Abstract
It is widely believed that functional mammalian microRNA (miRNA) recognition sequences are located preferentially in the 3' untranslated region (3'UTR) of target mRNAs. Nonetheless, putative miRNA target sites within coding regions have been found at lower frequency in genome-wide studies, and several have been genetically validated. To account for these findings, it has been proposed that translation may inhibit miRNA access to target sites. Here we identify a naturally occurring viral miRNA target that, owing to the compact nature of the viral transcriptome, is situated naturally in the coding region of one transcript and in the 3′UTR of an overlapping mRNA. Examination of the expression of these mRNAs reveals that the cognate miRNA can inhibit expression in both contexts, but inhibition is more potent when the target site is in the UTR. Similarly, forced translation of the target site in the UTR diminished, but did not abolish, its down-regulation by the miRNA. These data affirm that miRNAs can exert regulatory effects on targets within coding regions; however, the dampening of these effects by translation likely accounts for the observed selection for target sites in the 3′UTRs.
Keywords: ORF targeting, target prediction, Kaposi's sarcoma-associated herpesvirus, KSHV ORF56/ORF57(MTA), splicing
MicroRNAs (miRNAs) are small [18-22 nucleotides (nt)] noncoding RNAs that are strongly implicated in the posttranscriptional regulation of mammalian gene expression (1–4). This regulation is mediated by basepairing interactions between the miRNA [following its incorporation into an Argonaute-containing RNA-induced silencing complex (RISC)] and its target mRNAs. In general, only limited basepairing is observed between miRNAs and their targets, with the major complementarity involving a 7–8 nt region at the 5′ end of the miRNA, termed the seed sequence (5). Other factors are likely also involved in target selection (5–9) though their exact nature is still under investigation. MiRNA target recognition mediates down-regulation of gene expression, due to translational repression, enhancement of RNA degradation, or both (10–17). Experimental studies indicate that any given miRNA may regulate 100–200 transcripts (10, 18). Because human cells are thought to encode hundreds of miRNA genes, the potential global effects of such regulation are substantial, even though the magnitude of the down-regulation of any given target is often modest.
Genome-wide analyses of miRNA target selection indicate that most miRNA recognition sites are located in the 3′UTR of the targeted message. One of the most powerful approaches has involved microarray profiling of mRNAs whose levels are specifically down-regulated by miRNA expression, with analysis of the distribution of seed-homologous regions in the targeted mRNAs (10). These studies show substantial enrichment for seed homologies in the 3′UTR of putative mRNA targets. Subsequent proteomic studies that have directly examined polypeptides down-regulated by miRNA expression (11, 16) have reached similar conclusions. Supporting these conclusions are studies that have mapped many miRNA sites to 3′UTRs (19). Based on these findings, it has been proposed that translation may inhibit access of RISC to miRNA target sites. In fact, using reporter constructs, Gu et al. recently reported that translation of a miRNA target virtually nullified its ability to be down-regulated by that miRNA (20).
However, computational and experimental genome-wide analyses also suggest the existence of a minority class of potential targets within open reading frames (ORFs) (7, 10, 11, 14, 16, 21–28), and a handful of miRNA recognition sites have been mapped to the coding regions of transcripts (29–34), leading to the suggestion that sites within coding regions can be accessed by miRNAs. However, because the accessibility of a given seed-matched sequence to the corresponding miRNA may be influenced by the target site context and local secondary structures near the target sites (5, 7–9), genome-wide studies cannot directly prove that the bias in 3′UTR targeting is attributable to translation.
In the course of studies of miRNA regulation of virus replication, we encountered an interesting example of a natural miRNA recognition site that resides in the coding region of one viral RNA and the 3′ UTR of another—a consequence of the fact that in compact viral genomes many genes are expressed from nested sets of transcripts bearing different 5′ ends but a common 3′ polyA signal. In many such sets, the 3′ UTR of the larger upstream transcript contains an ORF which is preferentially translated from the smaller downstream mRNA. This arrangement affords a unique opportunity to examine the question of the role of translation on miRNA function in a natural context. Here we present a detailed analysis of this case, which sheds light on the role of translation in affecting the accessibility of natural miRNA recognition sites to the RISC.
Results
The Experimental System.
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a large DNA virus that is etiologically linked to Kaposi’s sarcoma, the leading neoplasm of untreated AIDS patients (35). Like all herpesviruses, KSHV can induce either a latent or a lytic infection. During latency, viral gene expression is drastically restricted, with only a handful of viral mRNAs and proteins being produced; this is the default pathway following de novo infection of most cells. However, under the appropriate circumstances, latently infected cells can be induced to undergo lytic replication, in which all viral genes are expressed in a temporally regulated cascade that leads to production of progeny virions.
The latency program of KSHV encodes 12 pre-miRNAs, which can engender ca. 20 mature viral miRNAs (36–41). Because these viral miRNAs are expressed in latency, when few other viral transcripts are expressed, interest has largely been focused on their potential cellular mRNA targets. However, the long half-lives of miRNAs in RISC indicates that all of these miRNAs continue to be present during lytic replication; moreover, several of their pri-miRNAs are further transcriptionally up-regulated during the lytic cycle, leading to enhanced accumulation of these miRNAs as replication proceeds. Accordingly, we undertook a series of experiments to determine if viral miRNAs might have additional targets in the lytic viral transcriptome. The first of these studies revealed that the lytic switch protein RTA, the master regulator of the lytic cycle, is specifically targeted by one viral miRNA, miR-K9-5p, which down-regulates RTA expression and stabilizes latency (42). In the present experiments, we asked if the known KSHV proteins governing lytic viral DNA replication could also be subject to regulation by viral miRNAs. Table S1 lists the seven viral genes selected for this analysis, their known roles in viral replication (43), and the nature of potential seed matches with the indicated virus-encoded miRNAs, as judged by sequence inspection of their 3′ UTRs. As can be seen, numerous seed homologies were predicted in 4 of the 7 UTRs (Table S1).
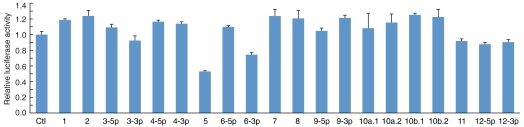
To determine which of these sequences actually contained functional miRNA recognition sites (Fig. S1A), each UTR was cloned downstream of a Renilla luciferase (LUC) reporter in the vector depicted in Fig. S1B (The vector bears a linked constitutive Firefly LUC reporter to normalize for transfection efficiency). For each chimera, the construct was cotransfected into HEK293 cells with each of the 20 KSHV miRNAs (presented as synthetic RNA duplexes); 42 h later, LUC activity was measured, corrected for transfection efficiency, and normalized relative to the levels expressed in the presence of a control miRNA. For six of the UTR chimeras, we observed no down-regulation by any of the 20 viral miRNAs—this included the viral DNA polymerase (ORF9) UTR, for which 14 potential seed-matched sequences had been proposed bioinformatically. However, one UTR, that of the DNA primase gene (ORF56) did mediate down-regulation—in this case, by two viral miRNAs, miR-K5 and miR-K6-3p (Fig. 1). The two miRNAs were predicted to target different sites within the ORF56 UTR (see below); when both miRNAs were cotransfected with the ORF56 UTR reporter, additive repression of luciferase activity was observed (Fig. S2). As expected, repression by miR-K5 was ablated by mutations introduced into its seed sequence (Fig. S2).
Fig. 1.
KSHV miR-K5 and miR-K6-3p down-regulate the expression of Renilla LUC reporter fused to genomic sequence of ORF56 3′UTR. HEK293 cells were cotransfected with each reporter construct (ORF56 3′UTR and VEC) along with a negative control miRNA (Ctl) or each of the indicated 20 KSHV miRNAs. Transfection conditions, dual luciferase assays, and calculation of relative luciferase activities were carried out as described in Methods. Mean values and error bars (standard deviation, SD) were derived from triplicate independent transfections.
Mapping the Recognition Sequences for miR-K5 and miR-K6-3p.
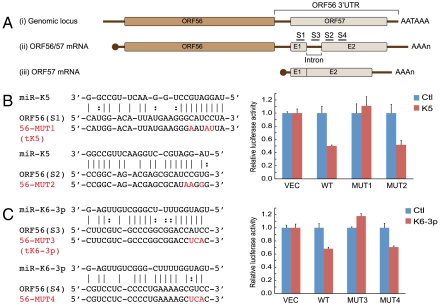
Fig. 2A [line (i)] depicts the genomic organization of the region surrounding ORF56. ORF56 (primase) is immediately 5′ to the coding region for ORF57, which encodes a posttranscriptional regulator known as MTA (for mRNA transcript accumulation). The gene for ORF57/MTA includes a small intron, and is followed by a short 3′UTR bearing a canonical polyadenylation signal. Fig. 2A [lines (ii) and (iii)] shows the structure of the known transcripts from this region, which have been mapped to high resolution (44, 45). As ORF56 has no adjacent polyA signal, its mRNA includes all of ORF57 in its 3′UTR. For unknown reasons, the intron in this context is very inefficiently spliced; therefore, in ORF56 mRNA, intronic sequences persist in the 3′UTR of over 90% of messages (Fig. S3B). Because there is no IRES (internal ribosome entry site) in this transcript, ORF57 sequences are not translated in this context. Instead, a separate promoter embedded within ORF56 directs the production of a monocistronic RNA which is the principal source of ORF57 protein [Fig. 2A, line (iii)]. Interestingly, in this context the intron is very efficiently spliced, with > 90% of the monocistronic ORF57 mRNA being spliced (Fig. S3B), while the unspliced RNA disrupts the MTA open reading frame.
Fig. 2.
miR-K5 and miR-K6-3p target ORF56 3′UTR. (A) ORF56/57 genomic locus and mRNA transcripts derived from this locus. As depicted in line (i), ORF57 coding sequence resides in ORF56 3′UTR. Transcription of this locus gives rise to an inefficiently spliced ORF56/57 transcript (line ii) and an efficiently spliced ORF57 transcript (line iii). E1 and E2 (Exon 1 and 2) represent ORF57 coding sequence. AATAAA denotes the polyadenylation signal; AAAn denotes the polyadenylation tail of mRNAs. S1–S4 indicates the bioinformatically predicted miR-K5 and miR-K6-3p binding sites. (B) (C) Identification of miR-K5 (B) and miR-K6-3p (C) target sites in ORF56 3′UTR. The predicted miR-K5 and miR-K6-3p binding sites in ORF56 3′UTR (S1, 2, 3, and 4) and target site mutants (56-MUT1, 2, 3, and 4) are shown on the left. Solid lines in the alignments denote perfect Watson-Crick basepairing; dotted lines in the alignments denote G:U wobble basepairing. Mutated nucleotides in the target sites are shown in red. On the right, HEK293 cells were cotransfected with VEC, or each of ORF56 3′UTR reporter constructs (WT and 56-MUT1-4) along with the indicated miRNAs. Mean values and error bars (SD) were derived from quadruplicate independent transfections.
Examination of the sequence of the ORF56/primase 3′UTR with several miRNA target prediction programs suggested two likely targets for miR-K5—one in Exon 1 of the ORF57 gene, and one in Exon 2 of the gene. The sequences of these target sites, and their predicted basepairing with miR-K5, are shown in Fig. 2B. Site 1 (in Exon 1) has a perfect 8mer seed match. Site 2 (in Exon 2) has extensive seed homology, but requires a bulged nt in the mRNA. For each site, we engineered mutations in the mRNA to disrupt seed pairing (the exact mutations are depicted in Fig. 2B). These mutations were constructed in the UTR of the Renilla LUC-ORF56 3′UTR chimera; this plasmid, or its wild type (WT) version, was then cotransfected into 293 cells with a control miRNA or miR-K5, and luciferase assayed as before. As shown in the bar graph of Fig. 2B, mutation of Site 1 (in Exon 1) ablated down-regulation of Renilla LUC by miR-K5, while mutations in Site 2 (in Exon 2) had no effect. The phenotype of the mutation in Exon 1 not only indicates that this site is recognized by miR-K5, but also suggests it is the only functional recognition site for this miRNA in the ORF56 3′UTR.
We took a similar approach to map the target site(s) for miR-K6-3p in the ORF56 3′UTR. The locations of the top two target sites predicted computationally are shown in Fig. 2A. One site (termed Site 3) resides within the intron of the ORF57 gene, while the other (Site 4) maps to Exon 2 of ORF57. Fig. 2C shows that only the mutation in Site 3 abolishes miR-K6-3p action, indicating the target site for this miRNA is in the intron of ORF57. Because this intron is very inefficiently spliced in ORF56 mRNA, the site is available for miR-K6-3p regulation in this context. This site contains a perfect 6mer seed match and five contiguous Watson-Crick pairs to bases 11–15 of miR-K6-3p (Fig. 2C) [classified as a 3′-supplementary site (5, 7)], which may explain enhanced miRNA targeting efficacy in this case.
MicroRNA Regulation of ORF56 Protein.
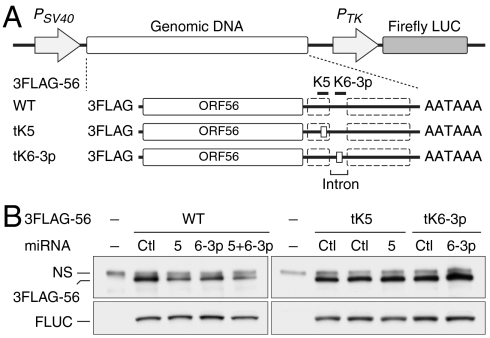
Because the above results were obtained with a reporter construct, we sought to validate that it truly recapitulates the regulation of the genomic KSHV locus. Accordingly, genomic KSHV DNA spanning the WT ORF56 and 57 genes was cloned into a plasmid under SV40 promoter control, and ORF56 was FLAG-tagged to allow detection of its protein product. (A downstream Firefly luciferase gene under separate promoter control allowed normalization for transfection efficiency, Fig. 3A, WT). Into this vector, we also cloned the aforementioned mutations in the target sites for miR-K5 and miR-K6-3p (Fig. 4A, mutants tK5 and tK6-3p). Each plasmid was cotransfected into 293 cells along with miR-K5, K6-3p or both, and the level of ORF56 expression was determined by immunoprecipitation followed by immunoblotting with anti-FLAG antibody. Fig. 3B shows that, just as in the LUC reporter, (i) miR-K5 down-regulates ORF56/primase expression more potently than miR-K6-3p; (ii) the two together display more down-regulation than either alone; and (iii) mutation of each target site abolishes down-regulation by the corresponding miRNA.
Fig. 3.
miR-K5 and miR-K6-3p repress ORF56 protein expression. (A) Genomic DNA expression constructs consist of a 3FLAG tag and the region spanning ORF56 coding region and its entire 3′UTR (solid line between ORF56 and polyadenylation sequence, AATAAA). Defective miRNA binding sites are illustrated as small boxes in the 3′UTR of 3FLAG-56 tK5 and tK6-3p (tK5 and tK6-3p are described in Fig. 2 B and C). ORF57 coding sequence (dashed boxes) and the intron are indicated. (B) Immunoblot analyses of 3FLAG-56 (anti-FLAG immunoprecipitates) and transfection control Firefly luciferase (FLUC) in HEK293 cells either mock transfected (-), or cotransfected with the indicated plasmids and miRNAs for 48 h. Bands labeled NS represent proteins that are nonspecifically precipitated by anti-FLAG antibody. Quantitation of 3FLAG-56/FLUC is described and plotted in Fig. S4A.
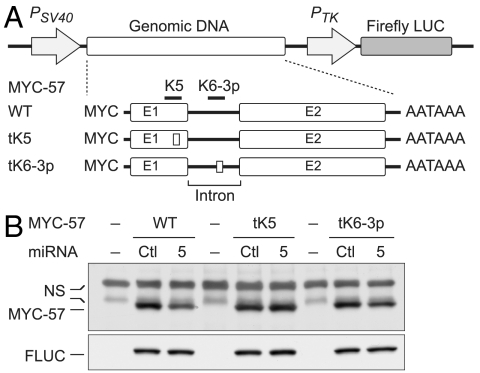
Fig. 4.
miR-K5 targets protein coding region of monocistronic ORF57 mRNA. (A) Genomic DNA expression constructs consist of a MYC tag and the region spanning ORF57 exons (E1 and E2), intron, and its entire 3′UTR (solid line between E2 and AATAAA). Defective miRNA binding sites are illustrated as small boxes within Exon 1 of MYC-57 tK5 (amino acid sequence encoded by tK5 is unchanged) and the intron of MYC-57 tK6-3p (tK5 and tK6-3p are described in Figs. 2 B and C). (B) Immunoblot analyses of MYC-57 and transfection control FLUC in HEK293 cells either mock transfected (-), or cotransfected with the indicated plasmids and miRNAs for 48 h. Bands labeled NS represent proteins that are nonspecifically cross-reactive with anti-MYC antibody. Quantitation of MYC-57/FLUC derived from three independent measurements is described and plotted in Fig. S4B.
MicroRNA Regulation of Monocistronic ORF57 mRNA.
The unique arrangement of ORF57 sequences in two different contexts—one in which they are translated and one in which they are not—allowed us an opportunity to learn about the impact of translation on miRNA accessibility in a natural context, without construction of engineered miRNA target sites into mutationally extended coding regions of experimental reporter genes. Accordingly, we constructed an expression vector expressing genomic ORF57 DNA as a monocistronic transcript. ORF57 was tagged with a MYC epitope, and the WT plasmid (MYC-57) shown to express ORF57 protein efficiently after transfection into 293 cells (Fig. 4). (We also verified that this transcript was efficiently spliced, just as in KSHV-infected cells; Fig. S3C). When this construct was cotransfected with miR-K5, clear down-regulation ORF57 expression was observed (Fig. 4B). Quantitation of this effect showed that when the target site was within a translated ORF57 gene, expression of the targeted gene product was down-regulated 36% by miR-K5 (Fig. S4B); by contrast, when the target site was in the 3′UTR, expression of the targeted gene product was diminished by 50% (Fig. S4A). This down-regulation was nonetheless completely abrogated by mutational ablation of the miR-K5 binding site in Exon 1 (while it was unaffected by mutation of the miR-K6-3p site in the same gene; Fig. 4B). (The mutations in the miR-K5 site were designed to alter miRNA recognition without changing the coding sequence of ORF 57). This result strongly indicates that miR-K5 action was clearly not abolished by translation.
Examination of miR-K5 Regulation by Translation in Experimental Contexts.
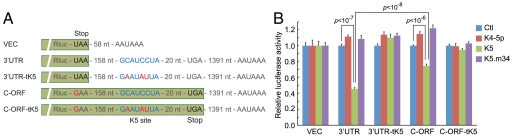
To gain further insight into the role of translation in influencing miRNA action, we generated several additional constructs in the LUC reporter bearing the 3′UTR of ORF56; these are diagrammed in Fig. 5A. In mutant C-ORF, the UAA stop codon of Renilla LUC was mutated to GAA; this allowed extension of translation to 20 nt 3′ to the miR-K5 binding site. Plasmid C-ORF-tK5 is an isogenic derivative of C-ORF in which the miR-K5 target site is disrupted by mutation. Each of these plasmids was cotransfected into 293 cells with either a control miRNA, an irrelevant KSHV miRNA (miR-K4-5p), miR-K5, or a mutant of miR-K5(m34) bearing two substitutions in the seed sequence. Forty-two hours later, LUC was assayed; for each construct, the results were normalized to the level produced by the same plasmid in the presence of the control miRNA. (This corrects for the differing specific activities of luciferases lacking or bearing the C-terminal extension). Fig. 5B shows the results of this experiment. As we observed in the authentic genomic construct (Figs. 3, 4), when the miR-K5 site was translated (C-ORF), clear Renilla LUC repression was observed, but it was reproducibly less in extent than when the site is not translated (3′UTR). In both situations, the repression is abolished by either seed sequence mutation in miR-K5 or mutation of the recognition sequence in the mRNA. As expected, a control, irrelevant KSHV miRNA (miR-K4-5p) did not affect expression from any of the tested constructs.
Fig. 5.
Translation attenuates, but does not block, miR-K5-mediated down-regulation. (A) Experimental constructs of the miR-K5 site embedded within the extended Renilla LUC (Rluc) in the isogenic dual luciferase reporter. VEC contains no ORF56 3′UTR. 3′UTR and C-ORF have an identical sequence, except at three nucleotides that disrupt the UAA stop codon of Rluc and two additional stop codons (not shown in the figure) preceding the miR-K5 site, thereby allowing extension of the ORF to 20 nucleotides 3′ to the miR-K5 site. 3′UTR-tK5 and C-ORF-tK5 are isogenic derivatives in which the miR-K5 site is mutationally inactived. “Stop” denotes stop codon. The miR-K5 site is in blue and mutated nucleotides are in red. (B) HEK293 cells were cotransfected with the indicated reporter constructs and miRNAs. Mean values and error bars (SD) were derived from quadruplicate independent transfections. Statistical significance is indicated (p value, two-tailed t test).
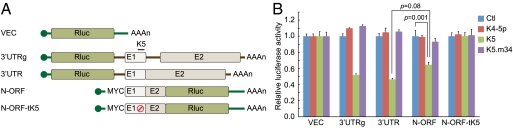
Because the translated miRNA target site in the above construct resides at the C terminus of the Renilla LUC protein, while in authentic ORF57 it is in the N terminus of the corresponding ORF, we designed an additional set of vectors to explore whether the position of the translated miRNA target site in the coding region affected its action. As depicted in Fig. 6A, construct N-ORF was generated by in-frame fusing Exon 1 and part of Exon 2 of ORF57 with Renilla LUC at its N terminus and an isogenic derivative in which the miR-K5 binding site was mutated (designated tK5) was also constructed. As before, LUC activities were analyzed from 293 cells cotransfected with either a control miRNA, an irrelevant KSHV miRNA (miR-K4-5p), miR-K5, or the miR-K5 seed mutant m34. Fig. 6B shows the results of this experiment. Again, N-ORF chimeric protein bearing translated miR-K5 recognition sequences displayed down-regulation by miR-K5, and this down-regulation was abolished by mutations in either the miRNA or its binding site. Furthermore, the degree of down-regulation was less than that observed when the same sites were present in the 3′UTR of the Renilla LUC gene. [Parenthetically, we note that miR-K5-mediated repression (an 8mer seed match) in a translated region is similar to or slightly greater than miR-K6-3p-mediated repression (a 6mer seed match with 3′-supplementary site in a nontranslated region; Figs. 1, 3, 4, 5, 6, and Fig. S4)]. We conclude that translation of these miRNA binding sites modestly reduces but does not abolish their ability to function in miRNA-mediated repression.
Fig. 6.
Translation attenuates miR-K5-mediated down-regulation. (A) Experimental constructs for dual luciferase assays. 3′UTRg (with an intron, used in Figs. 1 and 2) and 3′UTR (without an intron, used in Fig. 5) are described previously. In N-ORF, Exon 1 and part of Exon 2 were in-frame fused to Rluc that allows translating an ORF57-Rluc chimeric fusion protein. N-ORF-tK5 is an isogenic derivative of N-ORF in which the miR-K5 site is mutationally inactived, as illustrated by a red symbol at the mutated target site. (B) HEK293 cells were cotransfected with the indicated reporter constructs and miRNAs. Mean values and error bars (SD) were derived from quadruplicate independent transfections. Statistical significance is indicated (p value, two-tailed t test).
Discussion
In this work we have capitalized on the fact that the parsimonious genomic organization of some DNA viruses results in many cases in which the same sequence is represented in multiple RNA transcripts. In herpesviruses, which typically have more ORFs than polyA sites, this results in situations where entire coding regions can be found in the 3′UTR of other ORFs. Such coding regions are generally not translated from this position, and usually require specification of another 5′ end just upstream of its own AUG codon to allow for their expression. We identified a unique case in which one such coding region (here, for ORF57/MTA) happens to harbor a recognition site for a virus-encoded miRNA. In contrast to some earlier experiments, we find that this site can mediate repression by its cognate miRNA irrespective of whether it is being translated, though translation does appear to interfere with maximal miRNA repression.
Other data also point to the potential functionality of miRNA recognition sites within coding regions. As noted in the Introduction, a few such sites have been identified in cellular mRNAs, though in none of those cases was the efficiency of miRNA action compared in the presence or absence of translation (29–34). More recently, extensive genome-wide analyses of potential miRNA recognition sites were conducted using UV-cross linking and Argonaute immunoprecipitation coupled to deep cDNA sequencing of the precipitated products. Although this analysis does not directly determine functionality of the sites, it was remarkable that many RISC-associated sites recovered in this fashion were found to be in coding domains (21, 24, 27). [Similarly, a recent computational approach predicts that ORF targeting may be especially prevalent in Drosophila (32)]. Hafner et al. also found that mRNAs bearing such sites displayed lower levels of down-regulation than those whose sites were localized to the 3′ UTR (24). If this is so, it may explain why studies in which lowering of transcript abundance in the presence of a miRNA was used as a marker of miRNA regulation might have underrecognized mRNAs bearing translated targets. We also note that our results are very consistent with the genome-wide analyses of miRNA target distributions, which, although they show strong enrichment for 3′UTR localization, nonetheless consistently reveal a small enrichment for targets within ORFs (10, 11, 16).
Our results differ somewhat from those of another group who have experimentally examined the effects of translation on miRNA accessibility. Gu et al. (20) employed model reporter constructs in which mutations were engineered into the stop codons of reporter ORFs located upstream of two tandem 21 nt miRNA target sites; this allowed translation to proceed through the miRNA recognition sequence. This study found that such translation nearly completely abrogated miRNA regulation, even though siRNA action at such sequences was either unimpaired or only modestly affected. By contrast, we observed a more modest (though reproducible) decrease in the efficiency with which translated sites can be targeted.
What might account for these differences? We note that the miRNA target site we chose to study exists naturally within a coding region, and therefore may have undergone selection for properties that allow it to function in this locale. This is in contrast to the model constructs of Gu et al., in which sites that have never resided outside a UTR were experimentally repositioned within a coding region. Because we observe that translation reduces miRNA function even for our naturally translated site, it may well be that normally untranslated sites may be especially sensitive to this inhibitory influence. If so, once a sufficiently large lexicon of translated target sites is available, it will be interesting to see if they have sequence features that differentiate them from miRNA targets located in untranslated regions.
Another feature that may explain variation in the degree to which coding sequences may be accessed by the RISC machinery is translational efficiency. If translation indeed impairs access of miRNAs to their recognition sites, then factors that pause translation might enhance such access. Consistent with this, Gu et al. observed that introduction of nine consecutive rare codons into a model ORF enhanced miRNA-mediated inhibition (20). Accordingly, we examined codon usage in ORF57 and C-ORF. These analyses revealed that (i) neither ORF57 nor C-ORF harbors a stretch of consecutive rare codons preceding the miR-K5 target site; and (ii) the distribution of rare codons in that region does not differ significantly from that of randomly selected cellular and viral coding sequences. Nonetheless, it is not excluded that other features of ORF57 mRNA (e.g., secondary or tertiary structures) might influence translatability in a manner that could affect miRNA target accessibility.
In conclusion, we wish to emphasize that the differences observed between our study and that of Gu et al. are quantitative rather than qualitative. Both studies indicate that translation can adversely impact upon miRNA recognition, and help explain why target sites in coding regions are underrepresented relative to those in 3′UTRs. By examining a well controlled situation involving an unrearranged genomic locus in which the identical miRNA target is presented in both a translated and untranslated context, we believe the present study provides a clearer picture of the magnitude of such effects as they may be encountered in other natural genomic loci in vivo.
Methods
Plasmid Constructions.
The annotated KSHV genome sequence was obtained from NCBI Refseq: NC_009333 (Human Herpesvirus 8 type P). KSVH DNA fragments were PCR amplified from the body cavity lymphoma cell line BCBL-1 first-strand cDNA or genomic DNA and cloned into WT or engineered psiCHECK2 (Promega), a dual luciferase reporter vector. First-strand cDNA was synthesized by reverse transcription of total RNA (harvested at 24 h after induction of KSHV lytic replication in BCBL-1) using oligo(dT)20 primer (Invitrogen) and SuperScript III system (Invitrogen). Genomic DNA was isolated from uninduced cells using DNAzol (Molecular Research Center). PCR amplification was performed using Phusion High-Fidelity (HF) DNA polymerase (Finnzymes) or PfuUltra HF DNA polymerase AD (Stratagene). Nucleotide substitutions were introduced by QuikChange Lightning Site-Directed Mutagenesis or Multi Site-Directed Mutagenesis Kit (Stratagene). Isogenic tK5 DNA sequences bear a defective miR-K5 binding site but the encoded amino acid sequence is unchanged. All constructs were verified by DNA sequencing. For complete details of plasmid constructions, see SI Text: Methods.
Dual Luciferase Reporter Assays.
Experiments were carried out in a 24-well format. In brief, HEK293 cells (1.1 × 105) were seeded in antibiotic-free medium 24 h before transfection. Transfections were performed using 1 uL Lipofectamine 2000 (Invitrogen), 150 ng dual luciferase reporter plasmids, and a final concentration of 10 nM synthetic miRNA mimics (called PremiRs, double-stranded RNA molecules obtained from Applied Biosystems; PremiRs are chemically modified to ensure the correct miRNA strand is incorporated into the RNA-induced silencing complex. Mature miRNA sequences are listed in SI Text: Methods). At 42–43 h after transfection, cell lysates were prepared by passive lysis buffer (Promega) and dual luciferase assays (Promega) were performed using the 20/20n Luminometer (Turner Biosystems). Relative luciferase activities were calculated as described below. First, Renilla luciferase (Rluc) activity was normalized to Firefly luciferase (Fluc) transfection control, referred as Rluc/Fluc. For each reporter construct (3′UTR, C-ORF, or N-ORF), the Rluc/Fluc value obtained from each miRNA cotransfection was normalized to the value obtained from empty vector (VEC) cotransfected with the same miRNA. This normalization is to correct nonspecific up-regulation or down-regulation of Rluc expression in the presence of a miRNA. The normalized Rluc/Fluc value was then normalized to the value obtained from the same reporter construct cotransfected with control (Ctl) miRNA, which was set at 1. Mean values, error bars (standard deviation), and Student’s t-Test (two-tailed test) were calculated from three or four independent transfections as indicated.
Whole-Cell Extracts, Immunoprecipitation, and Western Blotting.
To express MYC-57, HEK293 cells in a well of 6-well dishes were transfected with 1.0 ug plasmid and 10 nM miRNA (final concentration) using 5 uL Lipofectamine 2000. For immunoprecipitation of 3FLAG-56, cells in a 60-mm dish were transfected with 1.5 ug plasmid and 10 nM miRNA using 10 uL Lipofectamine 2000. At 48 h after transfection, cells were lysed in NETT [150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl, pH7.5, 0.5% Triton X-100, a protease inhibitor cocktail (Sigma, P8340), 2 mM PMSF, 10 mM β-glycerophosphate, 5 mM NaF, and 2 mM Na3VO4] by three freeze-thaw cycles. Insoluble cell debris was removed by centrifugation and soluble whole-cell extracts were quantified using Bradford protein assay (BioRad). To immunoprecipitate 3FLAG-56, extracts were first adjusted to a final concentration of 1 ug/uL in NETT and precleared with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) at 4 °C for 30 min. The precleared extracts (400 ug) were incubated with anti-FLAG M2 affinity gel (Sigma) at 4 °C for 2 h. The bound gels were washed five times with NETT and the immunoprecipitaes were eluted with SDS-PAGE sample buffer. Western blotting procedure is described in SI Text: Methods. Band intensities of Western blot films were analyzed using ImageJ program (Version 1.43).
Bioinformatic miRNA Target Predictions and Seed Match Analyses.
The miRNA target prediction algorithms miRanda v1.0b (46), RNAhybrid (47), and RNA22 (48) were used to search candidate 3′UTRs for potential miRNA target sites. Microsoft Word software was used to search candidate 3′UTRs for 6 mer, 7mer-A1, 7mer-m8, and 8mer seed matches by finding the perfect reverse complementary sequence of miRNA seed (miRNA bases 2–7 or 2–8) and an adenosine opposite miRNA base 1.
Supplementary Material
Acknowledgments.
We thank members of the Ganem lab for helpful discussions. This work was supported by grants from the National Institutes of Health and Howard Hughes Medical Institute. D.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2010.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102033108/-/DCSupplemental.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 3.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen CB, et al. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson DG, et al. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 16.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 17.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 18.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 19.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 20.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 24.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forman JJ, Coller HA. The code within the code: MicroRNAs target coding regions. Cell Cycle. 2010;9:1533–1541. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen WF, Hu YL, Uttarwar L, Passegue E, Largman C. MicroRNA-126 regulates HOXA9 by binding to the homeobox. Mol Cell Biol. 2008;28:4609–4619. doi: 10.1128/MCB.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 35.Ganem D. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest. 2010;120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai X, et al. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin YT, et al. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16:1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 40.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umbach JL, Cullen BR. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol. 2010;84:695–703. doi: 10.1128/JVI.02013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu FY, et al. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J Virol. 2001;75:1487–1506. doi: 10.1128/JVI.75.3.1487-1506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirshner JR, Lukac DM, Chang J, Ganem D. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Majerciak V, Yamanegi K, Zheng ZM. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J Virol. 2006;80:11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1.1–R1.14. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miranda KC, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.