3.5 min |
| Co 2+ | 14,700 ± 900 | 2.4 ± 0.4 | 0.6 × 107 | 20 h |
| Zn2+ | 1,300 ± 100 | 4.8 ± 0.5 | 0.02 × 107 | 8 h |
kcat and KM values were determined as described in Materials and Methods. Error represents SD from the mean of three independent measurements. Dissociation rates were determined by measuring the loss of Rpe activity during incubation of metal-loaded enzyme with an excess of the chelator DTPA at 23 °C. DTPA was shown to trap dissociated metals but not to extract them actively (SI Materials and Methods). Measurements represent the averages of three independent experiments.
Of these four metals, only Fe2+ and Zn2+ are found in E. coli in substantial concentrations under normal growth conditions. E. coli lacks a Co2+ importer, and Mn2+ is not actively imported during routine growth (24). It seemed likely that Fe2+ is the metal that provides activity to Rpe under normal physiological conditions.
Fenton Chemistry Is Responsible for the Inactivation of Rpe.
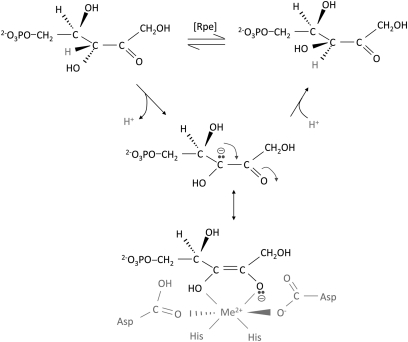
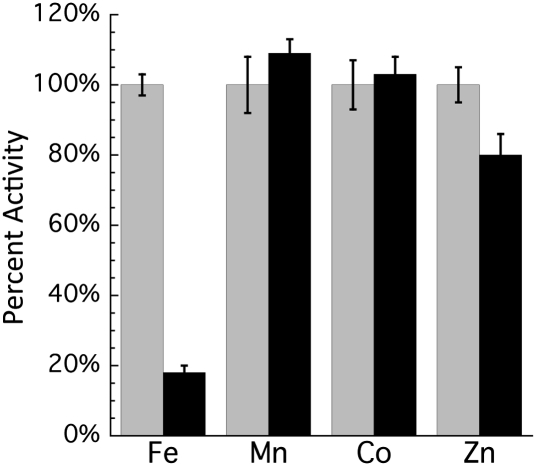
The proposed mechanism of Rpe implies that the metal will be exposed to solutes, including H2O2 (Fig. 3); therefore, we tested whether Fenton chemistry was responsible for loss of Rpe activity. The sensitivity to H2O2 of each of the metalloforms of Rpe was tested. When purified Rpe was loaded with Mn2+, Co2+, or Zn2+, there was no loss in activity when challenged with H2O2 (Fig. 4). However, when Rpe was metallated with Fe2+, the activity was quickly lost. Because the enzyme that is found in cell extracts is acutely sensitive to H2O2, we concluded that Fe2+ is indeed the native cofactor that provides activity in laboratory cultures.
Fig. 3.
Rpe active site structure and mechanism. Rpe interconverts ribulose-5-phosphate and xylulose-5-phosphate by abstracting a proton from one face of the C-3 carbon and then adding a proton to the opposite face. The metal coordinates substrate and stabilizes the intermediate oxyanion (25).
Fig. 4.
Sensitivity of Rpe metalloforms to H2O2 in vitro. Pure Rpe was metallated with the indicated metals. Activity then was measured before (gray bars) and after (black bars) treatment with 500 μM H2O2 for 2 min. Error bars represent SD from the mean of three independent measurements.
Reactions between H2O2 and Fe2+ initially generate ferryl radicals, which have a substantial lifetime before decomposing into Fe3+ and a hydroxyl radical (11, 26). Hydroxyl radicals react at nearly diffusion-limited rates with most biomolecules, including proteins, raising the possibility that H2O2 exposure might lead to covalent Rpe damage. We found, however, that when Fe-loaded Rpe was inactivated with H2O2, about 80% of the initial Rpe activity could be recovered by adding Fe2+ back to the enzyme (Fig. 5A). Reductants alone were insufficient, confirming that the oxidized Fe atom had dissociated from the enzyme. Similar results were obtained by the addition of Co2+ or Mn2+. Full activity was never recovered. Thus, we concluded that at most only a very minor fraction of the protein was critically and irreversibly damaged; instead, most of the activity loss was caused by the dissociation of Fe.
Fig. 5.
Reversibility of Rpe inactivation. (A) A single cycle of in vitro damage is mostly reversible. Pure Rpe was metallated with 100 μM Fe2+ before being treated with 200 μM H2O2 for 5 min. Catalase and DTPA were added to remove Fe and H2O2, and Rpe was assayed. Damaged enzyme then was reactivated by anaerobic incubation with 100 μM Fe2+ and 500 μM ascorbate. (B) Brief damage in vivo is mostly reversible. Hpx− cultures were grown in anaerobic medium, and chloramphenicol then was added to block further protein synthesis. Cells then were aerated and exposed to H2O2 (100 μM) for 10 min. Cultures were returned to the anaerobic chamber, treated with catalase, and assayed before (+H2O2) or after (+Fe2+) in vitro reactivation. In vivo reactivation was measured by incubating cells for an additional 20 min after the termination of H2O2 stress. (C) Protracted stress irreversibly damages Rpe in vivo. Anaerobic cells were diluted into aerobic minimal gluconate medium at time 0. At indicated time points Rpe activity was measured before and after in vitro reactivation with Co2+. (D) Repetitive Fenton chemistry irreversibly damages Rpe in vitro. Pure Rpe was metallated with Fe2+ or Co2+. After 10 min, samples were diluted into aerobic buffer containing Fe2+ and ascorbate to trigger cycles of H2O2 damage and remetallation by Fe2+. At indicated time points, aliquots were removed, damage was terminated by the addition of catalase and DTPA, and Co2+ was added to activate the remaining functional apoenzyme. Error bars represent the SD from the mean of three experiments. The enzyme is not inactivated if the active site is occupied by Co.
In vivo experiments showed a similar outcome. When Hpx− Δedd cells were exposed to a bolus of H2O2, activity quickly declined. After the H2O2 was removed from the culture, activity rebounded to about 70% of the original activity, even though new protein synthesis was blocked (Fig. 5B). The recovery of activity inside the cell reached the same level as when activating metals were added back to the lysate of the H2O2-treated cells.
Repeated Rounds of Inactivation Are Required to Damage Rpe Permanently.
The data suggested that when Fe-loaded Rpe is oxidized by H2O2, the catalytic Fe cofactor is oxidized and lost; however, despite the presumptive formation of a hydroxyl radical [Eqs. 1 and 2], only a small fraction of those events caused irreversible protein damage. Inactivation was predominantly caused by metal loss. In vivo, however, continual enzyme remetallation should occur. When Rpe activity was tracked during extended Hpx− growth in aerobic medium, activity quickly fell. At early time points most of the enzyme was in a nonmetallated but reactivatible form, because activity could be recovered by metal addition in vitro. However, the amount of recoverable activity declined progressively, suggesting that Rpe gradually accrued irreversible damage from multiple rounds of Fenton reactions (Fig. 5C).
To replicate cycles of damage and remetallation in vitro, purified Rpe was exposed to Fe, ascorbate, and oxygen in vitro. The oxidation of Fe2+ by oxygen ensured the constant presence of a low level of H2O2 (27), and the reduction of Fe by ascorbate regenerated Fe2+. At various time points the Fe and H2O2 were removed by diethylenetriaminepentaacetic acid (DTPA) and catalase, and the amount of functional Rpe polypeptide was determined by the addition of Co2+ before assay. When ascorbate was omitted, activity diminished only at first and thereafter remained constant at about 65% of the starting activity, consistent with a single cycle of enzyme damage. However, when ascorbate was added to enable the cyclical activation and oxidation of the enzyme, activity dropped steadily, down to 15% after 1 h and to about 3% after 2 h (Fig. 5D). Taken together, these results indicate that a single bolus of Fenton chemistry irreversibly damages only a fraction of the Rpe molecules. Complete inactivation of the population results from repeated cycles of the reaction.
Mn Protects Rpe from Damage by H2O2.
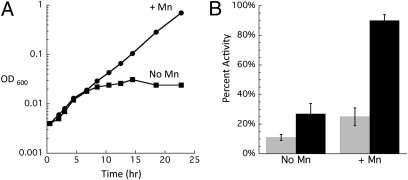
The induction of an Mn importer, MntH, has been shown to be a critical aspect of the response of E. coli to H2O2 (24, 28). We proposed recently that the role of Mn in the protection against H2O2 stress might be to replace Fe2+ in the mononuclear metal sites of enzymes, thereby diminishing the frequency of Fenton chemistry in those sites. To test this idea with respect to Rpe, Hpx− Δedd cells were grown aerobically on gluconate with or without Mn supplementation. As shown in Fig. 6A, the addition of Mn fully suppressed the aerobic growth phenotype in Hpx− Δedd cells. To verify that the protective effect was caused by the protection of Rpe, its activity was measured in samples harvested from cultures grown both with and without Mn supplements. Fig. 6B shows that the supplements boosted Rpe activity by several-fold at the 10-h time point, when unsupplemented cells began to lag. Because the dissociation rate of Mn in vitro is so fast, these assays almost certainly underrepresented the activity of Rpe containing Mn. Therefore, we sought to quantify the amount of functional Rpe polypeptide by providing Co2+ to the extracts. The samples from Mn-supplemented cultures showed nearly 100% activity, compared with less than 25% activity from untreated cultures (Fig. 6B). These data support the idea that Mn protects Rpe from H2O2 by replacing Fe2+ in the active site during H2O2 stress.
Fig. 6.
Mn supplements protect Rpe against irreversible damage in vivo. (A) After anaerobic preculture, cells were diluted at time 0 into aerobic minimal gluconate medium with or without 50 μM MnCl2. Data are representative of multiple growth experiments. (B) Cells were handled as in A except that aerobic inoculum was to 0.05 OD. At various times, aliquots were taken, and Rpe activity was measured before and after reactivation with Co. Time 0 is an anaerobic time point. Error bars represent the SD from the mean of three independent experiments.
Fe Must Be Accessible to Solvent for Enzymes to Be Vulnerable to H2O2.
The full range of enzymes that use Fe2+ as a cofactor in vivo—and therefore might be vulnerable to H2O2—is unknown. Presumably many enzymes that require divalent metal cofactors might be able to use Fe2+. However, Fenton chemistry requires the direct binding of Fe to H2O2, and so we suspected that only those enzymes that might have undercoordinated Fe in their active sites are potentially vulnerable. To test this idea, transketolase was examined. Transketolase requires a divalent metal to form a bridge between the polypeptide and a thiamine pyrophosphate (TPP) cofactor, but the metal does not participate in the reaction chemistry. The metal itself is buried beneath the bound cofactor and is not exposed to solvent (PDB code 1QGD). We determined that Fe2+ was able to activate transketolase as efficiently as did Mg2+. However, in contrast to Rpe, the addition of H2O2 did not affect the activity of the Fe2+/TPP-loaded enzyme (Fig. 7). These results demonstrate that enzymes that use Fe2+ as a metal cofactor are sensitive to H2O2 only if the metal site is exposed to solvent; the Fe2+ is fully coordinated, Fenton chemistry is blocked, and the enzyme is not vulnerable to H2O2. This feature presumably will circumvent damage to many enzymes, such as transketolase, that might occasionally or frequently bind Fe2+ as a divalent cofactor.
Fig. 7.
Transketolase and necessity of solvent exposure for metal sensitivity to chelation and to H2O2. Metals were added to pure transketolase (Sigma) before the addition of TPP and 500 μM H2O2. After mixing, samples were incubated for 5 min and then assayed as described in SI Materials and Methods.
Discussion
In addition to their facility at redox reactions, transition metals excel at surface chemistry, in which they directly coordinate substrates and thereby activate them for a wide variety of reaction types. Rpe is an example of the latter: Its mononuclear metal atom binds the carbonyl and carboxylate moieties of ribulose-5-phosphate, facilitating epimerization by stabilizing the deprotonated anionic intermediate. In addition, over the course of the reaction, the metal alternatively coordinates and releases the aspartyl residues that act as proton acceptors and donors to the substrate. Although a nontransition metal such as Mg can suffice in enzymes where its sole role is to neutralize charge, transition metals often are requisite when the metal environment must change during the reaction cycle, because they can tolerate changes in their coordination sphere with minimal activation energy.
Our study indicates that Fe is the transition metal that activates Rpe. This finding makes both ecological and evolutionary sense, because Fe typically is abundant in anaerobic habitats, including both the mammalian gut in which E. coli dwells and the anaerobic world in which Rpe originally evolved. Indeed, the service of ferrous Fe in nonredox enzymes probably is systematically underrecognized, because Fe is oxidized quickly and released when these enzymes are studied in aerobic buffers. Thus, although in vitro experiments suggest that Mn can activate scores of enzymes in E. coli (http://www.Ecocyc.org/), in reality Mn is scarcely imported under routine growth conditions (24). More likely, Fe is the cognate metal of many of these enzymes.
Fe Enzymes Are Targets of Oxidants.
Catalase and peroxidase activities are sufficient to cope with the H2O2 that is formed as a by-product of aerobic metabolism, but the diffusion of H2O2 into the cell from environmental sources can drive its steady-state levels above the threshold of toxicity (29). Recent studies from several laboratories indicate that protein damage is likely to determine the fate of oxidant-exposed cells (30–33). Rpe, and probably mononuclear Fe enzymes in general, now join Fe–S dehydratases as primary classes of enzymes that H2O2 damages. In both cases H2O2 directly oxidizes a solvent-exposed Fe atom that normally serves to bind substrate; the oxidized Fe atom then dissociates from the enzyme. Interestingly, the hydroxyl-like oxidants that are formed need not damage the active sites of either type of enzyme fatally—the enzymes can be reactivated repeatedly, both in vivo and in vitro. When H2O2 oxidizes Fe–S clusters, hydroxyl radical release is averted when the nascent ferryl radical abstracts a second electron from the cluster (12). We speculate that Rpe similarly might be spared covalent damage because the ferryl radical usually dissociates from the active site and does not release a hydroxyl radical until it enters the bulk solution. Upon reflection, it is probably an essential feature of Fe-containing enzymes that they are not irreversibly damaged upon each encounter with H2O2. Even in innocuous habitats wild-type E. coli contains about 20 nM steady-state H2O2 from endogenous sources (29); because the inactivation rates of these enzymes are likely 104–105 M−1 s−1 at physiological temperatures (Fig. 1D) (12), they are likely to be oxidized by H2O2 on the order of every 10 min. This situation would be intolerable were the polypeptide irreparably damaged each time.
Role of Alternative Metals in Fe Enzymes.
The reliance of metabolism upon Fe-cofactored enzymes presents problems during periods of either Fe starvation or oxidative stress. One apparent solution is to replace Fe with Mn. When the problem is Fe scarcity, the deactivation of the Fur repressor stimulates synthesis of MntH, the Mn importer (28). Presumably the resultant Mn influx allows mononuclear enzymes such as Rpe to continue to work. Because Mn does not share the reduction potential of Fe, the solution is slightly more complicated for redox enzymes: Key Fe-dependent enzymes, such as superoxide dismutase and ribonucleotide reductase, are replaced by distinct isozymes that bind and poise Mn at an appropriate potential (34, 35).
The induction of the Mn transporter is not sufficient to protect these enzymes from H2O2, however, if the H2O2-stressed cell still contains ample Fe that might yet outcompete Mn for enzyme binding. Thus, the induction of MntH by OxyR is supplemented by its induction of the Fur repressor (23), which inhibits expression of Fe importers, and of Dps, which sequesters intracellular Fe in a ferritin-like storage protein (9, 20–22). In collaboration these systems ensure that the Fe/Mn ratio tips toward the latter. Carbonylation assays suggest that many cellular proteins are protected in this way (24).
Although the evidence fits the idea that Mn protects Rpe by integrating as the cofactor metal, we were unable to demonstrate this notion directly: By the point of assay, Mn was not bound to the Rpe that was harvested from these cells. Thus, some uncertainty remains. Still, the rapid dissociation of this weakly bound metal is expected during extract preparation. In any case, it is clear that Mn import blocked enzyme damage and sustained activity. An alternative model for the antioxidant effects of Mn (36–41) is that it acts as a chemical scavenger of reactive oxygen species, including H2O2 (37, 39, 42–44). That model seems not to apply to E. coli, because direct measurements showed that Mn import did not affect H2O2 levels (24). Further, we found that excess Co, which also can activate Rpe but does not chemically degrade H2O2, was able to substitute for Mn in preserving the function of the pentose–phosphate pathway during H2O2 stress (Fig. S2). It seems possible that this Mn protection scheme operates constitutively in those lactic acid bacteria that produce H2O2 in high quantities, because these bacteria characteristically require millimolar levels of intracellular Mn for good growth (45). Work in the Daly laboratory has suggested that additional mechanisms of Mn-based protection also might occur in these organisms (36). It will be interesting to see whether these organisms have evolved enzymes that are better than E. coli Rpe at binding Mn, or if the requirement for such high intracellular Mn is a reflection of weak binding in these cells as well.
It has long been of interest to identify the mechanisms that ensure that enzymes bind their cognate metals. The example of E. coli experiencing H2O2 stress, during which cellular Mn2+ rises from 15 to 180 μM (24), indicates that metallation is substantially controlled by the relative concentrations of the competing metals. However, this example also points out that this level of control is not enough, because Zn, which binds more tightly to the Rpe active site than does either Mn or Fe, is abundant inside the cell. Indeed, when Mn-loaded Rpe was coincubated with Zn in vitro, activity steadily declined as the association/dissociation equilibrium of Mn was interrupted by the permanent binding of Zn (Fig. S3). The quick solvation of Mn and Fe from mononuclear enzymes may be the counterpoint to their facility at the exchange reactions that allow them to be good surface catalysts. Inside the cell competition from Zn presumably is suppressed by other intracellular ligands, such as the glutathione pool, that substantially sequester it. Whether the cell also employs systems that either actively load Fe/Mn into mononuclear enzymes such as Rpe or that facilitate the dissociation of inappropriately bound metals remains to be determined.
Materials and Methods
Reagents, strains, growth conditions, and assays used in this study are described in SI Materials and Methods. Strains used in this study are listed in Table S1.
Bacterial cultures were grown aerobically at 37 °C with vigorous shaking or anaerobically at 37 °C in a Coy anaerobic chamber. All enzyme assays were conducted under anaerobic conditions in sealed cuvettetes.
E. coli Rpe was purified by His tag using a variation of the protocol described by Akana et al. (25). Purification details are described in SI Materials and Methods. After purification, the His tag was removed. Because EDTA had been included in buffers, the as-isolated enzyme was inactive and recovered activity only upon the provision of metals.
For determinations of kinetic constants, purified Rpe was activated by 500 μM Co, Fe, Mn, or Zn for 10 min before anaerobic assays. Metal dissociation rates were determined by anaerobic incubation of the metal-loaded enzyme with an excess of DTPA; aliquots were withdrawn and assayed at indicated time points. To test sensitivity to H2O2, purified Rpe was activated for 5 min in anaerobic buffer containing 100 μM of each metal and then diluted into the Rpe assay mixture. The reaction was run for 6 min, and then H2O2 was added (final concentration of 500 μM). Rates were compared before and after H2O2 treatment.
Purified transketolase (Sigma) was metallated by 200 μM ferrous ammonium sulfate or MgCl2. After 2 min, TPP was added to a concentration of 2 mM for 2 min before the addition of H2O2 (final concentration of 500 μM). Samples then were incubated for an additional 2 min before being assayed.
Complete details for each of the methods described here are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Grant GM049640 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100410108/-/DCSupplemental.
References
- 1.Loew O. A new enzyme of general occurrence in organisms. Science. 1900;11:701–702. doi: 10.1126/science.11.279.701. [DOI] [PubMed] [Google Scholar]
- 2.Glass GA, et al. The respiratory burst oxidase of human neutrophils. Further studies of the purified enzyme. J Biol Chem. 1986;261:13247–13251. [PubMed] [Google Scholar]
- 3.Mehdy MC. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994;105:467–472. doi: 10.1104/pp.105.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmel-Harel O, Storz G. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 5.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 6.Choi H, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 7.Zheng M, et al. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rai P, Cole TD, Wemmer DE, Linn S. Localization of Fe(2+) at an RTGR sequence within a DNA duplex explains preferential cleavage by Fe(2+) and H2O2. J Mol Biol. 2001;312:1089–1101. doi: 10.1006/jmbi.2001.5010. [DOI] [PubMed] [Google Scholar]
- 11.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 12.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo CF, Mashino T, Fridovich I. α, β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 14.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J Biol Chem. 1991;266:1478–1483. [PubMed] [Google Scholar]
- 15.Gardner PR, Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J Biol Chem. 1991;266:19328–19333. [PubMed] [Google Scholar]
- 16.Liochev SI, Fridovich I. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc Natl Acad Sci USA. 1992;89:5892–5896. doi: 10.1073/pnas.89.13.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 18.Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese S, Wu A, Park S, Imlay KRC, Imlay JA. Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol Microbiol. 2007;64:822–830. doi: 10.1111/j.1365-2958.2007.05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 21.Ilari A, Ceci P, Ferrari D, Rossi GL, Chiancone E. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem. 2002;277:37619–37623. doi: 10.1074/jbc.M206186200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao G, et al. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 23.Zheng M, Doan B, Schneider TD, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akana J, et al. D-Ribulose 5-phosphate 3-epimerase: Functional and structural relationships to members of the ribulose-phosphate binding (β/α)8-barrel superfamily. Biochemistry. 2006;45:2493–2503. doi: 10.1021/bi052474m. [DOI] [PubMed] [Google Scholar]
- 26.Rush JD, Maskos Z, Koppenol WH. Distinction between hydroxyl radical and ferryl species. Methods Enzymol. 1990;186:148–156. doi: 10.1016/0076-6879(90)86104-4. [DOI] [PubMed] [Google Scholar]
- 27.Murakami K, Tsubouchi R, Fukayama M, Ogawa T, Yoshino M. Oxidative inactivation of reduced NADP-generating enzymes in E. coli: Iron-dependent inactivation with affinity cleavage of NADP-isocitrate dehydrogenase. Arch Microbiol. 2006;186:385–392. doi: 10.1007/s00203-006-0153-1. [DOI] [PubMed] [Google Scholar]
- 28.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+) J Bacteriol. 2002;184:3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daly MJ, et al. Protein oxidation implicate as the perimary determinant of bacterial radioresistance. PLoS Biol. 2007;5:769–779. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci USA. 2010;107:14373–14377. doi: 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosshard F, et al. Protein oxidation and aggregation in UVA-irradiated Escherichia coli cells as signs of accelerated cellular senescence. Environ Microbiol. 2010;12:2931–2945. doi: 10.1111/j.1462-2920.2010.02268.x. [DOI] [PubMed] [Google Scholar]
- 34.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07593.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol Microbiol. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 37.Seib KL, Tseng HJ, McEwan AG, Apicella MA, Jennings MP. Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: Distinctive systems for different lifestyles. J Infect Dis. 2004;190:136–147. doi: 10.1086/421299. [DOI] [PubMed] [Google Scholar]
- 38.Tseng HJ, McEwan AG, Paton JC, Jennings MP. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect Immun. 2002;70:1635–1639. doi: 10.1128/IAI.70.3.1635-1639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol. 2000;36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 40.Daly MJ, et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 41.Daly MJ, et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE. 2010;5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Runyen-Janecky L, Dazenski E, Hawkins S, Warner L. Role and regulation of the Shigella flexneri sit and MntH systems. Infect Immun. 2006;74:4666–4672. doi: 10.1128/IAI.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horsburgh MJ, Wharton SJ, Karavolos M, Foster SJ. Manganese: Elemental defence for a life with oxygen. Trends Microbiol. 2002;10:496–501. doi: 10.1016/s0966-842x(02)02462-9. [DOI] [PubMed] [Google Scholar]
- 44.Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 45.Archibald F. Manganese: its acquisition by and function in the lactic acid bacteria. Crit Rev Microbiol. 1986;13:63–109. doi: 10.3109/10408418609108735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.