Abstract
Most human cancers are aneuploid and have chromosomal instability, which contrasts to the inability of human cells to normally tolerate aneuploidy. Noting that aneuploidy in human breast cancer correlates with increased expression levels of the Mps1 checkpoint gene, we investigated whether these high levels of Mps1 contribute to the ability of breast cancer cells to tolerate this aneuploidy. Reducing Mps1 levels in cultured human breast cancer cells by RNAi resulted in aberrant mitoses, induction of apoptosis, and decreased ability of human breast cancer cells to grow as xenografts in nude mice. Remarkably, breast cancer cells that survive reductions in levels of Mps1 have relatively less aneuploidy, as measured by copies of specific chromosomes, compared with cells that have constitutively high levels of Mps1. Thus, high levels of Mps1 in breast cancer cells likely contribute to these cells tolerating aneuploidy.
Keywords: genomic instability
Most human cancers are aneuploid, with aberrant numbers as well as structures of chromosomes. This state of aneuploidy in cancer cells is closely linked to chromosomal instability (1–3), a dynamic process that allows distribution of variable chromosomal content among daughter cells. Paradoxically, although chromosomal instability and aneuploidy are hallmarks of cancer and generally regarded as contributors to carcinogenesis (2, 4), normal eukaryotic cells are highly intolerant of aneuploidy (3, 5, 6). A resolution of this paradox would thus likely require cancer cells to acquire an ability to tolerate, and even thrive, with chromosomal instability and associated aneuploidy.
Cellular and molecular mechanisms that could contribute to tolerance of aneuploidy have not yet been described, and even the mechanisms of chromosomal instability are incompletely understood. Experimental data show that chromosomal instability is functionally associated with defects in mitotic spindle checkpoints (7), allowing mitoses to proceed in settings of improper chromosomal alignment, kinetochore attachment, or kinetochore tension, thus leading to variable distribution of chromosomes among daughter cells. Although dysfunction of mitotic checkpoint genes would seem likely causes of chromosomal instability, efforts to attribute chromosomal instability in human cancers to defects in these checkpoint genes have been generally futile. For example, although early studies found mutations of the BubR1 gene in a few colorectal cancers (8), subsequent studies, including genome-wide sequencing of colorectal cancers and other common human cancers, have failed to demonstrate frequent mutations of this gene or related checkpoint genes (9, 10). Furthermore, although functionally defective or weakly expressed levels of the Mad2, BubR1, or Bub1 checkpoint proteins lead to chromosomal instability in cells and increased tumor development in genetically modified mice (11–13), expression levels of checkpoint genes are frequently increased, rather than decreased, in human cancers compared with those seen in comparable normal cells (14, 15). Thus, the occurrence of chromosomal instability in cancer cannot be readily explained by mutations in spindle checkpoint genes, and the increased expression levels of many of these checkpoint genes raises questions regarding potential functional roles of high levels of checkpoint proteins in cancer cells.
Some evidence for checkpoint genes having important functional roles in cancer cells comes from reports that reducing checkpoint gene expression leads to cell death in cancer cells (16, 17). In our previous studies (14), we noted that expression levels of the Mps1 gene (formerly known as TTK) are particularly elevated in breast cancer cells, compared with normal cells, and we therefore focused on Mps1 to determine (i) whether the high expression levels of this gene in human breast cancer are important for cellular function, (ii) how reductions of Mps1 in highly aneuploid breast cancer cells affect the viability of those cells, and (iii) whether elevated levels of this checkpoint gene contribute to the cancer cell's ability to tolerate aneuploidy.
Results
High Levels of Mps1 Correlate with High Histologic Grade in Breast Cancer.
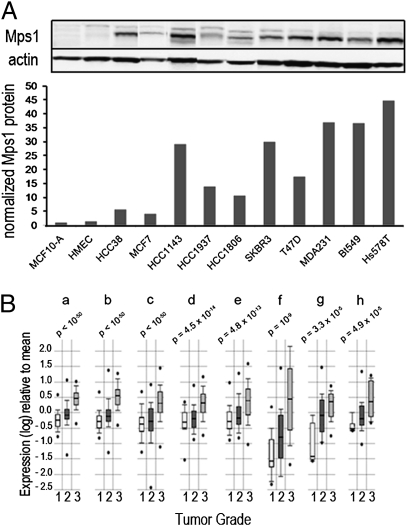
Our previous studies found that mRNA transcript levels of Mps1, the gene that encodes a protein kinase involved in the mitotic spindle checkpoint apparatus, are increased in aneuploid breast cancer cell lines and tumor tissues relative to normal breast epithelial cells or nonmalignant (and chromosomally stable) MCF10A cells (14). Using immunoblots to compare levels of this protein in cultured human breast cancer cells to those in the nonmalignant MCF10A and HMEC cells, we verified that Mps1 protein levels parallel mRNA levels and are also significantly elevated in breast cancer cells (Fig. 1A). To examine possible relationships between levels of Mps1 mRNA and phenotypic characteristics of breast cancer, we probed available annotated microarray databases for expression of Mps1 in samples of breast cancer tissues. Focusing first on histological grade, which is strongly associated with aneuploidy (18), we noted that data from eight different studies independently demonstrate strong positive correlations between levels of Mps1 mRNA and tumor grade (Fig. 1B). Examining data from studies with relevant annotation also demonstrated elevated Mps1 levels are positively correlated with poor survival (Fig. S1), with p53 mutations, and with the basal-like phenotype (Fig. S2), a class of breast cancer that has greater chromosomal copy number variation and genetic complexity than other subtypes of breast cancer (19–21).
Fig. 1.
Increased expression of Mps1 in human breast cancer. (A Upper) Immunoblot analysis of Mps1 protein levels in breast cancer cell lines; actin measured for loading control. (A Lower) The bar graph shows protein quantities normalized to those in MCF10A cells. (B) Correlations between Mps1 mRNA levels and tumor grade in human breast cancer tissues. Data from eight different annotated microarray datasets (a–h; refs. 41–47) were evaluated for correlation between Mps1 expression and histological grade. RNA expression within each set is expressed as log ratios for tumors of specified grades to mean level for all tumors within given set. Boxes represent lower and upper quartiles, whiskers represent one SD from mean, and dots represent ranges. P values (comparing Mps1 levels in grade 3 tumors with levels in grade 1 tumors) were calculated by using Student's t test.
Decreased Survival and Induction of Apoptosis in Breast Cancer Cells After Reduction of Mps1 Levels.
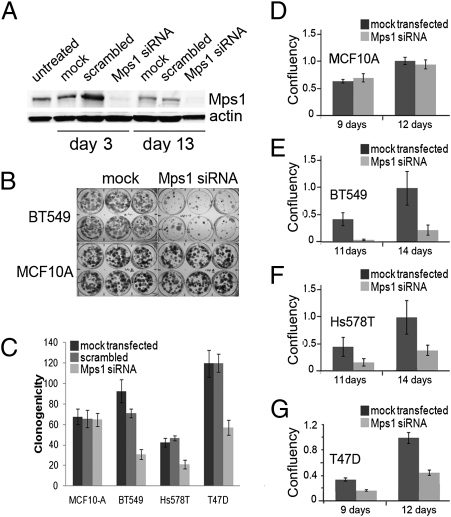
Because increasing expression of Mps1 through transfection of an inducible Mps1 construct does not significantly affect in vitro growth properties of MCF10A cells, we reasoned that high levels of Mps1 in breast cancers might function to maintain cancer cell homeostasis, rather than to serve as a classic oncogene driving the cancer cell phenotype. Consistent with this concept, reductions in Mps1 and BubR1 levels have been found to decrease viability of several human cancer cell lines (17). To explore the role of overexpressed Mps1 in breast cancer cells, we transiently transfected three different breast cancer cell lines and the nonmalignant MCF10A cell line with fluorescein-tagged siRNA constructs designed to reduce levels of Mps1. Evaluation of fluorescence in transfected cells was used to verify efficient transfection in all cell lines (Fig. S3A). Remarkably, although reduction of Mps1 levels in cells by 85% (Fig. 2A and Fig. S3B) had no immediate effect on cell viability (measured by Trypan blue exclusion immediately after transfection; Fig. S4), sustained reduction of Mps1 resulted in significantly decreased survival and growth (measured by clonogenicity and confluency) in the breast cancer cells (Fig. 2 B–G). Interestingly, reducing the levels of the BubR1 and Mad2 checkpoint proteins have also been reported to provoke cell death within six cell divisions (16), but the same investigators found that reductions of Mps1 alone were not sufficient to significantly affect survival of several human cancer cell lines that they tested (17). As our results with MCF-10A, BT549, T47D, and Hs578T cells would suggest, responses to reduced Mps1 are likely highly variable among different cell lines.
Fig. 2.
Viability of cultured breast cancer cells after reduction of Mps1 expression levels with siRNA. (A) Western blot analysis of BT549 cell extracts showing protein levels of parental control (transfection reagents only, no oligonucleotides), scrambled construct-transfected, and Mps1 siRNA-transfected immediately after transfection (day 3) and after 10 additional days of growth in fresh media (day 13). (B) Growth of colonies of BT549 and MCF10A cell lines showing reduced growth in siRNA-transfected cells. (C) Clonogenicity (colony forming efficiency) of MCF-10A cells and three breast cancer cell lines after reduction of Mps1 levels by siRNA, compared with mock-transfected and scrambled vector-transfected controls. Heights of bars represent mean numbers of colonies (500 cells plated), and error bars represent SDs (n = 6). (D–G) Confluency of culture dishes transfected with Mps1 siRNA after growth for numbers of days specified. Confluency was measured by percentages of plates occupied by cells using Image Pro Plus, and values reflect both survival and growth rates of surviving cells. Levels were normalized to controls at 12 d (D and G) or 14 d (E and F).
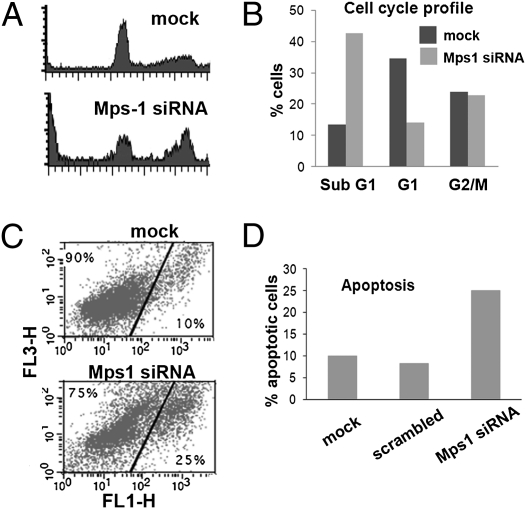
Flow cytometry of Mps1-depleted cells detected an increase in the sub-G1 population and increased staining for Annexin V in these breast cancer cells (Fig. 3), consistent with reductions of Mps1 protein causing cellular apoptosis rather than simply a reduction in cell growth rates. By contrast, growth and survival of MCF10A cells (with low baseline levels of Mps1) were not significantly affected by similarly effective Mps1 siRNA transfection (Fig. 2 B–D and Fig. S3A), and there was no significant increase in apoptosis in similarly treated MCF10A cells compared with untreated cells. This result indicates that the cancer cells and nontumorigenic MCF10A cells have differential requirements for Mps1, corresponding to their different constitutive expression levels.
Fig. 3.
Induction of apoptosis by reductions in Mps1 levels. (A) Flow cytometry measurements of cell-cycle distribution of propidium iodide-stained T47D cells. Cultures treated with Mps-1 siRNA for 3 d are compared with cells treated with the same transfection reagents but no siRNA vector (mock) (B) Plots showing distribution of T47D cells in sub-G1, G0/G1, and G2/M phase compartments. (C) Staining for propidium iodide (vertical axis) and Annexin V (horizontal axis) in T47D cells that are mock treated (Upper) or treated with Mps1 siRNA (Lower). Only nonnecrotic cells (without high PI staining) are shown. Cells with increased Annexin V staining are scored as apoptotic. Treatment of cells with scrambled sequence resulted in staining similar to mock treated. (D) Histogram comparing the percent of apoptotic cells in each of the different treatments.
Reduced Growth of Breast Cancer Xenografts After RNAi-Mediated Decreases in Mps1.
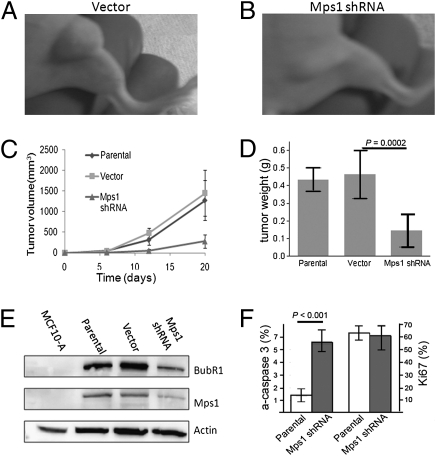
Although other investigators have shown that reductions in cellular levels of checkpoint proteins lead to decreased growth and increased cell death in vitro, the effects of such reductions of Mps1 on tumor growth in vivo had not been reported. To explore the effects of such reductions of Mps1 in vivo, athymic nude mice were inoculated with highly tumorigenic Hs578T breast cancer cells that had been stably transfected with a tetracycline/doxycycline-inducible construct, pENTR/H1/TO:Mps1 shRNA, or control empty vectors. This shRNA construct significantly reduces Mps1 mRNA levels in Hs578T and BT549 malignant cells when grown in the presence of 100 μg/mL tetracycline in vitro (Fig. S5). Although there was little difference between growth rates of tumors formed by the parental cell line or cells transfected with the empty control vector (Fig. 4 A, C, and D), the growth rates of xenografts bearing the Mps1-specific shRNA construct were significantly reduced in animals fed with doxycycline (Fig. 4 B–D for treatments beginning 1 d after inoculation and Fig. S6 for treatments beginning 12 d after inoculation).
Fig. 4.
Reduced Mps1 levels result in decreased in vivo growth of breast cancer xenografts. (A–C) Growth of mouse xenografts tumors of Hs578T cells with doxycycline-inducible Mps1 shRNA compared with xenografts with cells from parental (C) and vector (no dox) (A and C) controls. In this experiment, doxycylcine treatment began 24 h after inoculation of tumor cells. Error bars indicate SEM. (D) Weights of tumors explanted from animal killed 20 d after inoculation. Error bars indicate SEM, and P values were calculated by using Student's t test. (E) Western blot analysis of Mps1 protein and BubR1 protein using lysates from tumors of control (parental), vector, and Mps1 shRNA xenografts, compared with MCF10A cells as a frame of reference. (F) Quantitation of Mps1 and BubR1 proteins from tumors normalized to controls, with P values calculated by using Student's t test.
Notably, immunoblot analysis demonstrated that both Mps1 and BubR1 protein levels were reduced by ≈50–60% in samples procured from the tumors with doxycycline-induced shRNA (Fig. 4E), supporting the recently proposed role of Mps1 as a kinase that phosphorylates and stabilizes BubR1 (22). It is remarkable that the reductions in BubR1 levels in these tumors (≈60%) were significantly less extreme than the reductions of BubR1 induced in mouse models by haploinsufficiency or hypomorphic alleles of the BubR1 gene (23). Although these mice with hypomorphic BubR1 alleles did exhibit accelerated aging, even greatly reduced BubR1 (80%) was found to be compatible with embryogenesis and reasonably normal development. Thus, although our experimental system is limited with respect to reducing Mps1 (and BubR1) selectively in tumor cells, it would seem likely that similar levels of reduction of these genes would be generally tolerated by normal adult eukaryotic cells.
Explanted tumors from killed animals were also evaluated for levels of active caspase 3, a marker for cellular apoptosis, and Ki-67, a marker for cellular proliferation, by immunohisochemistry. Interestingly, although Mps1 shRNA did not affect proliferation, as measured by percentage of cells staining for Ki-67, it did markedly increase apoptosis within the tumors (Fig. 4F and Fig. S6). Thus, increased cellular apoptosis apparently contributes significantly to the reduced growth of tumors after induction of Mps1 shRNA.
Reduced Mps1 Levels Lead to Aberrant Mitoses in Breast Cancer Cells.
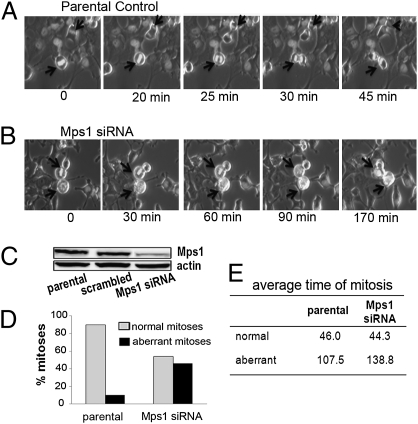
Mps1 has several critical roles in mitosis, including centrosome duplication (24), mitotic spindle assembly (25), and maintenance of the spindle assembly checkpoint (26, 27), and pharmacological or genetic inhibition of Mps1 function in various types of mammalian cells have generally resulted in accelerated mitosis (28, 29). To determine the effect of reduced Mps1 on mitotic progression in the malignant breast cancer cells used in our study, Hs578T cells stably expressing pBOS-H2B-GFP (30) were transiently transfected with Mps1 siRNA, or a scrambled siRNA control sequence, and examined by time-lapse fluorescence microscopy after 72 h of incubation. Control cells (either transfected with scrambled siRNA sequence or treated only with transfection reagents) progressed to anaphase within 20–25 min of initiating metaphase and completed mitosis (prometaphase to telophase) by 45 min (Fig. 5 A and D and Movie S1). Only a small subset of control Hs578T cells (<10%) underwent aberrant mitoses (Fig. 5 C and D). By contrast, whereas ≈50% of the Mps1-reduced Hs578T cells also progressed through mitosis with an average time of 46 min, nearly 50% (18 of 39) of cells in this treatment group exhibited aberrant and prolonged mitoses (Fig. 5 B–D). Of these aberrant mitoses (n = 18), six (33%) either entered prometaphase but failed to align their chromosomes or remained in metaphase with chromosomes aligned along the spindle for >60 min. These cells failed to enter anaphase but rather retrogressed to a prometaphase-like state with condensed unaligned chromosomes before undergoing apoptosis (Fig. 5 B and D and Movie S2). We also observed two aberrantly dividing cells (11%) that oscillated between prometaphase and metaphase before eventually dividing, three cells (17%) that bypassed metaphase and died, and seven cells (39%) that divided without cytokinesis, giving rise to multinucleate cells (Movie S3). The types of mitotic errors, summarized in Table 1, are consistent with reported findings of increased segregation errors, including misaligned chromosomes, lagging chromosomes, and anaphase bridges, in U2OS cells with kinase-dead Mps1 protein (28) or RNAi-depleted Mps1, although it is notable that these aberrant mitoses in U2OS cells occurred in cells that progressed through mitosis at an accelerated rate. Although cancer cells with different genetic backgrounds (e.g., Hs578T and U2OS) appear to have differences in how reduced Mps1 affects progression through mitosis, the resulting increases in aberrant mitoses appear to be similar. Because the levels of Mps1 after reduction by RNAi in our study approximate those seen constitutively in nonmalignant cells, our results suggest that high levels of Mps1 are required for aneuploid cancer cells, but not nonmalignant cells, to progress through mitosis normally.
Fig. 5.
Reduced Mps1 levels result in aberrant mitoses in breast cancer cells. Individual frames from time-lapse movies of Hs578T cells containing pBOS Histone H2B-GFP; untreated (parental) (A) and transiently transfected with Mps1 siRNA (B). Corresponding movies are available as Movies S1–S3. Arrows indicate dividing cells that undergo mitosis. (C) Western blot showing levels of Mps1 reduction in cells used for these time-lapse experiments. (D) Graph comparing normal to aberrant mitoses in untransfected controls (parental) and cells transiently transfected with Mps1 siRNA (Mps1). (E) Average times for both normal and aberrant mitoses in control and Mps1 siRNA- treated cells.
Table 1.
Mitotic aberrations induced by Mps1 siRNA
| Type of mitotic aberration | Ctrl | Mps1 siRNA |
| Prolonged oscillation between prometaphase and metaphase followed by apoptosis | 1 | 6 |
| Prolonged oscillation between prometaphase and metaphase, then division | 2 | 2 |
| Truncated metaphase, divides without alignment and dies | 0 | 3 |
| Successful polyploid division with normal timing | 1 | 0 |
| Mitosis without cytokinesis | 0 | 7 |
| Total number of aberrant mitoses/ total number mitoses examined | 4/39 | 18/39 |
Reduced Mps1 Levels Lead to Selective Survival of Cells with Less Aneuploidy.
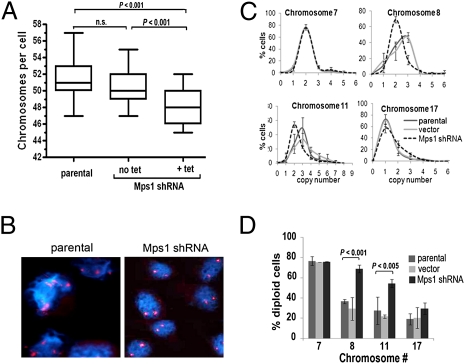
Although it has been shown that reducing levels or activity of checkpoint proteins leads to decreased cell survival (16, 17, 28), the characteristics of cancer cells that survive these reductions of checkpoint proteins have not been described to our knowledge. Considering the role of Mps1 in faithful transmission of chromosomes during cell division, we questioned whether high Mps1 expression plays a role in tolerance of cancer cells to aneuploidy, and whether reductions of Mps1 would thus selectively target the most highly aneuploid cells in a culture with genomic variability. To address these questions, we reduced Mps1 expression in low-density cultures of Hs578T cells by using a tetracycline-inducible shRNA construct, pENTR/H1/TO:Mps1 shRNA, and cultured these cells in the presence of 100 μg/mL tetracycline for 5 d. As noted above for the transfections with siRNA, this treatment with Mps1 shRNA led to significantly reduced cell viability. Conventional metaphase spreads showed that cells surviving reduction of Mps1 have fewer total numbers of chromosomes (47.97 ± 0.24) than parental cells (51.14 ± 0.26) or cells that had been transfected with vector only (50.50 ± 0.22) (Fig. 6A). Further evidence of a selective process favoring survival of cells with relatively lower levels of aneuploidy came from fluorescence in situ hybridization (FISH) studies measuring copy numbers of chromosomes 7, 8, 11, and 17 in surviving cells. Cells surviving shRNA-induced reductions in Mps1 protein levels exhibited a shift of the mode from three copies to two copies per cell for chromosomes 8 and 11, both of which are represented in control (parental) Hs578T cells by highly variable copy numbers and a mode of three copies, and a significantly reduced variability in the numbers of these chromosomes per cell (Fig. 6 B–D; P < 0.005). These results are notable in light of findings that normal eurkaryotic cells are particularly intolerant of extra copies of chromosomes (6) and, thus, the selective decrease in the numbers of Hs578T cells with more than two copies of chromosomes 8 and 11 likely reflects reduced tolerance of the Mps1-depleted cells for extra copies of these chromosomes.
Fig. 6.
Reduced Mps1 levels lead to selective survival of cells with less aneuploidy. (A) Numbers of chromosomes were counted in metaphase spreads of parental Hs578T cells (n = 83), cells transfected with Mps1 shRNA and no doxycycline induction (n = 70), and cells transfected with Mps1 shRNA induced by doxycycline (n = 62). Boxes represent lower and upper quartiles, and whiskers represent one SD from mean. P values were calculated by using Student's t test for each two-way comparison. (B) FISH of Hs578T expressing Mps1-shRNA showing cells probed for CEN 11 (red) in untransfected parental cells and cells with dox-induced pENTRH1/TO Mps1-shRNA (Mps1 shRNA) (C) Plots showing frequency distributions of copy numbers for chromosome 7, 8, 11, and 17 in parental Hs578T cells, cells with Mps1 shRNA and no doxycycline induction (vector), and cells transfected with Mps1 shRNA induced by doxycycline (Mps1 shRNA). Error bars, 1 SD. (D) Percent of cells diploid for chromosomes 7, 8, 11, and 17 in same treatment groups as C. Error bars, 1 SD; P values were calculated by using Student t test for two-way comparisons of cells with Mps1-siRNA and parental cells.
Normal eukaryotic cells are apparently more tolerant of chromosome losses than chromosomal gains (6), and we correspondingly did not see such marked changes in copy numbers for chromosome 17, which has a modal copy number of 1 in the control (parental) Hs578T cells. After transfection of Mps1 shRNA, we saw a modest but statistically insignificant increase in the percentage of cells with two copies of this chromosome (Fig. 6 C and D). Although this result could reflect the relatively greater tolerance of cells for loss of chromosomes (compared with gain of chromosomes), it should also be acknowledged that there are few options for restoring two chromosomes to daughter cells through unbalanced segregation of replicated copies of a single chromosome. Finally, we also examined copy numbers for chromosome 7, which has a modal copy number of two in the control Hs578T cells. In cells transfected with Hs578T siRNA, we observed no significant change in the modal copy number (Fig. 6B) or the percent of cells with two chromosomes (Fig. 6C), indicating that reductions of Mps1 levels lead to decreases in the numbers only for those specific chromosomes with variant copy numbers in the cancer cells.
Because reducing Mps1 levels results in reduced overall survival of Hs578T cells, the decrease in chromosome copy number variation among surviving cells likely reflects selection of cells based on a degree of aneuploidy, rather than a process involving elimination of chromosomes in interphase cells. These data would thus suggest that high levels of Mps1 in cancer cells function to support tolerance of aneuploidy, and reducing the levels of Mps1 diminishes the level of aneuploidy tolerated in these cells.
Discussion
Decreased expression or defective function of mitotic spindle checkpoint proteins, including Mps1, leads to chromosomal instability in experimental systems, and these findings might lead to anticipation of low levels of checkpoint proteins in human cancers. However, our analysis of breast cancer cell lines and data from multiple microarray gene expression datasets show aneuploid human breast cancers actually have significantly increased expression levels of Mps1. Moreover, high expression of Mps1 correlates strongly with the histological grade of breast cancers, suggesting that the most aneuploid of these cancers have the highest levels of Mps1 expression. Undertaking experiments to determine whether these increased levels of Mps1 are functionally significant in breast cancer cells, we found that reducing these levels of Mps1 in cancer cells with RNAi leads to apoptosis and decreased cell survival, specifically in those cells with constitutively elevated expression of this checkpoint protein.
One explanation for increased apoptosis in Mps1-reduced cultures would involve a role for Mps1 in blocking apoptotic pathways in these cells, because other checkpoint proteins have been implicated to have direct roles in apoptosis signaling. For example, Aurora B and Survivin, downstream components of the spindle checkpoint, have both been shown to affect antiapoptotic pathways by hyperphosphorylating Bcl2 and inhibiting Bax (31). By contrast, Mad2 has been implicated in proapoptotic pathways, based on findings that reducing levels of Mad2 by shRNA results in decreased levels of apoptosis and reduced Caspase 3 activity (31). In other studies, BubR1 stability after mitotic arrest has been shown to depend on caspase activity (32, 33), and it is thus possible that that high levels of Mps1 could indirectly inhibit apoptosis by hyperactivating BubR1 (22), explaining increased apoptosis in cells after reducing Mps1 levels.
Our experimental evidence, however, points to a role for high levels of Mps1 in providing stability and protection for aneuploid cells during mitosis. Reducing Mps1 levels in aneuploid cells increases the frequency of aberrant and catastrophic mitoses, and similar to the effects reported after reducing levels of the BubR1 and Mad2 checkpoint proteins (16), measurable loss of cell viability after several cell divisions. As evidenced by our analysis of surviving cells, this process appears to selectively affect aneuploid cancer cells and spares cells with relatively less aneuploidy. Although these results do not preclude Mps1 participating directly in antiapoptotic signaling, it appears that much of the function of increased Mps1 (and likely other checkpoint genes) in breast cancer cells involves tolerance of aneuploidy.
Clues for how high levels of Mps1 could lead to tolerance of aneuploidy in cancer cells might be found by examining the roles of checkpoint proteins in maintaining cell viability during mitosis. The spindle assembly checkpoint is activated by failure of chromosomes to make appropriate attachment to the mitotic spindle or generate sufficient tension at kinetochores (34), and experimental evidence from budding yeast indicates that Mps1 is required for the tension-induced activation of the mitotic checkpoint and error correction (35). It is not unreasonable to expect that aneuploid cancer cells would require high levels of Mps1 to satisfy tension requirements of the spindle checkpoint, and reducing these high Mps1 levels is apparently particularly disruptive to cancer cells, triggering mitotic arrest and cell death. The role of Mps1 in maintaining the efficiency of the mitotic spindle in both normal and cancerous cells might be related to its function in stabilizing the BubR1 protein (22), which is known to regulate kinetochore–microtubule interactions (36).
Our data do not provide any evidence to suggest that increased levels of Mps1 promote cell growth, including our experiments that found no phenotypic changes in nontumorigenic MCF10A cells after transfection with Mps1. Moreover, there is no evidence from studies in yeast to suggest that high levels of Mps1 could contribute directly to a malignant phenotype; in fact, excessive expression of the Mps1 protein in Saccharomyces cerevisiae actually causes cells to arrest in mitosis (37). Although increased levels of Mps1 or other checkpoint proteins apparently do not directly lead to stimulation of cell growth, this overexpression can, in some settings, contribute to chromosomal instability. For example, transgenic mice engineered to have overexpression of Mad2, a related checkpoint gene, have cells with widespread chromosomal instability and develop various types of neoplasms (38), but increased rates of cell division or other features of abnormal cell growth were not described in these animals. Interestingly, in this model, continued overexpression of Mad2 is not required for tumor maintenance, suggesting that the chromosomal instability was important for initiating carcinogenesis, but not maintaining the neoplastic phenotype (38). Thus, these data also indicate that high expression levels of checkpoint genes can contribute to chromosomal instability and aneuploidy—perhaps through tolerance of the aneuploidy—but do not function as typical oncogenes that activate cell growth pathways.
Targeting checkpoint genes has been proposed for cancer treatment (16, 17), and our results demonstrate the importance of high Mps1 expression on viability of aneuploid cancer cells, not only in cell culture, but also in vivo, where reducing Mps1 levels in xenografts of Hs578T cells resulted in marked inhibition of overall tumor growth as a result of increased apoptosis of tumor cells. Our observation that reducing Mps1 in cancer cells selectively targets those cells with relatively high levels of chromosomal variation could explain findings that cancer cell lines with elevated Mps1 are relatively resistant to taxanes (39), and targeting Mps1 might sensitize some cells to paclitaxel treatment. This sensitization is likely related to the roles of the checkpoint proteins in stability of kinetochore/microtubule attachment and error correction (28, 40) and offers insights into how targeting Mps1 in cancer cells with other pharmaceutical agents—particularly those that disrupt the mitotic spindle—could offer therapeutic strategies for cancer treatment.
Materials and Methods
Cell Culture and Viability Assays.
Cell cultures were purchased from American Type Culture Collection (ATCC) and cultured with serum and other additives according to ATCC recommendations. Viability was measured by using standard clonogenic assays by reseeding treated cells in six-well tissue culture plates at ≈200 cells per well, culturing for 8–14 d, and staining with 0.5% crystal violet. An area occupied by proliferating cells was calculated by using Image Pro Plus (Media Cybernetics).
Analysis of Microarray Data.
Microarray datasets (41–47) of breast cancer gene expression were probed for expression of Mps1 (TTK) by using Oncomine version 3 and correlated with histological grade, survival, p53 status, and BRCA1 mutations. A Student's two-class t test was used to determine significance of correlations, and Oncomine-generated box plots were used for visualization.
Methods for RNAi, metaphase spread analysis, FISH, immunohistochemisty, time lapse microscopy, Annexin V staining, and breast cancer xenografts used standard protocols, which are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Raluca Yonescu and Constance Griffin of the Cytogenetics Core (Johns Hopkins Kimmel Cancer Center) for assistance with preparations of metaphase spreads. This work was supported by National Cancer Institute Grants (NCI) P50 CA058184 and R01 CA101232. J.D. was also supported by NCI Training Grant T32CA067751.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007645108/-/DCSupplemental.
References
- 1.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pihan G, Doxsey SJ. Mutations and aneuploidy: Co-conspirators in cancer? Cancer Cell. 2003;4:89–94. doi: 10.1016/s1535-6108(03)00195-8. [DOI] [PubMed] [Google Scholar]
- 5.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 9.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 10.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 11.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 12.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 13.Schliekelman M, et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan B, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 15.Myrie KA, Percy MJ, Azim JN, Neeley CK, Petty EM. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett. 2000;152:193–199. doi: 10.1016/s0304-3835(00)00340-2. [DOI] [PubMed] [Google Scholar]
- 16.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owainati AA, et al. Tumour aneuploidy, prognostic parameters and survival in primary breast cancer. Br J Cancer. 1987;55:449–454. doi: 10.1038/bjc.1987.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Melchor L, et al. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–3175. doi: 10.1038/sj.onc.1210975. [DOI] [PubMed] [Google Scholar]
- 21.Chin SF, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, et al. Phosphorylation sites in BubR1 that regulate kinetochore attachment, tension, and mitotic exit. J Cell Biol. 2008;183:667–680. doi: 10.1083/jcb.200805163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 24.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones MH, et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 27.Vigneron S, et al. Kinetochore localization of spindle checkpoint proteins: Who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jelluma N, et al. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 2008;3:e2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 31.Vogel C, Hager C, Bastians H. Mechanisms of mitotic cell death induced by chemotherapy-mediated G2 checkpoint abrogation. Cancer Res. 2007;67:339–345. doi: 10.1158/0008-5472.CAN-06-2548. [DOI] [PubMed] [Google Scholar]
- 32.Kim M, et al. Caspase-mediated specific cleavage of BubR1 is a determinant of mitotic progression. Mol Cell Biol. 2005;25:9232–9248. doi: 10.1128/MCB.25.21.9232-9248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baek KH, et al. Caspases-dependent cleavage of mitotic checkpoint proteins in response to microtubule inhibitor. Oncol Res. 2005;15:161–168. doi: 10.3727/096504005776367906. [DOI] [PubMed] [Google Scholar]
- 34.Pinsky BA, Biggins S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Maure JF, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 37.Poddar A, Daniel JA, Daum JR, Burke DJ. Differential kinetochore requirements for establishment and maintenance of the spindle checkpoint are dependent on the mechanism of checkpoint activation in Saccharomyces cerevisiae. Cell Cycle. 2004;3:197–204. [PubMed] [Google Scholar]
- 38.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanton C, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 41.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivshina AV, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 43.Desmedt C, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 44.Sotiriou C, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 46.Ginestier C, et al. Distinct and complementary information provided by use of tissue and DNA microarrays in the study of breast tumor markers. Am J Pathol. 2002;161:1223–1233. doi: 10.1016/S0002-9440(10)64399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma XJ, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.