Abstract
Two known zebrafish dystrophin mutants, sapje and sapje-like (sapc/100), represent excellent small-animal models of human muscular dystrophy. Using these dystrophin-null zebrafish, we have screened the Prestwick chemical library for small molecules that modulate the muscle phenotype in these fish. With a quick and easy birefringence assay, we have identified seven small molecules that influence muscle pathology in dystrophin-null zebrafish without restoration of dystrophin expression. Three of seven candidate chemicals restored normal birefringence and increased survival of dystrophin-null fish. One chemical, aminophylline, which is known to be a nonselective phosphodiesterase (PDE) inhibitor, had the greatest ability to restore normal muscle structure and up-regulate the cAMP-dependent PKA pathway in treated dystrophin-deficient fish. Moreover, other PDE inhibitors also reduced the percentage of affected sapje fish. The identification of compounds, especially PDE inhibitors, that moderate the muscle phenotype in these dystrophin-null zebrafish validates the screening protocol described here and may lead to candidate molecules to be used as therapeutic interventions in human muscular dystrophy.
Keywords: phosphodiesterase inhibitor, chemical treatment
Muscular dystrophy is a disease in which the muscle forms normally at first but then degenerates faster than it can be repaired. The most common form of muscular dystrophy is Duchenne muscular dystrophy (DMD), representing more than 90% of the diagnosed cases. Mutations in the dystrophin gene were found to be the cause of both DMD and Becker muscular dystrophy (1, 2). Currently, prednisone is the only treatment option available for muscular dystrophy patients in the United States, although there are currently other options through approved clinical trials. Other treatments currently being tested or considered for treating muscular dystrophy include the small molecule PTC124, which promotes read-through of nonsense mutations (3), encouraging muscle development by myostatin down-regulation (4, 5), and the use of oligonucleotides to promote exon skipping to restore dystrophin expression (6).
Recently, a number of chemical and drug screens have been published using zebrafish embryos (7–11). It is possible to quickly produce large numbers of mutant offspring that can then be assayed in multiwell plates and treated with different chemicals to determine if disease progression is modulated. Many of these screens have been highly successful in disease modeling (7) and drug screening (8–10), making the zebrafish ideal for high-throughput whole-organism screening of candidate compounds. Chemical compounds of relatively small molecular weight can bind to specific proteins and alter their function, resulting in nonheritable phenotype changes.
In addition to their suitability for chemical screens, zebrafish also represent a good model to investigate genes involved in muscle development and degeneration, including human muscular dystrophy (12–18). The orthologs of many dystrophin–glycoprotein complex (DGC) components are expressed in zebrafish (19). As such, our laboratory has two available fish models of DMD with dystrophin deficiency, sapje (12) and sapje-like fish (15). The muscle degeneration phenotype in these mutant fish is transmitted in a recessive manner, such that 25% of the offspring show dystrophic features of skeletal muscle after 3 d postfertilization (dpf). This disorder results in a muscle pathology that can be detected by birefringence under polarizing light and usually results in death by 7–9 dpf. The goal of this study was to perform a chemical screen in zebrafish dystrophin mutants to identify potential compounds that might correct the pathology detected by birefringence. In this article, we show the results of screening for candidate chemicals that restore normal muscle structure using the dystrophin-null fish, sapje and sapje-like. The compounds identified impact a number of different pathways, including the recently reported pathway that is modulated by sildenafil (Viagra) (20), and each has the potential to ameliorate the symptoms of muscular dystrophy.
Results
First Screen.
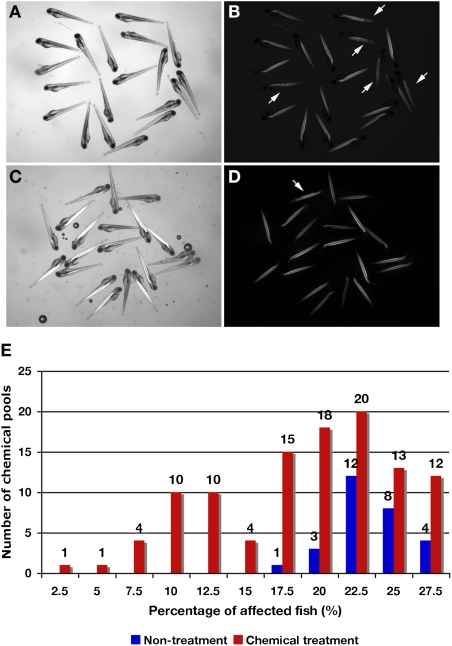
For the first screen (schematically outlined in Fig. S1), 140 chemical pools from a total of 1,120 chemicals in the Prestwick library were tested using embryos from matings of sapje heterozygous pairs. Thirty-two of the compound pools resulted in death of all of the embryos tested, whereas the remaining 108 pools had surviving fish (Table 1). Among the 108 groups that survived treatment, some contained pools of chemicals that seemed to influence the ratio of affected fish relative to normal-seeming fish (Fig. 1 A–D). In the nontreated fish used as controls, the distribution of mutant fish showing abnormal birefringence ranged between 17.5% and 27.5% (Fig. 1E). In the experimental groups tested with the chemical pools, the distribution of the affected fish ranged between 2.5% and 27.5%; 6 groups had less than 7.5% of mutant fish showing abnormal birefringence, and 16 groups had less than 10% (Fig. 1E). The six chemical pools (total of 48 chemicals) that had less than 7.5% of fish showing abnormal birefringence were selected as therapeutic candidates for restoring normal muscle function and were tested further in a secondary screening. The 256 chemicals comprising the 32 pools that caused death of all treated embryos were rescreened individually. However, there was no chemical exhibiting less than 7.5% of mutant fish with abnormal birefringence.
Table 1.
Survival of zebrafish after treatment with pools or individual chemicals from the Prestwick library
| Total | Dead | Alive | |
| Survival in first-screen pools | |||
| Chemical pools | 140 | 32 | 108 |
| Chemicals in Prestwick collection | 1,120 | 256 | 864 |
| Second screening in toxic pools of first-screen pools | |||
| Chemical | 256 | 34 | 222 |
Fig. 1.
Fish at 4 dpf from screening with the Prestwick library on heterozygous sapje fish pairs and the percentage of affected fish after chemical treatment with these chemical pools. Fish with nontreatment (A and B) and chemical treatment (C and D). (A and C) Bright image. (B and D) Birefringence image. In 20 embryos, five apparently affected fish are caused by a homozygous mutation in the zebrafish dystrophin gene, which is inherited in a recessive manner (25%; arrows). In fish treated with chemical pools, only one fish (5%; arrow) shows the muscle phenotype by birefringence. (E) Distribution of the percentage of affected fish treated with chemical pools and untreated fish. A total of 140 chemical pools (1,120 total chemicals) were tested. Treatment with 108 of the chemical pools was not lethal to the fish. Blue bars, untreated fish (n = 28; independent determination). Red bars, chemically treated fish with 108 chemical pools (in duplicate). Some chemical pools decreased the percentage of affected fish showing abnormal birefringence compared with untreated fish and might contain a compound able to restore normal muscle structure.
Second Screen.
In the second screen, pools were separated into individual chemicals for a total 48 molecules and tested to identify which individual compound influenced the ratio of fish showing abnormal birefringence. In this secondary screen, seven chemicals influenced muscle structure by decreasing the percentage of affected fish using both sapje (used in the first screen) and sapje-like fish, which has an independent dystrophin mutation (Table 2). These seven chemicals were divided into different groups: an antiinflammatory agent (chemical 1, epirizole), antiallergic agents (chemicals 2 and 3, homochlorcyclizine-dihydrochloride and conessine), a phosphodiesterase (PDE) inhibitor (chemical 4, aminophylline), an estradiol steroid (chemical 5, equilin), a chelating agent (chemical 6, pentetic acid), and a cardiotonic glycoside (chemical 7, proscillaridin A) as shown in Table 2.
Table 2.
List of candidate chemicals that influence the number of affected zebrafish in heterozygote dystrophin-deficient matings, with the percentage of affected fish indicated for each different mutant allele
| No. | Chemical name | Sapje (%) | Sapje-like (%) | MW | ||
| 1 | Epirizole | 10 | 15 | C11H14N4O2 | 234.25983 | Anti-inflammatory agent |
| 2 | Homochlorcyclizine dihydrochloride | 10 | 12.5 | C19H25Cl3N2 | 387.7835 | Anti-allergic agent |
| 3 | Conessine | 10 | 12.5 | C24H40N2 | 356.5998 | Anti-allergic agent |
| 4 | Aminophylline | 7.5 | 17.5 | C16H24N10O4 | 420.43428 | Nonselective PDE inhibitor |
| 5 | Equilin | 5 | 10 | C18H20O2 | 268.3589 | Group of steroid compounds |
| 6 | Pentetic acid | 7.5 | 10 | C14H23N3O10 | 393.35351 | Chelating agents |
| 7 | Proscillaridin A | 10 | 10 | C30H42O8 | 530.66444 | Cardiotonic agents |
MW, molecular weight.
Genotyping and Expression of Dystrophin in the Chemically Treated Fish.
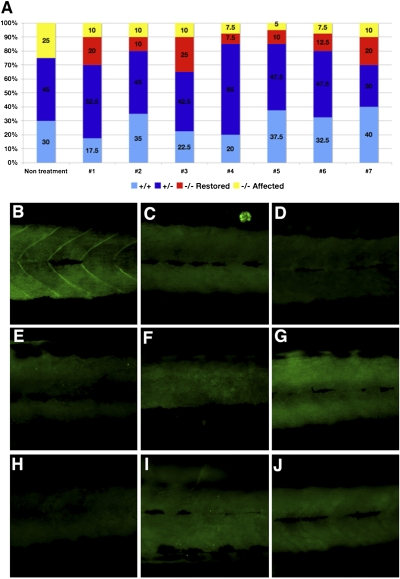
For all surviving fish treated with each of the seven compounds and showing apparent corrective results in the second screen, genotypes were determined using the genomic DNA extracted from heads of the individual fish (40 treated fish with each chemical). In the nontreated group of 4 dpf sapje heterozygous pair offspring, the resulting genotype percentages were 30% WT, 45% heterozygous, and 25% homozygous for dystrophin deficiency. All unaffected fish as detected by the birefringence assay were confirmed to be WT or heterozygous fish by genotyping. In the treated groups, some homozygous dystrophin-null fish had normal birefringence and thus, were considered phenotypically unaffected (Fig. 2A). Similar results were obtained in sapje-like fish, where some of the homozygous dystrophin-null fish had apparently normal birefringence reduction (Fig. S2). These results indicate that some of the dystrophin-null fish treated throughout the first 4 d of development failed to develop the abnormalities seen in some of their clutch mates.
Fig. 2.
Genotype and dystrophin expression of treated fish with seven candidate chemicals from the longer screen. (A) Genotyping of each of 40 fish treated with each of the seven potential candidate chemicals. Yellow, dystrophin-null affected fish (abnormal birefringence). Red, dystrophin-null unaffected fish (normal birefringence). Blue, heterozygous fish (normal birefringence). Light blue, WT fish (normal birefringence). In each of the seven chemically treated wells, some fish show normal birefringence despite genotyping results that show dystrophin-null fish. (B–J) Immunostaining with an anti-dystrophin antibody. (B) WT. (C) Dystrophin-null fish. (D–J) Dystrophin immunostaining of fish with restored muscle structure with each of the seven chemicals (chemicals 1–7 in Table 2). All restored fish with chemicals 1–7 showed no expression of dystrophin, similar to those of untreated dystrophin-null fish (C).
The expression of dystrophin was examined by immunostaining individual fish bodies with anti-dystrophin antibodies. WT fish showed positive staining of dystrophin in the myosepta (Fig. 2B). However, the homozygous dystrophin-null fish that showed no abnormality of birefringence showed no immunoreactivity with anti-dystrophin, and the same lack of staining was found in the nontreated dystrophin-null fish (Fig. 2C). Those treated dystrophin-null fish with normal birefringence did not restore dystrophin expression (Fig. 2 D–J).
Testing Candidate Chemicals from the Chemical Screen Using Dystrophin Morphants.
Six nanograms antisense morpholino oligonucleotides (MO) targeted to interfere with fish dystrophin translation were injected into one to two cell-stage WT embryos. The injected embryos showed reduced birefringence similar to that found in the dystrophin-null mutant sapje and sapje-like fish observed at 4 dpf. These embryos also showed markedly reduced expression of dystrophin in their myosepta. These morphants were used to confirm the effects of the seven candidate chemicals. Without chemical treatment, 28% of the morphants showed abnormal birefringence. However, treatment with each of the seven chemicals reduced the percentages of affected fish showing abnormal birefringence (Fig. S3).
Long-Term Culture Fish with Candidate Chemicals.
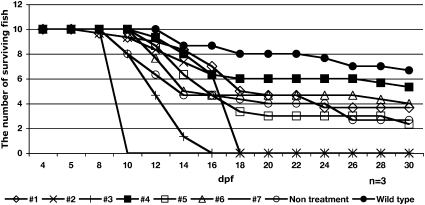
To determine if the treatment with a single compound could reverse the skeletal muscle phenotype after it was already present, affected fish (selected at 4 dpf by birefringence assay) were treated with the seven individual candidate chemicals (Table S1) from 4 to 30 dpf. The fish treated with chemical 4 were able to survive longer compared with the nontreated dystrophin-null fish (Fig. 3). Fish treated with chemicals 1 or 6 were able to survive for 30 d (Fig. 3). Other chemicals tested had no effect on the survival of affected fish (Fig. 3), and indeed, some were toxic to all fish. The motility of surviving affected fish was not relatively altered compared with that of WT.
Fig. 3.
Long-term treatment of dystrophin-null fish selected to have abnormal birefringence at 4 dpf with chemicals 1–7 from 4 to 30 dpf (n = 3). At 30 d, the average number of surviving fish is greater in chemical 4 (■) aminophylline-treated dystrophin-null fish compared with those in untreated dystrophin-null fish (○). Treated fish with chemicals 1 (◇), 2 (X), 3 (+), 4 (■), 5 ( ), 6 (△), 7 (—), and WT (●) are shown. Three chemicals proved toxic to zebrafish after prolonged incubation.
), 6 (△), 7 (—), and WT (●) are shown. Three chemicals proved toxic to zebrafish after prolonged incubation.
Twenty embryos from sapje heterozygous fish mates were incubated with each of the seven candidate chemicals from 1 to 30 dpf in triplicate. At 30 dpf, only the fish treated with three chemicals, 1, 4, and 6, had survived (Fig. S4). Interestingly, more fish treated with chemical 4 were able to survive compared with control fish. The fish treated with chemicals 1 and 6 were almost the same as control. The remaining four chemicals groups resulted in death of all three strains (+/+, +/−, and −/− dystrophin) (Fig. S4).
Recovered Skeletal Muscle of Dystrophin-Null Fish with Chemical 4, Aminophylline Treatment.
Fish treated with chemical 4 that survived to 30 dpf were sectioned, and they were stained by H&E and immunostained with anti-dystrophin, laminin, and myosin heavy-chain antibodies.
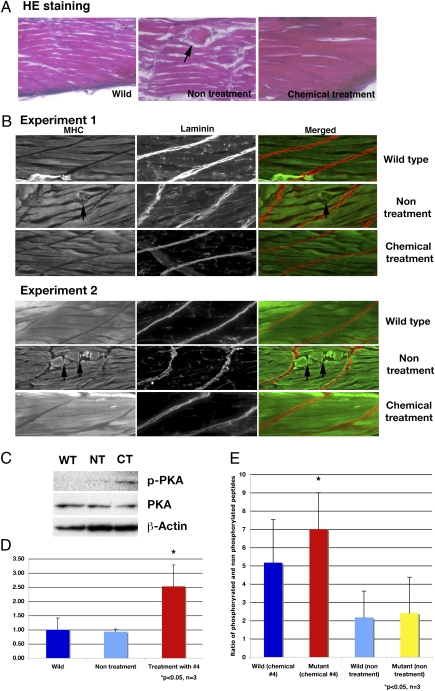
H&E staining showed that fish treated with chemical 4 had skeletal muscle structure restored to that similar to those of WT fish. In nontreated dystrophin-null fish, there was disorganized muscle structure (Fig. 4A Center, arrow).
Fig. 4.
Analysis of skeletal muscle of dystrophin-null fish that survive 30 d after treatment with chemical 4, aminophylline. (A) H&E staining of WT, nontreated, and chemical 4-treated fish. Nontreated dystrophin-null fish have disorganized structure (arrow) in skeletal muscle. The dystrophin-null fish with treated aminophylline have normal structure in skeletal muscle. (B) Immunostaining of WT, nontreated, and chemical 4-treated fish with anti-myosin heavy chain and anti-laminin antibody. Nontreated dystrophin-null fish have broken and disturbed structure (arrows) of skeletal muscle fibers. The treated dystrophin-null fish with aminophylline have normal structure in skeletal muscle. (C) Immunoblot of phosphorylated PKA and PKA at 30 dpf in surviving fish. WT, wild type; NT, nontreatment of dystrophin-null fish; CT, chemical 4-treated dystrophin-null fish. (D) Ratio of phosphorylated PKA and PKA in 30 dpf surviving fish (n = 3). Blue, WT; light blue, nontreatment of dystrophin-null fish; red, chemical 4-treated dystrophin-null fish. *P < 0.05 (vs. WT and nontreatment groups). Error bars indicate SDs. (E) Assay of cAMP-dependent PK activity (n = 3). Bars show the ratio of phosphorylated peptides and nonphosphorylated peptide. Blue, WT fish treated with chemical 4 (2.5 μg/mL) for 30 d. Red, affected sapje fish treated with chemical 4 (2.5 μg/mL). Light blue, nontreated WT fish. Yellow, nontreated affected sapje fish. *P < 0.05 (vs. nontreated WT and mutant fish). Error bars indicate SDs.
In the WT fish, myosin heavy chain was clearly expressed and highlighted that muscle fibers were normal (Fig. 4B). Interestingly, myosin heavy-chain staining of treated dystrophin-null fish, which survived until 30 dpf, showed normal myofiber structure, similar to that of the WT fish, whereas untreated dystrophin-null fish showed clear abnormalities of muscle (Fig. 4B Center). In the untreated dystrophin-null fish, the muscle fiber structure was disturbed, and some parts were abnormal (Fig. 4B, arrows). These results suggest that treatment with chemical 4 restored the muscle structure of these dystrophin-null fish, because they had abnormal structure when selected at 4 dpf before chemical treatment. This restoration of normal structure was found in each treated fish that survived to 30 d (n = 12).
PKA Expression and Activity Was Up-Regulated by Treatment with Chemical 4, Aminophylline.
Aminophylline is known to be a nonselective PDE inhibitor that increases the levels of intercellular cAMP, causing activation of cAMP-dependant PKA. To this end, the expression, phosphorylation, and activation of PKA in dystrophin-null fish treated with aminophylline for 25 d were examined. The expression of PKA was detected in all samples. More phosphorylated PKA was detected in aminophylline-treated fish compared with WT and untreated dystrophin-null fish (Fig. 4 C and D, P < 0.05). Moreover, the activity of PKA in aminophylline-treated fish was significantly increased compared with WT and untreated dystrophin-null fish (P < 0.05 in Fig. 4E). These results show that activated phosphorylated PKA and the activity of PKA were increased in aminophylline-treated fish compared with WT and untreated dystrophin-null fish, which suggests indirectly that intracellular cAMP is increased with aminophylline treatment.
Treatment of sapje Fish with a Series of PDE Inhibitors.
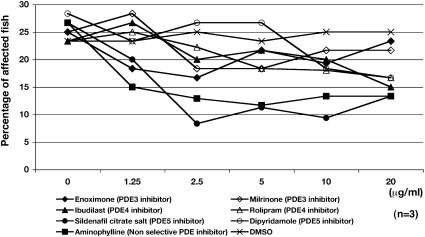
Aminophylline is a nonselective PDE inhibitor, and it is among a group of PDE inhibitors with different specificities (Table S1). To test whether other PDE inhibitors might also ameliorate dystrophic symptoms in sapje fish, 20 embryos from matings of heterozygous sapje fish were treated with a series of PDE inhibitors from 1 to 4 dpf (in triplicate). At 4 dpf, the percentage of affected fish was examined by birefringence assay. The results are plotted (Fig. 5): enoximone (PDE3 inhibitor), milrinone (PDE3 inhibitor), ibudilast (PDE4 inhibitor), rolipram (PDE4 inhibitor), sildenafil citrate salt (PDE5 inhibitor), dipyridamole (PDE5 inhibitor), aminophylline (nonselective PDE inhibitor), and DMSO (vehicle). Interestingly, sildenafil citrate, a PDE5 inhibitor (Fig. 5, closed circle), strongly decreased the percentage of fish showing abnormal birefringence, similar to that of chemical 4 (aminophylline) (Fig. 5, closed square). Other PDE5 and PDE4 inhibitors also had an effect in high concentrations (20 μg/mL). However, PDE3 inhibitors, enoximone and milrinone (Fig. 5, closed and opened diamonds), exhibited no influence on the percentage of affected sapje fish. Sildenafil citrate salt has previously been shown by others to influence the phenotype of mdx mice (21).
Fig. 5.
Treatment of dystrophin-null fish with a series of different PDE inhibitors. Twenty embryos from matings of heterozygous sapje fish are treated with a series of PDE inhibitors from 1 to 4 dpf (in triplicate at varying concentrations). At 4 dpf, the percentage of fish exhibiting abnormal muscle structure as examined by birefringence is shown. Enoximone (PDE3 inhibitor; ◆), milrinone (PDE3 inhibitor; ◇), ibudilast (PDE4 inhibitor; ▲), rolipram (PDE4 inhibitor; △), sildenafil citrate salt (PDE5 inhibitor; ●), dipyridamole (PDE5 inhibitor; ○), aminophylline (nonselective PDE inhibitor; ■), and DMSO (vehicle; X). Both aminophylline and sildenafil citrate clearly decreased the percentage of dystrophin-null fish showing abnormal birefringence.
Discussion
Fish with mutations in the zebrafish dystrophin gene (sapje and sapje-like mutants) are good models for studies of DMD (12, 15). Moreover, they are ideally suited for use in chemical screens to select drug candidates capable of correcting the muscle phenotype. Their muscle phenotypes are easily detectable by a highly accurate birefringence assay, eliminating the need for sacrifice of chemically treated fish before studies are completed. These fish are also small enough to be permeable to small molecules and can be assayed in large numbers. The small molecules used here were from the Prestwick library of bioreaticve human-approved use compounds. Using a two-tiered screening strategy, first, pooling compounds and then, screening individual compounds, resulted in identification of seven individual chemicals that decreased the percentage of phenotypically affected fish, which suggests that these seven chemicals might rescue the muscle phenotype found in the dystrophin-null fish.
Each of the seven chemicals increased the percentage of fish with normal birefringence. Genotyping of dystrophin mutations in these treated fish indicated that dystrophin-null fish were among those with normal birefringence. This suggests that these seven chemicals can prevent the onset of abnormal muscle structure in these dystrophin-null fish and result in an apparently normal fish with a dystrophin mutation. The immunohistochemical results of the dystrophin expression showed that the chemical treatment did not restore dystrophin expression. Recent reports propose that some chemicals may cause exon skipping in mutant dystrophin cDNA and recover the expression of truncated dystrophin (5). However, the seven chemicals examined here did not restore dystrophin expression, which suggests that expression of other proteins might be influenced by treatment with these seven chemicals and that they may be the potential therapeutic pathways. These pathways may also have additional compounds that influence their expression.
The muscle structure of aminophylline-treated dystrophin-null fish appeared normal at 30 dpf compared with that in dystrophin-null fish without chemical treatment (Fig. 4 A and B). These results indicate that action of these chemicals may be able to restore dystrophin-null muscle to normal muscle without restoring the dystrophin expression. Aminophylline, a nonselective PDE inhibitor, has been traditionally used for the treatment of asthma. It has been shown that aminophylline has numerous antiinflammatory effects, including the inhibition of inflammatory mediators and activation of NF-κB (22–25). Previously, other groups have reported that a PDE5 inhibitor restores mdx mouse muscle to normal (20, 21, 26). It is generally known that PDE inhibitors cause an increase in intracellular cAMP and/or cGMP. One of pathways up-regulated by increasing the amount of cAMP is the PKA pathway (27). Our results indicate that the activity of PKA is clearly up-regulated in aminophylline-treated dystrophin-null fish. PKA has some interesting target proteins for supporting muscle structure, such as cAMP response element-binding (CREB) (28), and skeletal muscle Ca2+ channels (29). Up-regulation of expression of these target proteins by PKA activation might modulate the progression of phenotypes in skeletal muscle.
We have already looked into other PDF inhibitors through our identification of aminophylline in this screen as targets for therapy. Two of these inhibitors, sildenafil and tadalafil, have been independently identified by others (20, 26), thus validating our screening strategy as a way of finding pathways that might influence the skeletal muscle abnormalities in dystrophin-null fish. The other compounds identified in our screen target additional pathways and represent additional targets for therapy. These can be easily characterized in our zebrafish model of DMD.
This analysis may indicate common and specific pathways of action of these candidate drugs. These candidate drugs will be tested in other strains of muscular dystrophy in laminin-deficient zebrafish. We have also shown the usefulness of the dystrophin-null zebrafish in screens of compounds that might ameliorate symptoms. Thousands more compounds are now available for further screening, something we have already started.
Materials and Methods
Fish and Fish Culture.
Two fish models of DMD with dystrophin deficiency used for chemical screening were the sapje (stop codon in exon 4) and sapje-like (splice site mutation in exon 62) (12, 15) mutants. The muscle degeneration phenotype in these mutant fish is transmitted in a recessive manner such that 25% of the offspring show degenerative muscle symptoms after 3 dpf (Fig. 1A). Twenty pairs of heterozygous sapje fish or sapje-like mutants were mated, and fertilized eggs were cultured at 28.5 °C. Zebrafish embryos were collected and raised at 28.5 °C according to standard procedures (30) and standard criteria (31) under the guidelines of our Institutional Animal Care and Use Committee.
Small Molecular Library.
The Prestwick chemical library (Harvard Institute of Chemistry and Cell Biology) was used as the source of small molecules for these experiments. This library contains 1,120 small molecules composed of 90% marketed drugs and 10% bioactive alkaloids or related substances. The active compounds in the library were selected for their high chemical and pharmacological diversity, as well as their known bioavailability and safety in humans.
Detection of Muscle Phenotype by Birefringence Assay.
Abnormal birefringence of muscle was observed and analyzed by placing anesthetized embryos on a glass-polarizing filter and subsequently covering them with a second polarizing filter as described in ref. 18.
Screening the Prestwick Library for Compounds That Decrease the Percentage of Affected Zebrafish.
The library was first screened in pooled groups of eight compounds in duplicate as described in SI Text (Fig. S1). After the identification of pools that decreased the number of affected fish, each individual compound in these pools was screened as outlined in Fig. S1.
Test of Candidate Chemicals in Dystrophin Morphant Fish Treated with Antisense MO Injection.
Antisense MO (Gene Tools) targeted to interfere with fish dystrophin translation was designed using the 5′ sequence around the putative translation start site of the zebrafish dystrophin mRNA (14). The morpholino sequences for dystrophin were MO: 5′-TTGAGTCCTTTAATCCTACAATTTT-3′; 6 ng morpholinos were injected into the yolk of one- to two-cell stage embryos as described in ref. 18. At 1 dpf, 20 injected embryos were arrayed in a 24-well plate and were treated with individual chemicals (1–7).
Histology and Immunohistochemistry.
Immunohistochemical staining of whole-mutant and morphant fish bodies was done as described in ref. 18. Embryos were incubated separately with anti-dystrophin (1:25; Sigma) or anti-laminin (1:25; Sigma) antibodies at 4 °C overnight. For immunohistochemical staining of fish muscle sections, fish were frozen in a cold acetone. Fish muscle samples were sectioned with a cryostat (HM505E; Microm) at a 10-μm thickness. Fish muscle sections were incubated with anti-dystrophin (1:25; Sigma), anti-myosin heavy chain (1:25, F59; Hybridoma Bank), or anti-laminin (1:25; Sigma) antibodies at 4 °C overnight. After washing three times, sections were incubated with secondary antibodies and examined as described in ref. 18.
Frozen sections were obtained as for immunohistochemistry and stained with H&E. For H&E staining of frozen muscle sections, sections were stained with Mayer’s hematoxylin solution (Fluka) for 10 min and 1% eosin solution (Sigma) for 30 s after washing with water.
Long-Term Treatment of sapje Fish with Candidate Chemicals.
Pairs of heterozygous sapje fish were mated, and fertilized eggs were cultured at 28.5 °C. Zebrafish embryos were collected and raised at 28.5 °C according to standard procedures and criteria. For long-term treatment of dystrophin-deficient fish, mutant fish showing abnormal birefringence were identified under the dissection scope at 4 dpf and placed in a new plate to be treated with candidate chemicals 1–7 from 4 to 30 dpf; 10 affected and 10 unaffected embryos were arrayed in 24-well plates and cultured in 1 mL fish water containing individual chemicals at 28.5 °C starting at 4 dpf. At 5 dpf, treated fish were cultured at room temperature in 100 mL fish water containing individual chemicals (2.5 μg/mL) until 30 dpf at room temperature. The number of surviving fish was counted and recorded every other day, and at 30 dpf, their genotypes were determined.
For long-term treatment of fish from 1 to 30 dpf, WT, heterozygous, and dystrophin-null fish from heterozygous fish matings were treated with candidate chemicals 1–7. The 20 embryos were arrayed in 24-well plates and cultured in 1 mL fish water containing individual chemicals at 1 dpf. At 5 dpf, they were cultured at room temperature in 100 mL fish water containing individual chemical (2.5 μg/mL) and monitored to 30 dpf. The number of surviving fish at every other day intervals was determined, and the genotypes of surviving fish were determined at 30 dpf.
Genotyping sapje and sapje-Like Fish.
Genomic DNA extracted from chemically treated fish was used as the PCR template. With primer sets for genotyping the specific mutations in the dystrophin gene of sapje or sapje-like fish (for sapje: forward primer 5′-CTGGTTACATTCTGAGAGACTTTC-3′ and reverse primer 5′-AGCCAGCTGAACCAATTAACTCAC-3′; for sapje-like: forward primer 5′-TCTGAGTCAGCTGACCACAGCC-3′ and reverse primer 5′- ATGTGCCTGACATCAACATGTGG -3′), PCR was performed at 52 °C with 35 cycles. The Molecular Genetics Core Facility at Children’s Hospital Boston sequenced PCR products after PCR purification using a standard kit (Qiagen).
Western Blotting.
Embryos or fish were homogenized in Tris-buffered saline (TBS) containing 4 M Urea, 2% SDS, 2% CHAPS, protease inhibitors, and phosphatase inhibitors (Roche). Proteins were analyzed with Western blot as described in ref. 18. Blotted proteins were incubated with primary antibody, anti-PKA C (1:100; Cell Signaling Technology), anti-phospho-PKA C (Thr197, 1:100; Cell Signaling Technology), or anti–β-actin (1:500; Sigma).
PKA Assay.
To assay the activity of PKA, the PepTag Assay for Non-Radioactive Detection of cAMP-Dependent Protein Kinase (Promega) was used following the manufacturer’s protocol. Extracted samples were incubated with PepTag A1 peptide and buffer and separated by eletrophoresis on 0.8% agarose gels. After excising the negatively charged phosphorylated and positively charged nonphosphorylated bands from the gel, the gels were melted at 95 °C. To compare the ratio of phosphorylated peptide to nonphosphorylated peptide among samples, the concentrations were measured at 570 nm by nanodrop after the addition of gel solubilization solution.
Supplementary Material
Acknowledgments
We acknowledge members of the L.M.K. laboratory for their experimental advice and help in performing these experiments and drafting this manuscript. In addition, we are grateful to Chris Lawrence and Jason Best, who managed our fish facility, and the Institute of Chemistry and Cell Biology at Harvard Medical School. L.M.K. was an investigator with The Howard Hughes Medical Institute. This work was supported by National Institute of Neurological Disorders and Stroke Grant 5P50NS040828-09 and the Muscular Dystrophy Association. All sequencing was accomplished in the Intellectual and Developmental Disabilities Research Center Molecular Core Laboratory supported by National Institute of Child Health and Human Development Grant 2P30HD018655-26.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1102116108/-/DCSupplemental.
References
- 1.Monaco AP, et al. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 2.Burghes AH, et al. A cDNA clone from the Duchenne/Becker muscular dystrophy gene. Nature. 1987;328:434–437. doi: 10.1038/328434a0. [DOI] [PubMed] [Google Scholar]
- 3.Welch EM, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 4.Bogdanovich S, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 5.Lu QL, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 6.van Deutekom JC, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 7.Dooley K, Zon LI. Zebrafish: A model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 8.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 9.Pichler FB, et al. Chemical discovery and global gene expression analysis in zebrafish. Nat Biotechnol. 2003;21:879–883. doi: 10.1038/nbt852. [DOI] [PubMed] [Google Scholar]
- 10.Stern HM, et al. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1:366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- 11.Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci USA. 2000;97:12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassett DI, Currie PD. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum Mol Genet. 2003;12:R265–R270. doi: 10.1093/hmg/ddg279. [DOI] [PubMed] [Google Scholar]
- 13.Guyon JR, et al. Delta-sarcoglycan is required for early zebrafish muscle organization. Exp Cell Res. 2005;304:105–115. doi: 10.1016/j.yexcr.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Guyon JR, et al. The dystrophin associated protein complex in zebrafish. Hum Mol Genet. 2003;12:601–615. [PubMed] [Google Scholar]
- 15.Guyon JR, et al. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum Mol Genet. 2009;18:202–211. doi: 10.1093/hmg/ddn337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore CJ, Goh HT, Hewitt JE. Genes required for functional glycosylation of dystroglycan are conserved in zebrafish. Genomics. 2008;92:159–167. doi: 10.1016/j.ygeno.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Thornhill P, Bassett D, Lochmüller H, Bushby K, Straub V. Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP) Brain. 2008;131:1551–1561. doi: 10.1093/brain/awn078. [DOI] [PubMed] [Google Scholar]
- 18.Kawahara G, Guyon JR, Nakamura Y, Kunkel LM. Zebrafish models for human FKRP muscular dystrophies. Hum Mol Genet. 2010;19:623–633. doi: 10.1093/hmg/ddp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steffen LS, et al. Zebrafish orthologs of human muscular dystrophy genes. BMC Genomics. 2007;8:79. doi: 10.1186/1471-2164-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamo CM, et al. Sildenafil reverses cardiac dysfunction in the mdx mouse model of Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 2010;107:19079–19083. doi: 10.1073/pnas.1013077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi YM, et al. Sarcolemma-localized nNOS is required to maintain activity after mild exercise. Nature. 2008;456:511–515. doi: 10.1038/nature07414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnes PJ. Theophylline: New perspectives for an old drug. Am J Respir Crit Care Med. 2003;167:813–818. doi: 10.1164/rccm.200210-1142PP. [DOI] [PubMed] [Google Scholar]
- 23.Luo WJ, Ling X, Huang RM. Effects of aminophylline on cytokines and pulmonary function in patients undergoing valve replacement. Eur J Cardiothorac Surg. 2004;25:766–771. doi: 10.1016/j.ejcts.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan P, et al. Anti-inflammatory effects of low-dose oral theophylline in atopic asthma. Lancet. 1994;343:1006–1008. doi: 10.1016/s0140-6736(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 25.Mahomed AG, Theron AJ, Anderson R, Feldman C. Anti-oxidative effects of theophylline on human neutrophils involve cyclic nucleotides and protein kinase A. Inflammation. 1998;22:545–557. doi: 10.1023/a:1022306328960. [DOI] [PubMed] [Google Scholar]
- 26.Asai A, et al. Primary role of functional ischemia, quantitative evidence for the two-hit mechanism, and phosphodiesterase-5 inhibitor therapy in mouse muscular dystrophy. PLoS ONE. 2007;2:e806. doi: 10.1371/journal.pone.0000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willoughby D, Wong W, Schaack J, Scott JD, Cooper DM. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 2006;25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen AE, Ginty DD, Fan CM. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BD, Scheuer T, Catterall WA. Convergent regulation of skeletal muscle Ca2+ channels by dystrophin, the actin cytoskeleton, and cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:4191–4196. doi: 10.1073/pnas.0409695102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusslein-Volhard C, Dahm R. Zebrafish: A Practical Approach. Oxford: Oxford University Press; 2002. [Google Scholar]
- 31.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.