Abstract
The zona pellucida (ZP) domain is a bipartite protein structural element comprised of ZP-N and ZP-C regions. Most notable for its ability to mediate protein polymerization, many ZP proteins polymerize and assemble into long fibrils that form specialized extracellular matrices. Other ZP proteins (namely, betaglycan and endoglin) do not polymerize but serve as important membrane coreceptors for ligands in the transforming growth factor-β (TGF-β) superfamily. Here, we present the 2.0-Å resolution crystal structure of the betaglycan ZP-C region in combination with a downstream region known as the external hydrophobic patch (EHP). Similar to the ZP-N region, the ZP-C region also adopts an immunoglobulin-like fold, despite sharing no sequence homology and possessing different disulfide linkages. The EHP region, which was previously thought to be external to the ZP region, is integral to the ZP-C domain and corresponds to the ZP-C G strand. Our structure also indicates that the critical maturation cleavage of ZP proteins, a process that activates nascent ZP proteins for polymerization, occurs within the immunoglobulin domain at the FG loop. Nonpolymerizing ZP proteins such as betaglycan and endoglin do not contain this cleavage site. Finally, our structure suggests that the AB loop and the convex surface pocket are regions important for TGF-β ligand binding.
Keywords: reproductive biology, signaling, inhibin
The zona pellucida (ZP) domain is a structural element found in the extracellular region of many eukaryotic proteins (1, 2). This structural element is characterized by a set of eight highly conserved cysteines and predicted to exist as a bipartite structure corresponding to ZP-N and ZP-C regions tethered by a linker (3, 4). The crystal structure of the ZP-N region has been solved and shown to adopt an immunoglobulin-like fold (5), suggesting that the ZP-C region is likely also a distinct domain by itself and that the so-called ZP domain, the collective structural element comprised of distinct ZP-N and ZP-C domains, may be more aptly referred to as the ZP region.
The ZP region is important for protein polymerization and protein–protein interactions (6, 7). Many ZP proteins polymerize to become long fibrils that form specialized extracellular matrices. The best-known example is the pericellular matrix known as zona pellucida or vitelline envelope that surrounds mammalian and nonmammalian oocytes, respectively, to ensure species-specific fertilization and prevent polyspermy. This matrix is composed from homologues of ZP1, ZP2, ZP3, and in some species, ZP4 proteins, all of which contain a single ZP region in the ectodomain (1). In Drosophila melanogaster, two ZP proteins, Piopio and Dumpy, contribute to the apical extracellular matrix and play crucial roles in tracheal development and tubulogenesis (8). In mammals, other well-characterized polymerizing ZP proteins include uromodulin (also known as Tamm–Horsfall protein) that likely forms the water-impermeable layer around the thick ascending loop of Henle in the kidney (9) and α-/β-tectorins that help form the tectorial membrane in the inner ear (10–12).
In addition to protein polymerization, the ZP region is also important for binding to ligands within the transforming growth factor-β (TGF-β) superfamily. Two important TGF-β coreceptors, betaglycan and endoglin, contain a ZP region in their ectodomain. In betaglycan, there are two distinct TGF-β binding sites: one in the membrane-distal region that shares sequence homology with endoglin (E region) (13) and one in the membrane-proximal ZP region (14). However, these two betaglycan binding sites differ in their ligand specificities. Whereas the TGF-β ligands TGF-β1/2/3, BMP-2/4/7, and GDF-5 engage both E and ZP regions, the TGF-β ligand inhibin binds only to the ZP region (15, 16). Recent studies have further localized the TGF-β binding site in the ZP region to the ZP-C domain (17, 18). The membrane-bound betaglycan ZP-C domain is necessary and sufficient for binding and mediating inhibin signaling (17), and antibody raised against the ZP-C domain neutralizes the betaglycan-dependent bioactivity of inhibin (18).
Several important questions pertaining to the mechanisms underlying ZP region function remain unresolved. With respect to protein polymerization, studies in the past several years have demonstrated that proteolytic cleavage of nascent membrane-tethered ZP protein into the pericellular space is linked to its polymerization ability, an important regulatory mechanism given that premature polymerization inside the cell would have devastating effects. Specifically, a region known as the external hydrophobic patch (EHP), located downstream from the consensus ZP region, has been shown to be important for ZP protein secretion and preventing premature polymerization (19–21). However, the structural basis for this coupling of proteolytic cleavage and polymerization has not been clear. With respect to TGF-β signaling, recombinant mutagenesis screens in the betaglycan ZP-C domain have identified several residues important for binding to TGF-β ligands inhibin and TGF-β1. Specifically, betaglycan V614 and surrounding residues in the ZP-C domain have been shown to be universally important for both inhibin and TGF-β1 binding (17). To clarify these observations, we have determined the crystal structure of the betaglycan ZP-C domain and revealed the critical TGF-β binding surface on betaglycan. The structure also reveals that the EHP region, previously thought to be ancillary to the ZP region, is an integral part of the ZP-C domain and redefines the C-terminus of the ZP region, in agreement with a recently reported ZP3 protein structure (22). The structural results suggest a strand-displacement model for ZP-mediated protein polymerization.
Results and Discussion
EHP Region Is Necessary for Soluble Betaglycan Expression.
The consensus ZP region in rat betaglycan spans residues 456–733, with residues 591–733 corresponding to the ZP-C portion. Residues downstream from the ZP region, referred to as the EHP in polymerizing ZP proteins (19–21), correspond to amino acids 746–755. Expression of the consensus ZP region, lacking the EHP, resulted in misfolded and aggregated protein (Fig. S1A). The combined betaglycan ZP-C and EHP regions were expressed, based on biochemical data (14, 15, 17) and secondary structure prediction (23), providing homogeneous monomeric protein in solution (Fig. S1B). Constructs of the entire betaglycan ectodomain and the full ZP region, containing the EHP, also produced homogenous, nonaggregated protein (Fig. S1 C and D). The ZP-C/EHP protein crystallized readily, and the structure was determined by the multiple isomorphous replacement with anomalous scattering method (Table S1).
Structure of the Betaglycan ZP-C Domain.
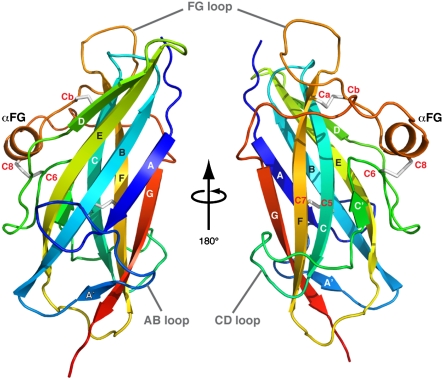
The betaglycan ZP-C and EHP regions collectively form the ZP-C domain and adopt an immunoglobulin-like β-sandwich fold (Fig. 1). Seven major β-strands form the classical A-B-E-D and C-F-G β-sheets. There are three extended and ordered loops, each containing a smaller secondary structure insertion: strand A′, strand C′, and helix αFG are found in the AB, CD, and FG loops, respectively. The AB and CD loops are kept ordered by a combination of hydrophobic interactions, polar contacts, and a disulfide bond. The FG loop, however, does not contact the hydrophobic core and is restrained by two separate disulfide bonds.
Fig. 1.
Structure of betaglycan ZP-C Domain. The ZP-C domain shown as rainbow-colored ribbon representation from blue (N-terminus) to red (C-terminus). Conserved cysteine residues are labeled in red, and the disulfide bonds are shown as gray sticks.
There has been disagreement regarding the correct disulfide linkages in the ZP-C region (24–26). ZP proteins are classified as either type I or type II based on the number of conserved cysteines and their disulfide linkage patterns (1) (Figs. S2A and S3). Type I ZP proteins have four conserved cysteines in the ZP-C region (C5, C6, C7, C8), whereas type II ZP proteins, including betaglycan, have six (C5, C6, C7, Ca, Cb, C8). In one model, the type I and type II ZP-C domains have been proposed to have very different disulfide bond arrangements: C5–C7, C6–C8 for type I and C5–C6, C7–Ca, Cb–C8 for type II (24, 25). However, it has also been suggested that the type II arrangement is similar to type I, with the additional two cysteines (Ca–Cb) forming an additional disulfide linkage (26). The betaglycan ZP-C structure conforms to this latter model; the earlier model for type II ZP-C domains would require significant distortions of the immunoglobulin-like architecture.
The ZP region does not have high sequence conservation with the exception of the invariant cysteines, but structure-based sequence alignment of ZP-C domains from several well-characterized ZP proteins shows good sequence conservation in strands A, B, C, E, F, G, which collectively form the hydrophobic core (Fig. S2B). This pattern of sequence conservation is consistent with other proteins known to adopt canonical immunoglobulin folds (27). However, the ZP-C D strand as well as the AB and FG loops are poorly conserved and vary in length.
Comparison of the ZP-N and ZP-C Domain Structures.
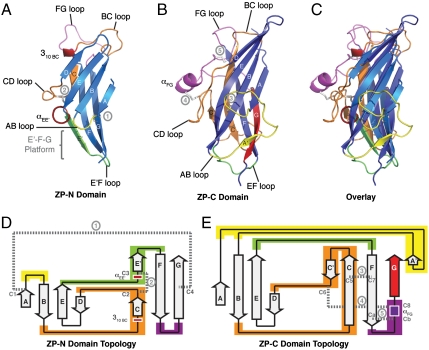
There are some remarkable similarities between the available ZP-N domain (5) and the current ZP-C domain structures despite sharing no sequence homology and possessing different disulfide linkages (Fig. 2). Specifically, both ZP-N and ZP-C domains adopt immunoglobulin-like β-sandwich folds, which is consistent with previous studies that suggest the ZP region to be a bipartite motif rich in β-structure with minimal α-helical content (1).
Fig. 2.
Structural comparison of ZP-N and ZP-C immunoglobulin domains. (A–C) Ribbon representation of (A) mouse ZP3 ZP-N domain, (B) rat betaglycan ZP-C domain, and (C) structural overlay of the two domains. (D and E) Topology representation of (D) ZP-N and (E) ZP-C domains. Corresponding ZP-N and ZP-C regions exhibiting major structural differences are similarly colored: AB loop (yellow), BC loop/C strand/CD loop (orange), EF loop (green), FG loop (magenta). The disulfide linkages are shown as gray sticks in the ribbon representation and dashed lines in the topology representation. Circled numbers in gray denote corresponding disulfide bonds between the ribbon and topology representations.
There are also some important distinctions between the ZP-N and ZP-C domains. The ZP-C domain incorporates more residues than the ZP-N domain, with most of this difference attributed to the extensive AB, CD, and FG loop elaborations found on the ZP-C domain. Specifically, the length of the ZP-N domains range from 87 (hENG) to 124 (rBG/hBG) residues, and the ZP-C domains range from 138 (hENG) to 202 (hZP3) residues. A number of structural features on the ZP-N and ZP-C domains are also different. The ZP-N domain exhibits a unique E′ strand and a pairing of two unequally sized β-sheets that results in an extended E′-F-G sheet platform (Fig. 2A, Bottom), which contains two conserved tyrosine residues implicated in ZP polymerization (5). In contrast, the ZP-C domain has two symmetrically sized β-sheets covering the central hydrophobic core and lacks an inter-β-sheet disulfide bond (Fig. 2).
Binding of Inhibin to the Betaglycan ZP-C Domain.
The ZP-C domain is thought to encompass the sole binding site for the TGF-β ligand inhibin and one of two binding sites for the TGF-β ligands TGF-β1/2/3, BMP-2/4/7, and GDF-5 on the betaglycan ectodomain (15, 16, 28). However, previous studies on the betaglycan:inhibin interaction were performed in cell-based binding assays using membrane-anchored betaglycan (15, 17). To verify that the ZP-C domain incorporates the only inhibin binding site on betaglycan in a cell-free format, we compared the ability of inhibin to bind to purified betaglycan full-length ectodomain, full ZP region, and ZP-C domain (Fig. S4A). All three soluble betaglycan fragments, when immobilized in equal molar amounts, bind to soluble inhibin with similar affinity, suggesting that the ZP-C domain contains the only genuine binding element within betaglycan.
Single amino acid substitutions in full-length betaglycan have implicated S608, G610, V614, A615, E616, H619, and V620 as important for TGF-β ligand binding (17) (Fig. S2B, residues highlighted in green). In particular, V614 was identified as one of the most critical residues for TGF-β ligand binding to the membrane-proximal ZP region site: The V614Y mutation abolished inhibin binding, failed to potentiate inhibin signaling, and significantly disrupted TGF-β1 binding with the ligand presumably relegated to binding only via the distinct E-region binding site (17). On the ZP-C domain, this stretch of residues from 608–620 maps mostly onto the AB loop, with V614 pointing inward and lining a small hydrophobic pocket (Fig. S4B).
In addition to the AB loop region, F635 and I637 on strand C have also been implicated with TGF-β ligand binding because conservative F635L and I637V mutations resulted in pronounced disruption to inhibin binding (17). Similar to the V614 residue, both F635 and I637 point toward the hydrophobic core and help line a pocket. In this case, F635 and I637 collectively form the floor of the large basic pocket on the ZP-C convex surface (Fig. S4C). Because the AB loop region and the large basic pocket do not form a single contiguous surface, it may be possible that one of these sites indirectly facilitates ligand binding via an accessory protein such as other membrane receptors within the TGF-β family.
The present work advances our understanding of betaglycan function in several ways. First, the structure of betaglycan ZP-C domain reveals the correct domain boundaries for the ZP region, enabling further biochemical characterization of the betaglycan ZP region in TGF-β signaling. Second, the TGF-β ligand interaction sites on the betaglycan ZP-C domain have been mapped. Finally, we demonstrate using purified proteins that the full-length betaglycan ectodomain, the full ZP region, and the ZP-C domain all bind to inhibin with comparable affinity and suggest that the betaglycan extracellular region contains at least three distinct domains.
Structural Features of the ZP-C Domain FG Loop.
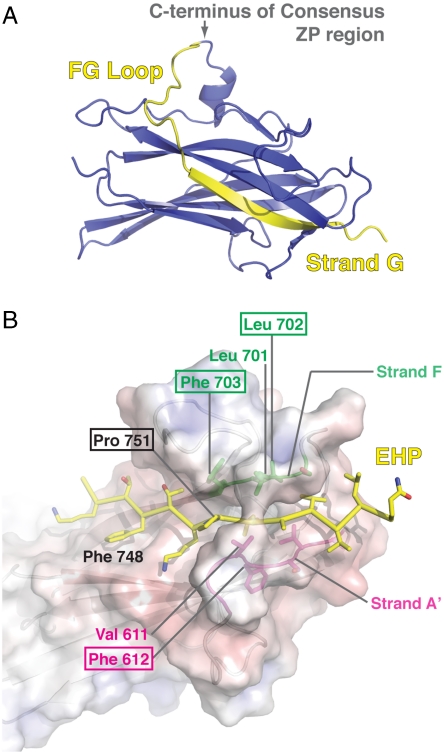
One significant finding from the betaglycan ZP-C structure is that the predicted consensus ZP-C region ends at the FG loop and that the EHP forms the final β-strand (G) that completes the ZP-C domain fold (Fig. 3A), an observation also made in the ZP3 protein structure (22). The conserved P751, a functionally important residue in the EHP (20, 21), makes hydrophobic contacts with the exposed F703 on strand F, is preceded by a stretch of charged residues that cover the N-terminal surface of this region, and directs the ensuing stretch of aliphatic residues to fit within the hydrophobic groove lined by strands A′ and F (Fig. 3B).
Fig. 3.
Structural features of the EHP region. (A) The consensus ZP portion (blue) and the EHP region (yellow) are depicted in ribbon representation of the ZP-C domain. (B) Key contact residues of the EHP region are indicated and colored according to their location: the EHP region (yellow), the F strand (green), and the A′ strand (magenta). Boxed amino acids indicate conserved residues. The surface representation is colored by Poisson–Boltzmann electrostatic calculations contoured from -10 (red) to 10 (blue) kT/e.
There are two notable features that map to the ZP-C FG loop. First, there is a consensus furin-like cleavage site on the FG loop of many ZP proteins (Fig. S2B, red box). Uromodulin does not have the consensus RXXR cleavage sequence, but is cleaved within this loop at 586-RFRS-589 (29). In contrast, betaglycan and endoglin, which do not polymerize even as soluble proteins (28, 30–32), lack a protease cleavage site in the FG loop. The second feature is a long insertion in the FG loop of ZP3 that contains four additional cysteines (Fig. S2B) and incorporates several O-linked glycans implicated for species-specific sperm binding (33, 34), although recent data in transgenic mice have also suggested that carbohydrates on the ZP3 ZP-C FG loop are not essential for sperm binding (35).
Mapping of ZP-C Domain Mutations and ZP-N/ZP-C Linker Boundaries.
Targeted mutation screens in other ZP proteins have also identified several naturally occurring missense mutations in the ZP-C domain that lead to autosomal dominant disorders. Specifically, ZP-C mutations in endoglin lead to the vascular disorder hereditary hemorrhagic telangiectasia. ZP-C mutations in uromodulin and α-tectorin lead to hyperuricemia and deafness, respectively (Figs. S2B and S5; Table S2 and references cited therein).
In addition to the EHP, the linker between ZP-N and ZP-C domains is thought to contain a sequence, referred to as the internal hydrophobic patch (IHP), that also regulates ZP protein polymerization (20, 21). We have redefined the ZP-N/ZP-C linker region boundaries based on the betaglycan ZP-C structure and revealed that the interdomain linker from polymerizing ZP proteins is generally long (from 24–28 residues, as shown in Fig. S2B, to 65 residues in Drosophila Piopio). However, there is disagreement over the IHP location: The position proposed for the human uromodulin IHP (430-DMKVSLK-436) (21) is N-terminal to the one proposed for the mouse ZP3 IHP (165-SEEKLAFSLR-174) (20). Surprisingly, the proposed mZP3 IHP maps onto the ZP-C A strand instead of the linker region; mZP3 F171, proposed as a key IHP residue (20), corresponds to the buried betaglycan F594 residue on the ZP-C A strand. Mutations in the uromodulin IHP region (D430L and L435S), which do map to the linker, result in premature polymerization and the appearance of intracellular filaments (21), suggesting that the linker region is inhibitory to polymerization.
Strand-Displacement Model for ZP-mediated Protein Polymerization.
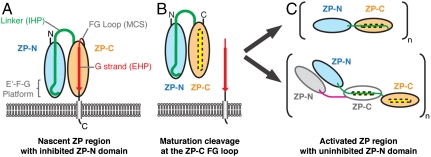
A modified model for the maturation and polymerization of ZP proteins emerges from the structural determination of the ZP-C domain (Fig. 4). It has been hypothesized that the IHP and EHP regions interact, maintaining ZP proteins in an inactive state (1, 20). The ZP-C and ZP3 (22) structures show that the EHP is, unexpectedly, an integral part of the immunoglobulin domain fold and that cleavage of ZP proteins at the FG loop releases a partial immunoglobulin domain with an exposed hydrophobic groove and core (referred to as ZP-CΔEHP, Fig. 4B). The incomplete ZP-C domain (ZP-CΔEHP) could promote protein polymerization through two potential mechanisms. In the first mechanism, the exposed groove in ZP-CΔEHP could bind the IHP sequence present in the interdomain linker, leading to reorganization of the ZP region and exposure of polymerizing elements on the ZP-N domain, like the E′-F-G platform. In this model, reorganization within the ZP region, promoted by loss of the ZP-C G strand, would provide polymerization-competent ZP-N domains (7) by releasing them from an inhibitory interaction with the IHP (Fig. 4C, Upper). Alternatively, the ZP-CΔEHP domain could bind to IHP linker residues from another ZP-CΔEHP protein, leading to direct polymerization through IHP:ZP-CΔEHP interactions (Fig. 4C, Lower). These two polymerization mechanisms are not mutually exclusive and could provide a means to driving fibril polymerization and cross-linking mediated by both ZP-N∶ZP-N and IHP∶ZP-CΔEHP interactions, through both homotypic and heterotypic interactions. In either event, the loss of the ZP-C G strand can be seen as a catalyzing event for polymerization that is revealed by the betaglycan ZP-C structure.
Fig. 4.
Model of ZP-mediated polymerization. (A) The nascent ZP protein, with ZP-N (blue) and ZP-C (orange) domains, is kept inactive by an inhibitory linker that contains the IHP sequence (green). Nascent ZP proteins are tethered to the membrane either by a transmembrane domain (gray) or by a glycosylphosphatidylinositol anchor (not shown). In polymerizing ZP proteins, the ZP-C FG loop has a maturation cleavage site (MCS), and the ZP-C G strand constitutes the EHP region (red). (B) Proteolytic cleavage at the ZP-C FG loop releases the ZP-C G strand and uncovers a hydrophobic groove (yellow) on the resulting ZP-CΔEHP domain. (C) Internal reorganization within the ZP region enables ZP-N∶ZP-N interactions (Upper) or allows the linker to bind to another activated ZP protein via IHP∶ZP-CΔEHP interactions (Lower).
This model would explain the coupling of proteolytic processing and polymerization in ZP proteins that is critical to the formation of the zona pellucida and other ZP-mediated structures. Structural features within the IHP, the extended ZP-C loops, and other motifs outside the ZP region may all contribute to the specificity of ZP-mediated polymerization. Interestingly, the immune system provides a precedent for this proposed ZP polymerization model. The pre-B cell receptor protein VpreB lacks the G strand of the immoglobulin domain fold, which is donated by the λ5 protein upon dimerization and folding of the two proteins (36). For ZP proteins, loss of the G strand upon proteolysis, followed by binding of the IHP segment, could provide a strong thermodynamic driving force to promote a lower energy, polymerized state. During review of this work, the crystal structure of the chicken ZP3 ZP region was reported (22), and it is consistent with key elements of the proposed strand-displacement polymerization model (Fig. 4), including the maturation cleavage site at the ZP-C FG loop, the displacement of the ZP-C G strand, and the resulting partial ZP-CΔEHP domain. These observations provide a framework for future research into other ZP proteins with diverse and important physiological roles in reproductive, auditory, renal, and vascular biology.
Materials and Methods
Production and Purification of Betaglycan.
Residues 591–763 (ZP-C domain), 456–733 (consensus ZP region), 448–763 (full ZP region), and 24–781 (full-length ectodomain) from rat betaglycan were cloned into pBACgus-3 (Novagen) containing N-terminal 6His- and S-tags, expressed in insect cells, and purified by Ni-NTA metal affinity (Qiagen) and Superdex 75/200 (GE Healthcare) size exclusion chromatography. The identities of the purified betaglycan proteins were verified by both N-terminal sequencing and tandem mass spectrometry analyses at the Stanford protein and nucleic acid facility.
Crystallization of the Betaglycan ZP-C Domain.
Purified betaglycan ZP-C protein, treated with recombinant enterokinase to remove the 6His- and S-tags, was concentrated to 10 mg/mL in 20 mM HEPES, 150 mM NaCl, pH 7.4 and crystallized by the sitting drop vapor diffusion method in 16–18% PEG-6000, 0.1 M citrate pH 5.0, 0.9–1.0 M LiCl2. Crystals were harvested in 20% PEG-6000, 0.1 M citrate pH 5.0, 1.0 M LiCl2. For freezing, the crystals were soaked in the harvest buffer containing 20% glycerol for 0.5–2 min, and then flash frozen in liquid nitrogen. For heavy atom derivatives, the crystal is harvested and soaked in cryoprotective buffer containing 20 mM K2PtCl4 or 20 mM K2OsO4 for 12 h or 1.0 M NaBr for 30 s prior to flash-freezing with liquid nitrogen. Data collection and processing information are described in detail in Table S1.
Data Collection and Structure Determination.
All data were processed using the HKL2000 suite (37). Phases were calculated from K2PtCl4, K2OsO4, and NaBr derivatives by multiple isomorphous replacement with anomalous scattering using the program Sharp (38). Density-modified maps, derived from DM in the CCP4 suite (39), were calculated with two ZP-C molecules per asymmetric unit. Initial model building was done using ARP/warp (40). Subsequently, iterative rounds of coordinate and B-factor refinement were done with Refmac (41) or Phenix (42), interspersed with manual model building in Coot (43). The final model was refined to 2.0 Å with Rwork and Rfree values of 18.6 and 24.5%, respectively. The details of model building and model refinement are provided in SI Materials and Methods and Table S1.
Supplementary Material
Acknowledgments.
The authors thank members of the Jardetzky and Woodruff laboratories for helpful discussions. The authors thank Pete Dunten at the Stanford Synchrotron Radiation Laboratory as well as James Holton and George Meigs at the Advanced Light Source for assistance with X-ray data collection. S.J.L. was supported in part by a predoctoral fellowship from the Canadian Institutes of Health Research and a Malkin Scholar Award from Northwestern University. This work was supported by the National Institutes of Health Grants U54HD041857 and R01HD37096 to T.S.J. and T.K.W.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3QW9).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010689108/-/DCSupplemental.
References
- 1.Jovine L, Darie CC, Litscher ES, Wassarman PM. Zona pellucida domain proteins. Annu Rev Biochem. 2005;74:83–114. doi: 10.1146/annurev.biochem.74.082803.133039. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-beta type III receptor. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 3.Llorca O, Trujillo A, Blanco FJ, Bernabeu C. Structural model of human endoglin, a transmembrane receptor responsible for hereditary hemorrhagic telangiectasia. J Mol Biol. 2007;365:694–705. doi: 10.1016/j.jmb.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Yonezawa N, Nakano M. Identification of the carboxyl termini of porcine zona pellucida glycoproteins ZPB and ZPC. Biochem Biophys Res Commun. 2003;307:877–882. doi: 10.1016/s0006-291x(03)01297-x. [DOI] [PubMed] [Google Scholar]
- 5.Monné M, Han L, Schwend T, Burendahl S, Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- 6.Jovine L, Qi H, Williams Z, Litscher E, Wassarman PM. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat Cell Biol. 2002;4:457–461. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- 7.Jovine L, Janssen WG, Litscher ES, Wassarman PM. The PLAC1-homology region of the ZP domain is sufficient for protein polymerisation. BMC Biochem. 2006;7:11. doi: 10.1186/1471-2091-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaźwińska A, Ribeiro C, Affolter M. Epithelial tube morphogenesis during Drosophila tracheal development requires Piopio, a luminal ZP protein. Nat Cell Biol. 2003;5:895–901. doi: 10.1038/ncb1049. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Muchmore A. Tamm–Horsfall protein—uromodulin (1950–1990) Kidney Int. 1990;37:1395–1401. doi: 10.1038/ki.1990.128. [DOI] [PubMed] [Google Scholar]
- 10.Legan PK, Rau A, Keen JN, Richardson GP. The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J Biol Chem. 1997;272:8791–8801. doi: 10.1074/jbc.272.13.8791. [DOI] [PubMed] [Google Scholar]
- 11.Legan PK, et al. A deafness mutation isolates a second role for the tectorial membrane in hearing. Nat Neurosci. 2005;8:1035–1042. doi: 10.1038/nn1496. [DOI] [PubMed] [Google Scholar]
- 12.Verhoeven K, et al. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet. 1998;19:60–62. doi: 10.1038/ng0598-60. [DOI] [PubMed] [Google Scholar]
- 13.López-Casillas F, Payne HM, Andres JL, Massagué J. Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: Mapping of ligand binding and GAG attachment sites. J Cell Biol. 1994;124:557–568. doi: 10.1083/jcb.124.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepin MC, Beauchemin M, Plamondon J, O'Connor-McCourt MD. Mapping of the ligand binding domain of the transforming growth factor beta receptor type III by deletion mutagenesis. Proc Natl Acad Sci USA. 1994;91:6997–7001. doi: 10.1073/pnas.91.15.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esparza-López J, et al. Ligand binding and functional properties of betaglycan, a co-receptor of the transforming growth factor-beta superfamily. Specialized binding regions for transforming growth factor-beta and inhibin A. J Biol Chem. 2001;276:14588–14596. doi: 10.1074/jbc.M008866200. [DOI] [PubMed] [Google Scholar]
- 16.Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- 17.Wiater E, Harrison CA, Lewis KA, Gray PC, Vale WW. Identification of distinct inhibin and transforming growth factor beta-binding sites on betaglycan: Functional separation of betaglycan co-receptor actions. J Biol Chem. 2006;281:17011–17022. doi: 10.1074/jbc.M601459200. [DOI] [PubMed] [Google Scholar]
- 18.Wiater E, et al. Endogenous betaglycan is essential for high-potency inhibin antagonism in gonadotropes. Mol Endocrinol. 2009;23:1033–1042. doi: 10.1210/me.2009-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao M, et al. Mutation of a conserved hydrophobic patch prevents incorporation of ZP3 into the zona pellucida surrounding mouse eggs. Mol Cell Biol. 2003;23:8982–8991. doi: 10.1128/MCB.23.24.8982-8991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jovine L, Qi H, Williams Z, Litscher ES, Wassarman PM. A duplicated motif controls assembly of zona pellucida domain proteins. Proc Natl Acad Sci USA. 2004;101:5922–5927. doi: 10.1073/pnas.0401600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaeffer C, Santambrogio S, Perucca S, Casari G, Rampoldi L. Analysis of uromodulin polymerization provides new insights into the mechanisms regulating ZP domain-mediated protein assembly. Mol Biol Cell. 2009;20:589–599. doi: 10.1091/mbc.E08-08-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han L, et al. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 23.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boja ES, Hoodbhoy T, Fales HM, Dean J. Structural characterization of native mouse zona pellucida proteins using mass spectrometry. J Biol Chem. 2003;278:34189–34202. doi: 10.1074/jbc.M304026200. [DOI] [PubMed] [Google Scholar]
- 25.Darie CC, Biniossek ML, Jovine L, Litscher ES, Wassarman PM. Structural characterization of fish egg vitelline envelope proteins by mass spectrometry. Biochemistry. 2004;43:7459–7478. doi: 10.1021/bi0495937. [DOI] [PubMed] [Google Scholar]
- 26.Kanai S, et al. Disulfide linkage patterns of pig zona pellucida glycoproteins ZP3 and ZP4. Mol Reprod Dev. 2008;75:847–856. doi: 10.1002/mrd.20836. [DOI] [PubMed] [Google Scholar]
- 27.Halaby DM, Poupon A, Mornon J. The immunoglobulin fold family: Sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563–571. doi: 10.1093/protein/12.7.563. [DOI] [PubMed] [Google Scholar]
- 28.Vilchis-Landeros MM, Montiel JL, Mendoza V, Mendoza-Hernández G, López-Casillas F. Recombinant soluble betaglycan is a potent and isoform-selective transforming growth factor-beta neutralizing agent. Biochem J. 2001;355:215–222. doi: 10.1042/0264-6021:3550215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santambrogio S, et al. Urinary uromodulin carries an intact ZP domain generated by a conserved C-terminal proteolytic cleavage. Biochem Biophys Res Commun. 2008;370:410–413. doi: 10.1016/j.bbrc.2008.03.099. [DOI] [PubMed] [Google Scholar]
- 30.Andres JL, Stanley K, Cheifetz S, Massagué J. Membrane-anchored and soluble forms of betaglycan, a polymorphic proteoglycan that binds transforming growth factor-beta. J Cell Biol. 1989;109:3137–3145. doi: 10.1083/jcb.109.6.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesha S, et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med. 2006;12:642–649. doi: 10.1038/nm1429. [DOI] [PubMed] [Google Scholar]
- 32.Raab U, et al. Expression of normal and truncated forms of human endoglin. Biochem J. 1999;339:579–588. [PMC free article] [PubMed] [Google Scholar]
- 33.Florman HM, Wassarman PM. O-linked oligosaccharides of mouse egg ZP3 account for its sperm receptor activity. Cell. 1985;41:313–324. doi: 10.1016/0092-8674(85)90084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Litscher ES, Wassarman PM. Inactivation of the mouse sperm receptor, mZP3, by site-directed mutagenesis of individual serine residues located at the combining site for sperm. Proc Natl Acad Sci USA. 1998;95:6193–6197. doi: 10.1073/pnas.95.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg's zona pellucida protein. Science. 2010;329:216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankovich AJ, et al. Structural insight into pre-B cell receptor function. Science. 2007;316:291–294. doi: 10.1126/science.1139412. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 38.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 39.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.