Abstract
Although the biological function of fruiting is the production and dissemination of seeds, humans have developed seedless fruits in a number of plant species to facilitate consumption. Here we describe a unique spontaneous seedless mutant (Thai seedless; Ts) of Annona squamosa (sugar apple), a member of the early-divergent magnoliid angiosperm clade. Ovules (seed precursors) of the mutant lack the outer of two normal integuments, a phenocopy of the inner no outer (ino) mutant of Arabidopsis thaliana. Cloning of the INO ortholog from A. squamosa confirmed conservation of the outer integument-specific expression pattern of this gene between the two species. All regions of the gene were detectable in wild-type A. squamosa and in other members of this genus. However, no region of the INO gene could be detected in Ts plants, indicating apparent deletion of the INO locus. These results provide a case of a candidate gene approach revealing the apparent molecular basis of a useful agronomic trait (seedless fruit) in a crop species, and indicate conservation of the role of a critical regulator of ovule development between eudicots and more ancient lineages of angiosperms. The outer integument is one synapomorphy of angiosperms separating them from other extant seed plants, and the results suggest that the evolution of this structure was contemporaneous with the derivation of INO from ancestral YABBY genes. Thus, a unique lateral structure appears to have coevolved with a novel gene family member essential for the structure's formation.
Keywords: cherimoya, stenospermy, parthenocarpy
Fruits form through an intimate developmental collaboration between ovules (the developmental precursors to seeds) and carpels (the precursors to the body of the fruit). For centuries, humans have attempted to interrupt this association to develop seedless fruits to make them easier to consume. This is a difficult task, because fruit development is usually arrested in the absence of seed formation (1). In addition, most mutations resulting in seedless fruits disappear unnoticed because sexual reproduction is precluded and intentional vegetative propagation is required to maintain seedless genotypes [as noted by Darwin (2)]. Examples of species where varieties producing seedless fruits have been selected and popularized include watermelons, grapes, Citrus, pineapples, and bananas. Two main mechanisms are responsible for the formation of seedless fruits (1): (i) parthenocarpy, where the fruit develops in the absence of fertilization, as in cultivated pineapples, some Citrus cultivars, and bananas; and (ii) stenospermy, where pollination and fertilization are required, but embryos either do not form or they abort before completion of seed formation, as in seedless watermelons and many seedless grapes (3). In both cases, the plant must have an inherent or acquired ability to sustain fruit development in the absence of seed formation.
Mutations leading to failure in seed formation have been studied in the model system Arabidopsis thaliana, where complete absence of seeds is commonly associated with defects in ovule development (for a review, see ref. 4). Stenospermy has not been observed in such mutants, as the pistils remain small and do not develop into fruits following pollination. For example, the Arabidopsis inner no outer (ino) mutant exhibits a near-complete failure in seed formation due to an absence of the outer of the two integuments normally present in the ovule (5, 6). Following pollination, the fruits do not expand beyond the size achieved at anthesis, even in those rare cases where one or two seeds are formed in this mutant (5). Parthenocarpy is observed in Arabidopsis plants homozygous for the fruit without fertilization/auxin response factor 8 (fwf/arf8) mutation, but this mutant has a normal seed set when pollinated (7). The conservation of the roles of these genes and hence their participation in seedlessness and fruit formation in other species has not been demonstrated, although conservation of the expression pattern of INO orthologs has been observed in some other species (8, 9).
Here we describe a unique case of seedless fruit production in a mutant of Annona squamosa based on a failure to form seeds due to a defect in ovule development. Annona species are members of the Annonaceae, the largest living family among the magnoliid group of early-divergent angiosperms. Cherimoya (A. cherimola), sugar apple (A. squamosa), or soursop (A. muricata) were used as a food source by pre-Columbian cultures in Central and South America (10). Cultivation has continued to the present day and may be poised for expansion in developing countries with subtropical climates. Annona fruits have a meat with a sherbet-like texture and a flavor that has been compared with a mixture of banana and pineapple (11), and was noted by author Mark Twain as “the most delicious fruit known to men” (12). Like the wild-type banana, Annona fruits include multiple large (∼2-cm) seeds (Fig. 1). Annona fruits do not normally develop parthenocarpically, and in commercial production hand pollination is commonly used to ensure complete fertilization and full development of all of the carpels in a flower to produce large uniform fruit. A spontaneous mutant of A. squamosa, Thai seedless (Ts), produces fully seedless normal-sized fruits (Fig. 1) and provides the potential to introduce this feature into Annona fruit production. We determined the developmental cause of seedlessness in Ts and, using information from studies on Arabidopsis, show that the mutant is associated with the deletion of the A. squamosa INO gene. This indicates conservation between crown eudicots and basal angiosperms of a genetic pathway regulating ovule development.
Fig. 1.
A. squamosa fruits. Wild-type broken open (A), and Ts broken open (B) and sliced (C). (Scale bars, 2 cm.)
Results and Discussion
Ovule Defects in Seedless A. squamosa Mutant Phenocopy an Arabidopsis Mutant.
Fruit development in Ts plants requires pollination. Rapid growth of the carpels started 6 d after pollination, and the degree of carpel development 10 d after pollination was similar to that observed in seeded A. cherimola (13). Unpollinated Ts flowers arrested development and began to drop 7 d after anthesis, and these observations did not differ in 2 consecutive years. These results indicate a case of stenospermy, and imply that failure in seed development in Ts plants results from ovule or embryo abortion. Pollen from Ts plants was fully fertile when used to pollinate wild-type A. cherimola, demonstrating that infertility did not result from polyploidy or other severe chromosomal aberrations.
To determine the developmental cause of ovule or embryo abortion in Ts, we compared carpel and ovule structures between Ts and wild-type A. squamosa and A. cherimola. The carpel features of wild-type and Ts A. squamosa were indistinguishable from those recently reported for A. cherimola (13). However, whereas both wild-type Annona species had anatropous (recurved), bitegmic ovules (Fig. 2A) typical of the majority of angiosperms, Ts had orthotropous (erect), unitegmic ovules (Fig. 2B). The single integument present was anatomically similar to the inner integument of wild-type A. squamosa, indicating an apparent loss of the outer integument (Fig. 2 A and B).
Fig. 2.
Ts phenocopies the Arabidopsis ino mutant. Stained ovule sections show anatropous, bitegmic ovules in wild-type A. squamosa (A) and Arabidopsis (C), and orthotropous, unitegmic ovules in Ts A. squamosa (B) and Arabidopsis ino (D) where only the inner integument is present. [Scale bar, 40 μm (A and B); 30 μm (C); 15 μm (D).] ii, inner integument; oi, outer integument; n, nucellus.
Studies on ovule development in model species, particularly Arabidopsis thaliana, have provided information on genes responsible for the growth and identity of different ovule structures (4). The well-characterized Arabidopsis ino mutant (5, 6) exhibited effects on ovule development that are nearly identical to those observed in Ts A. squamosa. Wild-type Arabidopsis ovules are bitegmic and anatropous (14) (Fig. 2C), whereas ino mutant ovules have a single integument and are erect due to the absence of the outer integument (5, 6) (Fig. 2D). On the basis of the similarity of effects on ovule development, we hypothesized that seedlessness in Ts could result from disruption of an A. squamosa ortholog of INO.
Expression of INO Genes Is Conserved Between A. squamosa and Arabidopsis.
INO is a member of the small YABBY gene family that encodes putative transcription factors associated with polarity determination in lateral organs (15, 16). To test our hypothesis about the nature of the Ts mutant, we used a combination of PCR-based methods to amplify INO-related sequences from A. squamosa and A. cherimola. Only a single gene was isolated from each species, and in phylogenetic analysis these genes fell clearly into a clade exclusively containing INO orthologs from other angiosperms (Fig. S1). Thus, we had isolated Annona orthologs of INO, and it appears that each Annona species includes only a single INO ortholog.
In Arabidopsis, INO is expressed exclusively in the abaxial (away from the ovule apex) side of the outer integument (15, 17) (Fig. 3). This pattern is conserved in Impatiens (8), and outer-integument expression was also observed for a Nymphaea INO ortholog (9). Using an A. cherimola INO cDNA probe on floral tissues of A. squamosa and A. cherimola, we observed hybridization only in the ovules (Fig. 3 A and B) and in a pattern that directly paralleled the pattern observed in Arabidopsis (Fig. 3 C and D). As in Arabidopsis, in both Annona species, expression initiated in the abaxial half of the outer-integument primordium at the earliest stages of development of this structure (Fig. 3), and was present only in the abaxial layer of the outer integument during subsequent growth. Thus, the pattern of INO expression in Annona is consistent with conservation of the function of this gene in Arabidopsis. As observed for the ino mutant in Arabidopsis (15), the absence of the outer integument in Ts was associated with an absence of an INO in situ hybridization signal (Fig. 3F).
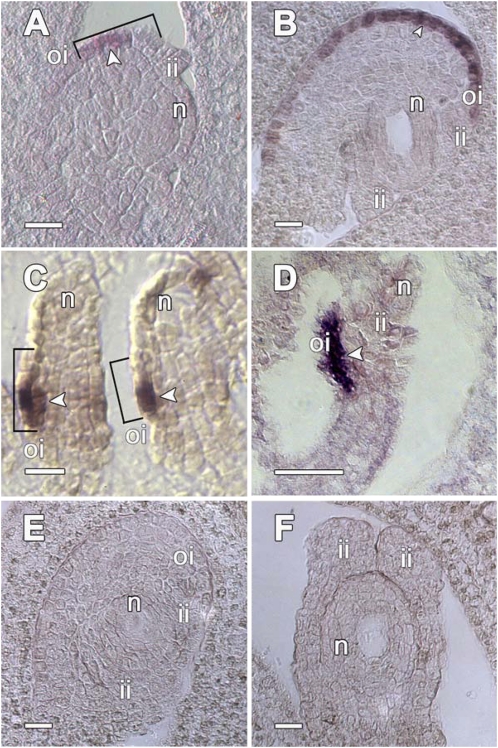
Fig. 3.
INO expression pattern is conserved between A. squamosa and Arabidopsis. (A–D) INO mRNA was detected by in situ hybridization (arrowheads) with antisense probes only in the outer integument in early-stage (A) and developing (B) ovules of wild-type A. squamosa in patterns similar to those observed at corresponding stages of development in Arabidopsis ovules (C and D). Hybridization was not detected with a sense INO probe in wild-type A. squamosa (E), nor with an antisense probe in Ts ovules (F). [Scale bar, 20 μm (A, B, and D–F); 10 μm (C).]
Apparent Deletion of the INO Locus in Ts.
To determine whether there were alterations in the INO gene of the seedless variety, we attempted to isolate the INO sequence from Ts with a primer set designed from the first isolated INO fragment in A. cherimola and that had been successfully used for PCR of INO sequences from both A. squamosa and A. cherimola wild-type genotypes (Table S1). However, no amplification product was produced from Ts. Four additional pairs of primers were designed to separately amplify different regions of the INO gene or immediately flanking sequence (Fig. 4A). Amplification products were obtained from DNA prepared from wild-type A. squamosa and A. cherimola but not from Ts (Fig. 4). The conditions used allowed efficient amplification of fragments even in those cases where primers included one to four mismatches with the INO gene sequence from one or the other of these two wild-type samples. Thus, small deviations in Ts from the sequence of the wild-type A. squamosa sequence could not explain the failure to amplify five different regions of the gene. Control primers designed to a 1-aminocyclopropane-1-carboxylate synthase gene of atemoya (a cultivated A. squamosa x A. cherimola hybrid) (GenBank accession no. AF401757) amplified products from both wild-type and Ts A. squamosa (Fig. 4), even when in the same reactions that failed to produce INO gene products from Ts (Fig. S2). Similar results were obtained with four additional independently prepared samples of DNA from Ts plants. The INO primer pairs were further tested on four additional species that span the phylogenetic range of the Annona genus (18). All primer pairs within the coding region produced robust products from all additional species. The primer pairs for the 5′-untranslated and flanking region produced the expected products from all but A. muricata, a member of the group most distantly related to A. squamosa (18). This inability to amplify from the most divergent species is consistent with the expected most rapid divergence of this noncoding region of the gene. Thus, most of the primer pairs should be active in all Annona species, negating the possibility that the absence of amplification from Ts results from introgression of INO from another species. Amplification of INO fragments was also performed on DNA from F1 progeny of a cross between wild-type seeded A. cherimola and Ts A. squamosa, and the expected products were produced from DNA from all 181 such atemoya trees. On the basis of these results, we concluded that the INO gene in Ts has been deleted, although the unlikely possibility of multiple internal rearrangements sufficient to prevent amplification of any part of the gene cannot be excluded.
Fig. 4.
INO gene fragments could not be amplified from Ts but were readily amplified from wild-type A. squamosa and five other species. (A) Primers pairs (Table S1) were designed to the A. cherimola INO sequence to enable amplification of the indicated regions. (B) Primer pairs for regions delineated in A, as indicated by corresponding letters at the tops of the gels, were used to amplify sequences from genomic DNA from the species/genotypes indicated below the gels. All primer pairs amplified the expected fragments from wild-type (WT) A. squamosa and from five other species spanning the diversity of this genus, the only exception being the “a” fragment consisting only of 5′-flanking sequences that failed to amplify from A. muricata, one of the species most divergent from A. squamosa (18). These same INO-specific primer pairs failed to amplify fragments from Ts DNA, whereas primers specific to an Annona 1-aminocyclopropane-1-carboxylate synthase gene (ACC) readily amplified the expected fragment from the Ts DNA sample (lane ACC) even when in a reaction that included primers that failed to amplify an INO fragment (Fig. S2). No amplification products were produced when genomic DNA was omitted (lanes No DNA). Rollinia indicates Rollinia emarginata that is phylogenetically embedded within the genus Annona and has been reclassified as a member of this genus (18).
The most parsimonious explanation for the combination of observations of the similarity between the mutant phenotypes of Arabidopsis ino and Ts ovules, the conservation of expression of the INO orthologs between the two species, and the absence/disruption of the INO gene in Ts is that Ts is an ino mutant. Consistent with this, and the expected recessive nature of such a mutation, all analyzed F1 progeny of wild-type A. cherimola and Ts (78 plants) produced flowers with anatropous, bitegmic ovules that developed into normal seeds in the fruit. The fruits were similar to fruits from the same interspecific cross of wild-type plants. Due to the biology of Annona, it will be several years before the next (F2) generation will become reproductively competent to allow for segregation analysis of the seedless trait and the apparent INO gene deletion.
Effects of Ovule Development Defects on Seed and Fruit Formation.
Although the effects on ovule integuments appear similar between Ts A. squamosa and Arabidopsis ino mutants, the two differ in that Ts A. squamosa produces well-developed fruits following pollination whereas fruit development does not occur in Arabidopsis ino (5). In Arabidopsis ino the majority of ovules fail to form embryo sacs, and pollen tubes grow through the transmitting tract but rarely target to ovules (19). In contrast, in Ts A. squamosa, most of the ovules contained fully developed, but sometimes degenerating, embryo sacs at the time of pollen tube arrival (Fig. S3), because only 13% of the ovules (n = 39) did not show fully developed embryo sacs. Pollen tube growth was normal, with pollen tubes reaching the nucelli (Fig. S3). Following pollen tube penetration of the nucellus, development of the embryo was not observed. However, an incipient and early-degenerating endosperm was observed in 32% of the ovules (n = 41) in the Ts flowers (Fig. S3). Thus, in contrast to other species, primarily evolutionarily derived angiosperms (20), the outer integument does not appear to be essential for pollen tube targeting in A. squamosa. This may be related to the fact that the ovules of this species show an endostomal micropyle where the outer integument does not fully cover the inner integument and does not participate in the micropyle (Fig. 2A). Similarly, many other basal angiosperms also exhibit an endostomal micropyle with a markedly asymmetric and hood-shaped outer integument (21), and this has been proposed to be a plesiomorphic trait in angiosperms (9). Consequently, the outer integument and the INO gene could have acquired new functional constraints when the outer integument was recruited into pollen tube attraction and guidance. Interestingly, in both Vitis and Cucurbita (which have seedless varieties), the micropyle is also endostomal. In contrast to the differing roles in pollen tube attraction, in both Arabidopsis and Annona the outer integument appears to support embryo sac and/or embryo development, as also observed in other species (22).
The interruption of the reproductive program in response to the absence of the outer integument appears to occur earlier in Arabidopsis (during embryo sac formation) than in A. squamosa (during embryo and endosperm development). This further progress in the reproductive program, and the more effective targeting of pollen tubes to ovules, could provide sufficient signals for initiation of complete fruit development in A. squamosa. Alternatively, in Arabidopsis, fruit development is actively suppressed in the absence of fertilization by a mechanism that can only be overcome by mutation (7) or hormone treatments (23). This system of suppression may be a recent innovation, and the process of initiation of fruit development may be less stringently regulated in magnoliids, allowing for easier induction of fruit development independent of seed development. Although the simplest explanation for the overall Ts mutant phenotype is that the absence of the outer integument is the root cause of both the failure in seed formation and subsequent development of seedless fruit, it remains possible that these latter effects have a separate genetic cause. As many as four independent genetic changes appear to be necessary for stenospermy in grape (3). Although we show disruption or deletion of the INO locus in Ts, we are unable to estimate the extent of the disrupted/deleted region, and it could be very large. It is possible that loss of INO function is the cause of failure in seed formation, but that disruption or absence of one or more additional proximal genes is necessary for seedless fruit formation. Although we consider the single-gene hypothesis the most likely, only further genetic analysis will allow differentiation of these competing theories.
Conserved Genetic Pathway in Ovule Development.
Our results provide compelling evidence that the critical role of INO genes in ovule development is conserved between Arabidopsis, a crown eudicot, and A. squamosa, a member of the magnoliids—a clade branching early in the evolution of angiosperms (24). In combination with the conservation of the pattern of INO expression in a range of other taxa including Impatiens (8) and Nymphaea (9), this indicates that the critical role for INO in outer-integument formation may be common to all angiosperm groups. Further, the observed correlation between the conservation of gene sequence and expression patterns and the apparent conservation of function helps to support such a hypothesized connection in studies on other genes in other nonmodel systems where sequence and expression conservation are the only available evidence (e.g., 25). Notably, the presence of the outer integument is a synapomorphy of extant angiosperms (barring clearly derived states), differentiating angiosperms from extant gymnosperms (26, 27). Phylogenetic analysis (28) indicates that the divergence of INO from other YABBY genes occurred in the interval separating angiosperms from extant gymnosperms. Thus, the derivation of INO appears to be contemporaneous with the origin of the outer integument and may have been intimately associated with the evolution of this angiosperm-specific structure. The apparent conservation of a single INO function is in contrast to at least one other YABBY gene, CRABS CLAW, which is proposed to have roles in carpel development in a variety of species (25) but also, possibly through multiple neofunctionalizations, has diverse roles in nectary formation (28) and leaf development (29) in some species.
With evidence for the apparent molecular basis of a mutant in the magnoliids, our study presents a unique case of elucidation of the function of a gene through forward genetics in an early-branching angiosperm that is not a grass. Moreover, a potentially agronomically useful manifestation of the particular mutation is the production of seedless fruits. The current popularity of banana fruits derives from the availability of seedless varieties (30), and the presence of this trait in Annona could dramatically increase the popularity of Annona fruits for human consumption. The determination of the molecular basis of the Ts mutant can aid introgression of the trait into commercially desirable varieties by providing a method for identification of homozygous mutant seedless plants before fruit production in this slowly reproducing crop. In addition, the likely conservation of the function of INO in the genetic regulatory pathway of ovule development across angiosperms broadens the implications of these results, making it plausible that this gene could be used in approaches for the production of seedless fruits in a wide variety of angiosperm species (31).
Methods
Plant Growth, Pollination, and Tissue Collection.
Annona cherimola cultivar Campas and A. squamosa wild-type and Ts mutant were grown in the Annona germplasm collection at the Estación Experimental La Mayora, CSIC, Málaga, Spain. Pollination procedures and microscopic preparations were performed as described previously (13). Anthers and pollen of A. cherimola were collected from flowers at anther dehiscence to hand-pollinate Ts A. squamosa stigmas. Flowers of Ts A. squamosa were pollinated at the preanthesis stage at 1800–1900 hours during the first day of the flower cycle, and the flowers were then plugged with cotton to prevent further unwanted pollination. Six gynoecia of each flower were weighed and fixed at daily intervals during the first 3 wk following hand pollination. Three of the six flowers were fixed at each interval in formalin-acetic acid-alcohol (FAA), dehydrated in an ethanol series, and embedded in paraffin wax. Flowers of the 7-y-old F1 progeny (A. cherimola x Ts A. squamosa) were also fixed in FAA, dehydrated in an ethanol series, and embedded in paraffin wax. The remaining three flowers were fixed in 2.5% glutaraldehyde in 0.03 M phosphate buffer (32), dehydrated in an ethanol series, and embedded in Technovit 7100 (Kulzer).
Nucleic Acid Methods.
Genomic DNA was extracted from leaf tissues as described previously (33). INO orthologs from A. squamosa and A. cherimola genomic DNA were isolated as described previously (8). Genomic DNA sequence adjacent to the first isolated fragment in A. cherimola was isolated following the protocol of the Genome Walker Universal Kit (Clontech) using primers GW1.5a, GW1.5b, GW2.5a, GW2.5b, GW3.5a, and GW3.5b for the 5′ region and LMINO5, GW1.3b, GW2.3a, GW2.3b, GW3.3a, and GW3.3b for the 3′ region (Table S1). Total RNA was extracted using the Qiagen RNeasy Plant Mini Kit. cDNA was synthesized using AccuPower RT/PCR PreMix (Bioneer). Polymerase chain reactions were performed by standard methods using BioTaq (Bioline).
Microscopy.
Tissue preparation and in situ hybridization were performed as described previously (34), with the following modification. For antisense INO A. cherimola probe, pGEM-INO A. cherimola was linearized with BamHI and transcribed using T3 RNA polymerase. The slides of in situ hybridization were mounted with Crystal Mount (Fisher Scientific) and photographed under differential interface contrast using a Zeiss Axiophot camera attached to a Zeiss Axioplan microscope. For stained tissue sections, paraffin-embedded Annona material was sectioned at 10 μm. To observe pollen tubes and callose, paraffin-embedded materials were stained with 0.1% aniline blue in 0.1 N PO4K3 (35, 36). Resin-embedded Annona and Arabidopsis (Fig. S3) material was sectioned at 2 μm and stained with a combination of periodic acid-Schiff's reagent and toluidine blue (37) to observe insoluble carbohydrates and nuclei. All Annona materials were photographed under a Leica DM LB2 microscope with epifluorescent UV with an excitation filter 340–380 nm and a barrier filter LP 425 for aniline blue. The Arabidopsis photograph in Fig. 2C was taken using a Zeiss Axioplan microscope. The other Arabidopsis materials (Fig. 2D) were embedded in resin, sectioned, processed, and photographed as described previously (14).
Sequence Analyses.
Alignment and phylogenetic analyses were performed as previously described (8). Sequences of representative YABBY proteins were obtained from GenBank (http://www.ncbi.nlm.nih.gov/BLAST, version 2.2.10) and were aligned using Clustal X version 1.82 (38). Alignments were manually edited to eliminate less conserved regions using Se-Al (39). PAUP* version 4.0b10 (40) was used for phylogenetic analysis of the edited sequence alignments. Heuristic searches under maximum parsimony were performed using an adaptation (41) of the BLOSUM62 amino acid weighting matrix (42), 100 random addition replicates, the tree bisection reconnection algorithm, and saving all trees in each replicate. Support for nodes was determined using 1,000 bootstrap replicates of maximum parsimony heuristic searches with five random sequence addition replicates for each resampling.
Supplementary Material
Acknowledgments
We thank Dior Kelley for technical assistance, Dior Kelley, Kay Robinson-Beers, Jacinto Villanueva, and Olarn Tutawiroon for images, W. E. Friedman for comments on the manuscript, and Yolanda Verdún for help with nucleic acid methods. This work was supported by grants from the Spanish Ministry of Science and Innovation (Project Grants AGL2007-60130/AGR, AGL2009-12621, and AGL2010-2140), Instituto Nacional de Investigación Agraria y Alimentaria (RF2009-00010), GIE-Aragón 43, Junta de Andalucía (FEDER AGR2742), and the European Union under the INCODEV Program (Contract 015100) and from the US National Science Foundation (IOS-0920618). J.L. was supported by grants from Junta de Andalucía and the National Science Foundation Molecular and Organismic Research in Plant History (MORPH) Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU828032, GU828033, HM237356, and HM237357).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014514108/-/DCSupplemental.
References
- 1.Varoquaux F, Blanvillain R, Delseny M, Gallois P. Less is better: New approaches for seedless fruit production. Trends Biotechnol. 2000;18:233–242. doi: 10.1016/s0167-7799(00)01448-7. [DOI] [PubMed] [Google Scholar]
- 2.Darwin C. The Variation of Animals and Plants Under Domestication. London: Murray; 1868. [PMC free article] [PubMed] [Google Scholar]
- 3.Bouquet A, Danglot Y. Inheritance of seedlessness in grapevine (Vitis vinifera L) Vitis. 1996;35:35–42. [Google Scholar]
- 4.Skinner DJ, Hill TA, Gasser CS. Regulation of ovule development. Plant Cell. 2004;16(Suppl):S32–S45. doi: 10.1105/tpc.015933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics. 1997;145:1109–1124. doi: 10.1093/genetics/145.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaiser JC, Robinson-Beers K, Gasser CS. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell. 1995;7:333–345. doi: 10.1105/tpc.7.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivian-Smith A, Luo M, Chaudhury A, Koltunow A. Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development. 2001;128:2321–2331. doi: 10.1242/dev.128.12.2321. [DOI] [PubMed] [Google Scholar]
- 8.McAbee JM, Kuzoff RK, Gasser CS. Mechanisms of derived unitegmy among Impatiens species. Plant Cell. 2005;17:1674–1684. doi: 10.1105/tpc.104.029207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada T, Ito M, Kato M. Expression pattern of INNER NO OUTER homologue in Nymphaea (water lily family, Nymphaeaceae) Dev Genes Evol. 2003;213:510–513. doi: 10.1007/s00427-003-0350-8. [DOI] [PubMed] [Google Scholar]
- 10.Popenoe H. Lost Crops of the Incas: Little Known Plants of the Andes with Promise of Worldwide Cultivation. Washington, DC: Natl Acad Press; 1989. [Google Scholar]
- 11.Schroeder CA. The cherimoya in South Africa. Calif Avocado Soc. 1971;54:129–132. [Google Scholar]
- 12.Twain M. Sacramento Daily Union. October 25, 1866 [Google Scholar]
- 13.Lora J, Hormaza JI, Herrero M. The progamic phase of an early-divergent angiosperm, Annona cherimola (Annonaceae) Ann Bot (Lond) 2010;105:221–231. doi: 10.1093/aob/mcp276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson-Beers K, Pruitt RE, Gasser CS. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell. 1992;4:1237–1249. doi: 10.1105/tpc.4.10.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva JM, et al. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegfried KR, et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- 17.Balasubramanian S, Schneitz K. NOZZLE regulates proximal-distal pattern formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development. 2000;127:4227–4238. doi: 10.1242/dev.127.19.4227. [DOI] [PubMed] [Google Scholar]
- 18.Chatrou LW, et al. Flanking regions of monomorphic microsatellite loci provide a new source of data for plant species-level phylogenetics. Mol Phylogenet Evol. 2009;53:726–733. doi: 10.1016/j.ympev.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Skinner DJ, Gasser CS. Expression-based discovery of candidate ovule development regulators through transcriptional profiling of ovule mutants. BMC Plant Biol. 2009;9:29. doi: 10.1186/1471-2229-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero M. Changes in the ovary related to pollen tube guidance. Ann Bot (Lond) 2000;85:79–85. [Google Scholar]
- 21.Soltis D, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 22.Arbeloa A, Herrero M. Development of the ovular structures in peach. New Phytol. 1991;118:527–533. [Google Scholar]
- 23.Vivian-Smith A, Koltunow AM. Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 1999;121:437–451. doi: 10.1104/pp.121.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angiosperm Phylogeny Group III An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- 25.Fourquin C, Vinauger-Douard M, Fogliani B, Dumas C, Scutt CP. Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc Natl Acad Sci USA. 2005;102:4649–4654. doi: 10.1073/pnas.0409577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endress PK, Igersheim A. Gynoecium structure and evolution in basal angiosperms. Int J Plant Sci. 2000;161:S211–S223. doi: 10.1086/314241. [DOI] [PubMed] [Google Scholar]
- 27.Doyle JA. Seed ferns and the origin of angiosperms. J Torrey Bot Soc. 2006;133:169–209. [Google Scholar]
- 28.Lee JY, et al. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005;132:5021–5032. doi: 10.1242/dev.02067. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi T, et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Ann Bot (Lond) 2007;100:1073–1084. doi: 10.1093/aob/mcm191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinelli F, et al. Gene regulation in parthenocarpic tomato fruit. J Exp Bot. 2009;60:3873–3890. doi: 10.1093/jxb/erp227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabatini DD, Bensch K, Barrnett RJ. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Escribano P, Viruel MA, Hormaza JI. Characterization and cross-species amplification of microsatellite markers in cherimoya (Annona cherimola Mill., Annonaceae) Mol Ecol Notes. 2004;4:746–748. [Google Scholar]
- 34.Kelley DR, Skinner DJ, Gasser CS. Roles of polarity determinants in ovule development. Plant J. 2009;57:1054–1064. doi: 10.1111/j.1365-313X.2008.03752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Currier HB. Callose substance in plant cells. Am J Bot. 1957;44:478–488. [Google Scholar]
- 36.Linskens H. Regarding a specific staining of the transmitting tract in the style and the number of callose plugs following selfing and outcrossing. Naturwissenschaften. 1957;44:16. (in German) [Google Scholar]
- 37.Feder N, O'Brien TP. Plant microtechnique: Some principles and new methods. Am J Bot. 1968;55:123–142. [Google Scholar]
- 38.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rambaut A. Oxford: Department of Zoology, Univ of Oxford; 2004. Se-Al Sequence Alignment Editor. Version 2.0 a11. [Google Scholar]
- 40.Swofford DL. Sunderland, MA: Sinauer; 2004. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.0. [Google Scholar]
- 41.Hill TA, Broadhvest J, Kuzoff RK, Gasser CS. Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GTPase. Genetics. 2006;174:707–718. doi: 10.1534/genetics.106.060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.