A central question in evolutionary genomics is how morphological differences between taxa arise from differences in gene repertoire. Answers require rigorous orthology assignments. Recent work (1) illuminated the mechanism of background adaptation in teleost fishes by investigating the function of agouti-related protein 2 (agrp2), a gene so far found only in teleosts.
Agouti family proteins antagonize melanocortin receptors, function in energy homeostasis, and regulate pigmentation. The authors (1) proposed a neurocrine axis in which zebrafish Agrp2 is expressed exclusively in the pineal gland, where it binds to the melanocortin 1 receptor and thereby, controls expression of melanin-concentrating hormones; this causes melanosome contraction in melanophores and adaptation to light backgrounds (1). These findings intriguingly implied that the acquisition of agrp2 in teleosts enabled the evolution of background adaptation by regulation through the pineal.
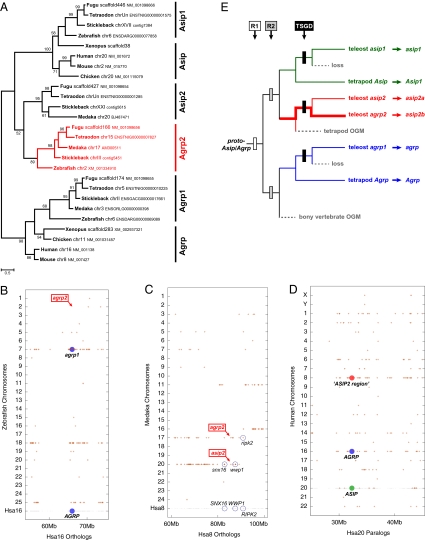
Tetrapods have two agouti family members [Agouti signaling protein (Asip) and Agrp], but teleosts have four (asip1, asip2, agrp1, and agrp2). Published studies (1, 2) vaguely suggest that agrp2 is a paralog of agrp1 generated during the teleost-specific genome duplication (TSGD). Phylogenetic analyses, however, remain ambiguous regarding vertebrate agouti gene relationships (2) (Fig. 1A).
Fig. 1.
Evolution of vertebrate agouti genes. (A) Maximum likelihood phylogeny of vertebrate agouti family proteins with current names (JTT+G+I model; 100 bootstrap replicates). (B–D) Conserved synteny dot plots derived from the Synteny Database (5). (B) The zebrafish agrp2 region on Dre2 (red box) shares conserved syntenies neither with the zebrafish agrp1 region (Dre7) nor with the human AGRP region (Hsa16). (C) The agrp2 and asip2 regions in medaka and other teleosts share conserved synteny with each other and with a region on human Hsa8, including several agrp2- and asip2-neighboring genes. (D) Analysis of the human genome shows that the ASIP region on Hsa20 shows more paralogous connections to the inferred ASIP2 region on Hsa8 than to the AGRP region on Hsa16 (43 vs. 25 genes, respectively). (E) Model for the evolution of the vertebrate agouti family by three rounds of genome duplication, including a revised gene nomenclature. The pineal-specific function of teleost agrp2 (new name: asip2b) could have evolved anywhere along the thick red line. OGM, ohnolog gone missing.
Our investigation of this gene family using synteny data clearly reveals that the teleost agrp2 chromosomal region shares syntenies neither with the teleost agrp1 region nor with the tetrapod Agrp region (Fig. 1B), contradicting the current gene nomenclature (2). In contrast, teleost agrp2 and asip2 regions show conserved synteny to a region on human chromosome 8 (Hsa8) (Fig. 1C). Furthermore, we find at least three regions of paralogy in the human genome (Fig. 1D)—Hsa16 (AGRP region), Hsa8, and Hsa20 (ASIP region)—most likely derived from a single Asip/Agrp region on ancestral vertebrate protochromosome B (3). The ASIP region on Hsa20 shares more paralogies with the region on Hsa8 (Fig. 1D). According to our model (Fig. 1E), an ancestral Asip/Agrp precursor gene was duplicated two times during the two rounds of vertebrate genome duplication (R1 and R2). After R2, Asip and Agrp were retained in all bony vertebrates, but a third ohnolog (4) went missing in the tetrapod lineage. This gene was retained, however, in the rayfin fish lineage, giving rise to agrp2 and asip2 after the TSGD.
Our results support a revision of agouti family nomenclature (Fig. 1E). Importantly, our model suggests that the genetic basis of teleost background adaptation may not root in the agouti family expansion during the TSGD but to R2, far deeper in vertebrate evolution (Fig. 1E). It is unlikely that zebrafish agrp2 (new name: asip2b) newly evolved its single pineal-specific function after the TSGD (as suggested by ref. 1), because asip2 (asip2a) is absent in zebrafish but present in other teleosts. The interesting findings of Zhang et al. (1) should, thus, spur research on the function of asip2a/b in other teleosts and agouti genes in basal rayfin fishes, basal lobefins like lungfish and coelacanth, background-adapting tetrapods, and more basal vertebrates in the light of our rigorous phylogenetic framework (Fig. 1E).
Acknowledgments
This work was supported by a Feodor Lynen Fellowship of the Alexander von Humboldt Society (to I.B.) and National Center for Research Resources at National Institutes of Health Grant R01RR020833 (to J.P.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Zhang C, et al. Pineal-specific agouti protein regulates teleost background adaptation. Proc Natl Acad Sci USA. 2010;107:20164–20171. doi: 10.1073/pnas.1014941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurokawa T, Murashita K, Uji S. Characterization and tissue distribution of multiple agouti-family genes in pufferfish, Takifugu rubripes. Peptides. 2006;27:3165–3175. doi: 10.1016/j.peptides.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe K. Robustness—it's not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- 5.Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]