Abstract
Although commensal bacteria are crucial in maintaining immune homeostasis of the intestine, the role of commensal bacteria in immune responses at other mucosal surfaces remains less clear. Here, we show that commensal microbiota composition critically regulates the generation of virus-specific CD4 and CD8 T cells and antibody responses following respiratory influenza virus infection. By using various antibiotic treatments, we found that neomycin-sensitive bacteria are associated with the induction of productive immune responses in the lung. Local or distal injection of Toll-like receptor (TLR) ligands could rescue the immune impairment in the antibiotic-treated mice. Intact microbiota provided signals leading to the expression of mRNA for pro–IL-1β and pro–IL-18 at steady state. Following influenza virus infection, inflammasome activation led to migration of dendritic cells (DCs) from the lung to the draining lymph node and T-cell priming. Our results reveal the importance of commensal microbiota in regulating immunity in the respiratory mucosa through the proper activation of inflammasomes.
Keywords: mucosal immunity, NLRP3, caspase-1, adaptive immunity
Viral respiratory infections cause severe morbidity and mortality in both humans and animals worldwide. Influenza virus is the major source of severe viral respiratory infections in adults, causing annual epidemics that result in severe morbidity and mortality involving 3 to 5 million people annually. The constant threat of the emergence of a novel influenza subtype engenders an even greater risk to society, as the recent pandemic with the swine flu (1) clearly demonstrated. Innate recognition of influenza virus through pattern recognition receptors (PRRs) plays a central role in the generation of adaptive immune responses. Recent studies highlighted the importance of the NOD-like receptor (NLR) activation of inflammasomes in antiviral defense (2). Work from our laboratory and others demonstrated that inflammasome activation and the downstream cytokines (3) play a key role in innate (4, 5) and adaptive (6) immune defense against influenza virus infection in vivo. Using a sublethal dose (10 pfu) of A/PR8 strain, we demonstrated that mice deficient in caspase-1 or apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) have diminished T- and B-cell responses and ultimately succumb to viral infection. Two other groups, using high lethal doses of influenza virus challenge, demonstrated that in the absence of the NLRP3 inflammasome, mice succumb to high doses of A/PR8 infection due to innate immune defects (4, 5). However, how inflammasomes are activated to elicit adaptive immunity following respiratory influenza infection in vivo remains unclear.
Commensal bacteria are essential in shaping intestinal immune responses in both health and disease (7, 8). Germ-free mice have underdeveloped gut-associated lymphoid tissues including Peyer's patches, isolated lymphoid follicles, and mesenteric lymph nodes (9). Commensal bacteria are sensed by the innate pattern-recognition receptors to maintain the homeostasis of intestinal epithelial cell turnover and integrity (10). Gut commensal microbiota also support intestinal immune homeostasis by regulating Tregs (11–13) and Th17 cells (14–17). In some instances, specific bacteria have been associated with immunological outcomes. Commensal bacteria, particularly the segmented filamentous bacteria, promote Th17 development in the intestine (14, 15). In addition to their beneficial effects in the intestine, recent studies highlight the importance of gut bacterial composition in a number of pathological conditions including diabetes (18) and obesity (19–22). Inflammatory bowel disease can develop as a result of the emergence of harmful intestinal bacteria (23). However, it remains unclear whether there is a role for microbiota in shaping the immune inductive function at a nonintestinal mucosal surface such as the lung.
Here, we examine the role of commensal bacteria in the initiation of adaptive immunity after respiratory infection with influenza virus. We demonstrate the requirement for intact commensal bacterial community in the establishment of Th1, CTL, and IgA responses to respiratory influenza virus infection. Notably, we found that neomycin-sensitive bacteria contributed to immunocompetence in the lung. This was in part mediated by providing signals for robust priming of pro–IL-1β and pro–IL-18 expression at steady state. Thus, our data reveal a key role for commensal bacteria in controlling adaptive immunity against a respiratory virus infection.
Results
Immune Responses to Respiratory Influenza Virus Infection Are Diminished by Antibiotic Treatment.
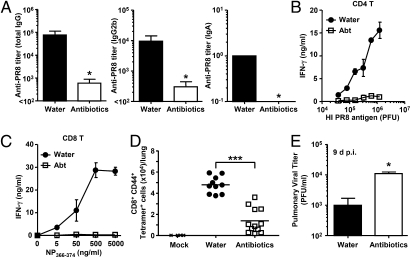
To determine the importance of commensal microbiota in immune responses within the respiratory tract, mice were subjected to a 4-wk oral administration of antibiotic combination, vancomycin, neomycin, metronidazole, and ampicillin (V/N/M/A) (10, 11, 24–26). This treatment resulted in significant changes in the composition of culturable commensal bacteria (Fig. S1). Antibiotic-treated mice were then infected intranasally with a sublethal dose (10 pfu) of A/PR8 influenza virus. Mice were kept on the V/N/M/A regimen for the entire duration of the experiments. Two weeks later, virion-specific Ig levels and T-cell responses were measured. Influenza virus-specific antibody titers (Fig. 1A) and CD4 T-cell responses (Fig. 1B) were significantly reduced in the antibiotic-treated group. Both cytokine secretion (Fig. 1 B and C) and the frequency of influenza virus-specific CTLs as determined by tetramer staining (Fig. 1D) were diminished in the antibiotic-treated mice. As a consequence, viral titer in the lung remained significantly elevated in the antibiotic-treated mice at day 9 postinfection (Fig. 1E). These data indicated that commensal microbiota composition profoundly affects the adaptive immune responses to respiratory influenza virus infection and that antibiotic treatment predisposes the mice to high viral replication in the lung.
Fig. 1.
Antibiotic-treated mice fail to induce acquired immunity to influenza virus infection. C57BL/6 mice were given ampicillin (1 g/L), vancomycin (500 mg/L), neomycin sulfate (1 g/L), and metronidazole (1 g/L) in drinking water for 4 wk before PR8 virus infection (10 pfu per mouse). Two weeks later, serum and nasal wash were collected and Ag-specific antibody titers were measured (A), and T-cells were isolated from spleen and restimulated with flu virion or NP peptide for 72 h, and IFN-γ production from CD4 T cells (B) and CD8 T cells (C) was measured. (D) Lymphocytes were collected from the lung of infected animals at 14 d postinfection and stained with flu-specific tetramer. (E) The lung washes of flu-infected mice were harvested at 9 d postinfection, and viral titers were measured by plaque assay. *P < 0.05 and ***P < 0.001 vs. water-fed group. Data represent the mean ± SD. Similar results were obtained from three separate experiments.
Selective Role of Commensal Microbiota in Immune Responses to Influenza Virus.
To examine whether the antibiotic treatment resulted in general immunodeficiency, we immunized these mice with ovalbumin (OVA) in complete Freund's adjuvant (CFA) in the footpad. Unlike lung infection with influenza virus (Fig. 1), immunization with ovalbumin in CFA led to normal Ig (Fig. S2A) and T-cell responses (Fig. S2B) in antibiotic-treated mice. Adaptive immunity following respiratory infection with influenza virus requires the inflammasome activation (6). This led us to consider the possibility that commensal microbiota may regulate immune responses that require (i) the activation of the inflammasomes, and/or (ii) respiratory infection. To address these possibilities, we took advantage of the fact that immunity to genital infection with herpes simplex virus type 2 (HSV-2) does not require inflammasome activation (27). Thus, mice treated with antibiotics were infected intranasally with HSV-2. In contrast to flu infection, we observed WT levels of CD4 and CD8 T-cell activation in response to intranasal infection with HSV-2 in antibiotic-treated mice (Fig. S2 C and D). Next, we examined whether intact commensal microbiota is required for immune response to a bacterial respiratory pathogen. We chose to study Legionella pneumophila, as it is a natural respiratory pathogen and is resistant to the antibiotics used in this study (28, 29). Upon intranasal infection with L. pneumophila, both CD4 and CD8 T-cell responses remained intact in antibiotic-treated mice (Fig. S2 E and F). Moreover, similar immune responses were generated in caspase-1–deficient mice, indicating that neither inflammasomes nor intact commensal bacteria is required for adaptive immune responses to L. pneumophila. These data suggested that the requirement for intact commensal bacteria is restricted to inflammasome-dependent priming and not a general characteristic unique to the respiratory mucosa or to viral infections.
Treg Suppression Is Not Responsible for Commensal Microbiota-Dependent Immunity Against Influenza Virus.
Several possible mechanisms could explain how commensal microbiota might support the generation of adaptive immunity to influenza virus in the lung (Fig. S3). First, commensal microbiota might contribute to inflammasome activation. Second, commensal bacteria-derived products could provide ligands for TLRs or other PRRs and enhance stimulation of adaptive immunity induced by influenza virus infection. Third, because commensal bacteria-associated DNA could block the conversion of Tregs within the gut mucosa (11), antibiotic treatment could result in an enhanced Treg-mediated suppression of adaptive immunity to influenza virus in the lung.
To test the third hypothesis, we measured the percentages of Foxp3+ Tregs in various tissues of water-fed vs. antibiotic-treated mice. As reported previously (11), Treg numbers were increased in the Peyer's patches but not in the spleen of antibiotic-treated mice (Fig. S4A). In addition, in lung and in the mediastinal LNs, we detected higher percentages of Foxp3+ CD4+ T cells in antibiotic-treated mice (Fig. S4A). Thus, we examined whether immune deficiency in the antibiotic-treated mice could be rescued by Treg depletion. To this end, Tregs were depleted by antibody against CD25 a few days before influenza virus infection in antibiotic-treated animals (Fig. S4B). This procedure has been shown previously to deplete Tregs transiently, during which critical suppressive function of the Tregs is mediated, resulting in enhancement of effector T-cell responses to various antigens (30, 31). Treg depletion was confirmed by flow cytometry (Fig. S4C). Our results showed that neither antibody (Fig. S4D) nor T-cell responses to influenza virus (Fig. S4E) were recovered by depletion of Tregs in antibiotic-treated mice. These data indicated that the antibiotic-mediated impairment in antiinfluenza immunity is not sufficiently explained by the effects of commensal bacteria on suppression of Treg conversion.
Rectal TLR Stimulation Restores Immune Response to Influenza Virus Infection in Antibiotic-Treated Mice.
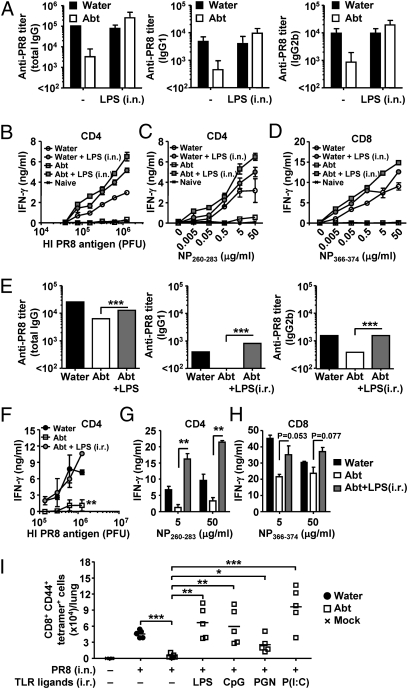
Even though the colon contains by far the highest number of commensal bacteria in the body, upper respiratory mucosa is also inhabited by numerous bacterial species. Thus, to examine whether a TLR agonist can restore immune responses to influenza virus infection in antibiotic-treated mice, and to determine whether the location of such TLR signals dictates the outcome of immune responses to influenza virus infection, we inoculated LPS either intranasally (local) (Fig. 2 A–D) or intrarectally (distal) (Fig. 2 E–H) at the time of influenza virus challenge in antibiotic-treated mice. We picked rectal inoculation to mimic the effect of the high levels of commensal bacteria present in the colon. Remarkably, both antibody and T-cell responses were completely restored by a single inoculation of LPS either locally or distally (Fig. 2). Next, we tested whether other TLR agonists exert similar effects. We found that rectal inoculation of CpG (TLR9 agonist), Poly I:C (TLR3 agonist), and to a lesser extent, peptidoglycan (TLR2 agonist), could restore immunity to influenza virus in the lung (Fig. 2I). These data indicated that a TLR stimulus is sufficient to restore immune responses in antibiotic-treated mice and further suggested that signals coming from distal commensal bacterial products (colon) may be sufficient to support immune priming in the lung.
Fig. 2.
Local and distal TLR stimulation restores immune response to influenza virus infection in antibiotic-treated mice. C57BL/6 mice were given antibiotics in drinking water for 4 wk before 10 pfu of PR8 viral infection with or without 2 μg of LPS injected intranasally (A–D) or intrarectally (E–H). Two weeks later, serum was collected and Ag-specific antibody titers were measured (A and E), and T cells were isolated from spleen and restimulated with flu virion (B and F) or NP peptide (C, D, G, and H) for 72 h, and IFN-γ production from CD4 T cells (B, C, F, and G) and CD8 T cells (D and H) was measured. (I) Water-fed and antibiotic-treated mice were infected intranasally with 10 pfu of PR8 virus with or without intrarectal injection of LPS (5 μg), CpG2216 (50 μg), peptidoglycan (20 μg), or Poly (I:C) (50 μg). Total numbers of influenza virus-specific CD8 T cells in the lung are shown. Data represent the mean ± SD, and are representative of at least three independent experiments (A–H) or are pooled from two independent experiments (I). *P < 0.05; **P < 0.01; ***P < 0.001.
Neomycin-Sensitive Commensal Bacteria Are Required for Immune Responses to Influenza Virus Infection.
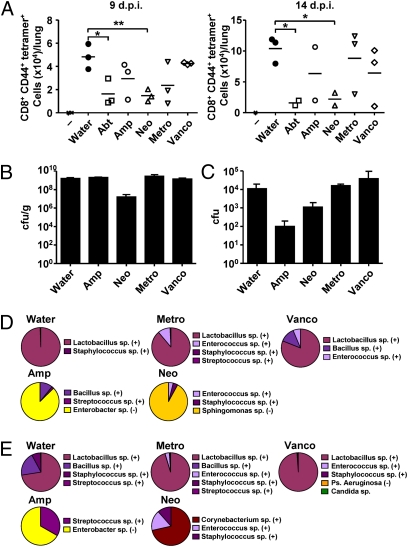
To better understand the bacterial classes responsible for endowing immune functions in the respiratory tract, we treated mice with individual antibiotics. Mice were treated with either combination of all four antibiotics (Abt) or individual antibiotics, ampicillin (Amp), vancomycin (Vanco), neomycin (Neo), or metronidazole (Metro). Remarkably, whereas treatment with ampicillin, vancomycin, or metronidazole alone had only a mild and variable effect on CTL responses after 9 or 14 d of influenza infection, neomycin treatment alone recapitulated the effects of the combination antibiotic treatment on the suppression of CTL responses (Fig. 3A). Metronidazole treatment still maintained CD8 T-cell responses in the lung. In contrast, oral neomycin treatment abolished CD8 T-cell responses in the lung. To determine the bacterial species that are affected by the various antibiotic treatments, stool and nasal wash samples from these mice were subjected to microbiological analyses. Culturable bacteria were enumerated and identified by a combination of biochemical analyses and 16S ribosomal sequencing (Fig. 3 B–E). In the antibiotic-untreated mice and in mice treated with oral vancomycin or metronidazole, Lactobacillus spp. (Gram positive) dominated the intestinal and nasal tracts. It has been reported that orally delivered neomycin is not absorbed through the gastrointestinal tract, thus selectively targeting aerobic bacteria in the intestinal lumen but not elsewhere (32). Consistently, oral neomycin treatment resulted in a significant decrease in the intestinal (Fig. 3B), but not nasal (Fig. 3C), bacterial load. Moreover, the stools of neomycin (aminoglycoside)-treated mice were dominated by Sphingomonas spp., which are known to be resistant to aminoglycoside antibiotics (33) (Fig. 3D). Oral ampicillin treatment diminished Gram-positive bacteria in both the intestine and in the nasal mucosa. Instead, both colonic and nasal mucosae were dominated by an outgrowth of Enterobacter spp., which are mostly ampicillin resistant (34) (Fig. 3 D and E). Collectively, these data indicated that oral treatment with neomycin impaired immune responses to respiratory influenza virus infection, which was associated with depletion of Gram-positive bacteria in the gut but not the nasal tract.
Fig. 3.
Effect of single antibiotic treatment on bacterial colonization and immune responses to respiratory influenza virus infection. C57BL/6 mice were each given single or four combinatorial antibiotics in drinking water for 4 wk before PR8 virus infection (10 pfu per mouse). Lymphocytes were collected from the lung of infected animals at 9 d or 14 d postinfection and stained with flu-specific tetramer (A). Bacterial load in the stool (B) and nasal wash (C) from antibiotic-treated mice (n = 3 per condition) were measured. Bacterial compositions in the stool (D) and nasal wash (E) from single antibiotic-treated mice are depicted. Purple and yellow tones denote Gram-positive and Gram-negative bacteria, respectively. Similar results were obtained from two to three separate experiments. *P < 0.05; **P < 0.01.
Commensal Bacterial Supply Signal 1 for IL-1β and IL-18 Secretion.
Next, we asked how commensal bacteria are required for optimal respiratory immune responses against influenza virus infection. Because intact commensal microbiota was required for adaptive immune responses to inflammasome-dependent (flu), but not inflammasome-independent pathogen (HSV-2, Legionella) (Fig. S2), we examined whether commensal microbiota might provide signals necessary for inflammasome-dependent cytokine secretion. Two distinct stimuli are necessary for the processing and secretion of IL-1β; the first stimulus (signal 1) to induce transcription and translation of pro–IL-1β; and the second (signal 2), to activate caspase-1 that processes IL-1β into mature form (35). The priming signal (signal 1), often provided through TLRs, leads to transcriptional activation of the genes encoding pro–IL-1β, pro–IL-18, and NLRP3 (36, 37). The second signal is mediated through the activation of the inflammasome complex, leading to the proteolytic activation of caspase-1. To test whether commensal microbiota might supply signals necessary for IL-1β and IL-18 secretion, we measured the levels of secreted IL-1β in the bronchoalveolar lavage (BAL) of mice infected with influenza that had been pretreated with the combination antibiotics. Notably, antibiotic treatment of WT mice impaired mRNA expression of pro–IL-1β, pro–IL-18, and NLRP3 and reduced secretion of mature IL-1β protein in the BAL to the levels of ASC-deficient mice following influenza infection (Fig. S5). These data indicated that intact commensal microbiota provides signals that support the expression of pro–IL-1β and pro–IL-18 (signal 1) even at the steady state. Upon influenza infection, virus-inflicted damage, including the activity of M2 ion channel (signal 2) (38), leads to the activation of inflammasomes and release of mature forms of IL-1β and IL-18.
Antibiotic Treatment Impairs DC Homeostasis and Migration by Reducing Priming Signals for Inflammasome-Dependent Cytokines.
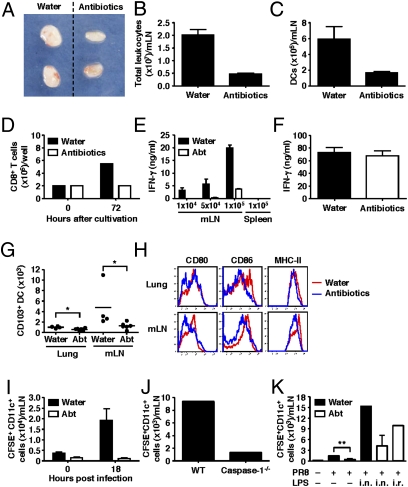
Because T-cell priming upon influenza virus infection is mediated by DCs in the mediastinal LNs (39), we examined the antigen presenting capacity of mLN DCs in antibiotic-treated mice. Mice were infected intranasally with a recombinant influenza virus encoding for the GP33-41 epitope of lymphocytic choriomeningitis virus (6). At steady state, the number of non-DCs in the lung, mLN, spleen, blood, and bone marrow were comparable in antibiotic-treated vs. -untreated mice. In contrast, gross lymph node size (Fig. 4A), cellularity (Fig. 4B), and DC numbers (Fig. 4C) in the mLN were considerably reduced in antibiotic-treated influenza virus-infected mice. Further, freshly isolated mLN DCs from antibiotic-treated mice, in the absence of exogenous peptide, induced no proliferation (Fig. 4D) or differentiation (Fig. 4E) of P14 CD8 TCR Tg T cells specific for the GP33-41 epitope presented on H-2Db. This defect was due to impairment of viral antigen peptide presentation by the mLN DCs from antibiotic-treated mice, because the same DCs were able to differentiate P14 naïve CD8 T cells after exogenous GP33 peptide addition (Fig. 4F).
Fig. 4.
Respiratory tract DCs fail to migrate to the draining LN and prime T-cell responses in antibiotic-treated mice. C57BL/6 mice were given antibiotics in drinking water for 4 wk before intranasal infection with 1,000 pfu of PR8-GP33 viruses. (A–C) Three days later, CD11c+ DCs were isolated from the mediastinal LN. (D–F) Naïve p14 tg CD8 T cells (2 × 105 cells per well) were cocultured with different numbers of CD11c+ DCs isolated from mLN of infected animals with (F) or without GP33 peptide (D and E) for 72 h. Splenic DCs from infected animals were used as negative control (E). IFN-γ production (E and F) and the number of CD8 T cells (D) were measured. The number of CD103+ DCs (G) and phenotype of total DC population (H) in the lung and mLN were measured in antibiotic-treated mice without influenza infection. (I and J) Water-fed, antibiotic-treated, and caspase-1–deficient mice were inoculated intranasally with CFSE. Six hours later, mice were infected with 1,000 pfu of PR8 viruses. Eighteen hours after infection, mediastinal LNs were collected. The numbers of CFSE+CD11c+DCs are shown. (K) LPS inoculation (intranasal or intrarectal) restored DC migration to the mLN following intranasal influenza virus infection. Data represent the mean ± SD. Similar results were obtained from three separate experiments.
We hypothesized that the reduction in antigen-presenting DCs in the antibiotic-treated mice is due to (i) reduction in the number of respiratory DCs, and/or (ii) the failure of DCs to migrate from the lung to the mLN. To determine the possible effect of antibiotics on respiratory DCs, flow cytometric analyses of the phenotype and number of lung DCs were carried out. Of note, we observed a significant reduction in the number of CD103+ DCs in the lung and mLNs of antibiotic-treated mice at the steady state (Fig. 4G). Respiratory tract CD103+ DCs are responsible for priming CD8 T-cell responses early after influenza virus infection (39, 40). In addition, expression levels of CD86, CD80, and MHC class II were lower in DCs from antibiotic-treated mice (Fig. 4H). Next, to determine the migration of respiratory tract DCs into the mLN upon influenza virus infection, mice were first inoculated with carboxyfluorescein succinimidyl ester (CFSE) intranasally and lung-migrant DCs were enumerated after 18 h post-infection in the mLN (Fig. 4I) (41). These data showed that commensal bacteria are required for lung DCs to migrate to the mLN. Consequently, P14 naïve CD8 T cells specific for the GP33 peptide failed to proliferate in the mLNs at 5 d after infection with either 10 pfu or 100 pfu of influenza virus in vivo (Fig. S6). Next, to test whether such a defect in lung DCs in the antibiotic-treated mice correlates with their failure to prime for inflammasome-dependent cytokines, we examined the ability of DCs to migrate from the lung to the mLN in caspase-1–deficient mice's response to flu infection. Migration of lung DCs to the mLN was severely impaired in the absence of caspase-1 (Fig. 4J). These data indicated that inflammasome activation, which is diminished by antibiotic treatment (Fig. S5), is required for migration of respiratory DCs to the mLNs. Finally, we examined whether the impaired immune responses observed in antibiotic-treated mice was directly linked to the diminished DC migration. To this end, we tested whether the TLR rescue of the influenza response in antibiotic-treated mice (Fig. 2) was colinked to DC migration from the lung to the mLN. Notably, LPS inoculation (intranasal or intrarectal) completely restored DC migration into the mLN after flu infection in antibiotic-treated mice (Fig. 4K). These data collectively indicated that alteration in the microbiota by antibiotics leads to the failure in the activation of inflammasome-dependent cytokine release and to the impairment of respiratory tract DC homeostasis and their migration to the draining LN. Such a defect in DC migration results in diminished T-cell responses following influenza infection.
Discussion
Commensal bacteria provide the mammalian hosts with essential functions well beyond the digestive system. Although their importance in the maintenance of immune homeostasis in the gut mucosa is widely appreciated (7), our study revealed that the commensal microbiota could regulate immune responses in the respiratory mucosa. We demonstrated that oral antibiotic treatments resulted in defective CD4 T-, CD8 T-, and B-cell immunity following intranasal infection with influenza virus. This was not due to global or local immune deficiencies in these mice, as antibiotic treatments did not impair immune responses against protein antigen injected with CFA or to respiratory infection with HSV-2 or Legionella pneumophila. Commensal bacteria-mediated support of the immune response against flu infection was also independent of their effect on Tregs, but was sufficiently restored by distal (rectal) inoculation of TLR agonists. In antibiotic-treated mice, synthesis of pro–IL-1β, pro–IL-18, and NLRP3 was impaired even at the steady state. Antibiotic treatment led to impaired respiratory DC distribution and activation status at steady state and reduced DC migration from the lung to the draining lymph nodes. Acute injection of LPS was able to overcome defective inflammasome activation and DC migration in antibiotic-treated mice. Of note, IL-1β is known to play a key role in the migration of Langerhans cells from the skin to the draining lymph node in a contact hypersensitivity model (42), suggesting that this cytokine may be a common inducer of DC mobilization.
Our data demonstrate that not all commensal bacteria could contribute equally to the immunocompetence in the lung. Oral neomycin, but not ampicillin, vancomycin, or metronidazole treatment consistently abolished CTL responses to influenza virus. Microbiological examination of the stool and nasal secretions from mice treated with a single antibiotic revealed that oral treatments with vancomycin or metronidazole made minimal changes to the composition or density of culturable endogenous microbiota, which consisted mainly of Lactobacillus spp. In contrast, oral neomycin treatment, which mainly targeted gut-resident bacteria, ablated the immune response to respiratory influenza infection. Interestingly, neomycin depleted all culturable Gram-positive bacteria in the colon, while maintaining those in the nasal tract. Oral ampicillin treatment had a systemic effect on depleting most of the Gram-positive bacteria, with an overgrowth of Enterobacter spp. in both the colonic and nasal mucosae. These data collectively indicated that commensal microbiota responsible for conferring an immunogenic environment in the lung are either the gut-resident Gram-positive bacteria and/or bacteria in the nasal tract. Of note, neomycin treatment resulted in a shift from mostly Lactobacillus spp. to Corynebacterium spp. in the nasal mucosa. It is intriguing that certain bacterial species are insufficient (Sphingomonas spp. in the gut and Corynebacterium spp. in the nasal tract) to promote immunocompetence against influenza virus infection. Perhaps these bacteria fail to prime basal levels of pro–IL-1β and pro–IL-18 expression. In the human intestine, anaerobic bacteria outnumber aerobic bacteria by a factor of 100–1,000 (43). The phyla Firmicutes (Gram positive) and Bacteroidetes (anaerobic Gram negative) are predominant in the human colon (44), whereas aerobes (facultative anaerobes), such as Escherichia, Enterobacter, Enterococcus, Klebsiella, Lactobacillus, and Proteus, remain a small minority (43). Interestingly, a systematic metagenomic analysis of total microbiota communities affected by similar oral antibiotic treatments in mice revealed that intestinal mucosa-associated Lactobacillus spp. are significantly reduced (45). However, future studies are needed to determine the bacteria responsible for conferring immunocompetence against influenza infection.
Our data revealed a link between commensal microbiota community and inflammasome-dependent cytokine activation. Currently, the precise mechanism by which gut commensal bacteria supports inflammasome activation in the lung is unknown. We speculate that products of a select group of commensal bacteria could trigger a variety of pattern recognition receptors to stimulate leukocytes either locally or systemically. Factors released by such leukocytes can support steady-state production of pro–IL-1β, pro–IL-18, and NLR proteins, thereby priming signal 1 for inflammasome-dependent cytokine activation. This idea is supported by our results that injection of TLR ligands can restore immune responses to influenza virus in antibiotic-treated mice. Commensal microbiota providing signals for PRRs has been shown by a recent study showing that peptidoglycan translocated from the gut microbiota to the systemic circulation is sensed by Nod1, resulting in enhanced systemic innate immunity mediated by neutrophils (46). We speculate that microbiome may be responsible for providing signal 1 for inflammasome-dependent cytokine activation in vivo under circumstances in which signal 2 (such as uric acid and alum) alone are capable of eliciting robust caspase-1–dependent IL-1β and IL-18 production.
In summary, our findings substantially expand the contribution of the microbiota in maintaining the immunological status of the host animal beyond the intestinal mucosa. Because antibiotic use is prevalent in the treatment of respiratory infections, our results imply a possible deleterious effect of such treatment in initiating proper immune responses to influenza virus. Conversely, it will be important to determine whether probiotic therapy can be explored for immune-stimulating effects during the flu season.
Materials and Methods
Antibiotic Treatment.
Mice were treated for 4 wk with ampicillin (1 g/L), vancomycin (500 mg/L), neomycin sulfate (1 g/L), and metronidazole (1 g/L) in drinking water as previously described (10, 11, 24–26). For some experiments, individual antibiotics (at the indicated concentration) were given in drinking water. Antibiotic-containing water was changed twice a week.
Virus Infection in Vivo.
A/PR8 virus (H1N1) and recombinant PR8 virus expressing the lymphocytic choriomeningitis virus (LCMV) glycoprotein epitope GP33-41 (PR8-GP33; a gift from S. Kaech, Yale University, New Haven, CT) used for all experiments was grown in allantoic cavities from 10- to 11-d-old fertile chicken eggs for 2 d at 35 °C. Viral titer was quantified by a standard plaque assay using Madin-Darby canine kidney cells and viral stock was stored at −80 °C. For intranasal infection, mice were fully anesthetized by i.p. injection of ketamine and xylazine and then infected by intranasal application of 20 μL of virus suspension (10–1,000 pfu of influenza virus or 106 pfu of HSV-2 in PBS). This procedure leads to upper and lower respiratory tract infection.
Nasal and Rectal Inoculation of TLR Agonist.
For intranasal inoculation, water-fed and antibiotic-treated mice were infected by intranasal application of 20 μL of virus suspension including 10 pfu of influenza virus and 2 μg of LPS. For intrarectal inoculation, water-fed and antibiotic-treated mice were inoculated by intrarectal application of 50 μL of PBS containing ultrapure LPS from Escherichia coli (InvivoGen; 5 μg), CpG2216 (Tri-Link Biotech; 50 μg), peptidoglycan from Bacillus subtilis (Fluka, now Sigma-Aldrich; 20 μg), or Poly (I:C) (InvivoGen; 50 μg) at the same time with intranasal infection with 10 pfu of influenza virus.
Supplementary Material
Acknowledgments
We thank S. Yu and B. Yordy for technical support and S. Shin and R. Medzhitov for helpful advice. This study was supported by grants from the National Institutes of Health (NIH) (AI054359, AI062428, and AI064705). T.I. was a recipient of the Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad. A.I. is a recipient of the Burroughs Wellcome Investigators in Pathogenesis of Infectious Disease. I.K.P. was supported by a NIH National Research Service Award (T32AI07019) from the Interdisciplinary Immunology Training Program in Yale University, Department of Immunobiology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019378108/-/DCSupplemental.
References
- 1.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgerald KA. NLR-containing inflammasomes: Central mediators of host defense and inflammation. Eur J Immunol. 2010;40:595–598. doi: 10.1002/eji.201040331. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz N, Kurrer M, Bachmann MF, Kopf M. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J Virol. 2005;79:6441–6448. doi: 10.1128/JVI.79.10.6441-6448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;16:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 8.Chervonsky A. Innate receptors and microbes in induction of autoimmunity. Curr Opin Immunol. 2009;21:641–647. doi: 10.1016/j.coi.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Mahony C, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 17.Niess JH, Leithäuser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 18.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 23.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaph C, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang SS, et al. An antibiotic-responsive mouse model of fulminant ulcerative colitis. PLoS Med. 2008;5:e41. doi: 10.1371/journal.pmed.0050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bashir ME, Louie S, Shi HN, Nagler-Anderson C. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172:6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 27.Sato A, Iwasaki A. Induction of antiviral immunity requires Toll-like receptor signaling in both stromal and dendritic cell compartments. Proc Natl Acad Sci USA. 2004;101:16274–16279. doi: 10.1073/pnas.0406268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu KP, Neu HC. Inactivation of beta-lactam antibiotics by Legionella pneumophila. Antimicrob Agents Chemother. 1979;16:561–564. doi: 10.1128/aac.16.5.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelstein PH. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981;14:298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onizuka S, et al. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res. 1999;59:3128–3133. [PubMed] [Google Scholar]
- 31.Pasare C, Medzhitov R. Toll-dependent control mechanisms of CD4 T cell activation. Immunity. 2004;21:733–741. doi: 10.1016/j.immuni.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Oudemans-van Straaten HM, van Saene HK, Zandstra DF. Selective decontamination of the digestive tract: Use of the correct antibiotics is crucial. Crit Care Med. 2003;31:334–335. doi: 10.1097/00003246-200301000-00067. [DOI] [PubMed] [Google Scholar]
- 33.Vanbroekhoven K, et al. Streptomycin as a selective agent to facilitate recovery and isolation of introduced and indigenous Sphingomonas from environmental samples. Environ Microbiol. 2004;6:1123–1136. doi: 10.1111/j.1462-2920.2004.00654.x. [DOI] [PubMed] [Google Scholar]
- 34.Kelesidis T, Karageorgopoulos DE, Kelesidis I, Falagas ME. Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: A systematic review of the evidence from microbiological and clinical studies. J Antimicrob Chemother. 2008;62:895–904. doi: 10.1093/jac/dkn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 36.Bauernfeind FG, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GeurtsvanKessel CH, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE. 2009;4:e4204. doi: 10.1371/journal.pone.0004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGill J, Van Rooijen N, Legge KL. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med. 2008;205:1635–1646. doi: 10.1084/jem.20080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths CE, Dearman RJ, Cumberbatch M, Kimber I. Cytokines and Langerhans cell mobilisation in mouse and man. Cytokine. 2005;32:67–70. doi: 10.1016/j.cyto.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 44.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.