Abstract
Background
Mediastinal gray zone lymphoma is a newly recognized entity with transitional morphological and immunophenotypic features between the nodular sclerosis subtype of Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma. Diagnostic criteria for mediastinal gray zone lymphoma are still challenging, and the optimal therapy is as yet undetermined. Epigenetic changes have been implicated in the loss of the B-cell program in classical Hodgkin’s lymphoma, and might provide a basis for the immunophenotypic alterations seen in mediastinal gray zone lymphoma.
Design and Methods
We performed a large-scale DNA methylation analysis of microdissected tumor cells to investigate the biological underpinnings of mediastinal gray zone lymphoma and its association with the related entities classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma, making comparisons with the presumptively less related diffuse large B-cell lymphoma.
Results
Principal component analysis demonstrated that mediastinal gray zone lymphoma has a distinct epigenetic profile intermediate between classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma but remarkably different from that of diffuse large B-cell lymphoma. Analysis of common hypo- and hypermethylated CpG targets in mediastinal gray zone lymphoma, classical Hodgkin’s lymphoma, primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma was performed and confirmed the findings of the principal component analysis. Based on the epigenetic profiles we were able to establish class prediction models utilizing genes such as HOXA5, MMP9, EPHA7 and DAPK1 which could distinguish between mediastinal gray zone lymphoma, classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma with a final combined prediction of 100%.
Conclusions
Our data confirm a close relationship between mediastinal gray zone lymphoma and both classical Hodgkin’s lymphoma and primary mediastinal large B-cell lymphoma. However, important differences were observed as well, allowing a clear distinction from both parent entities. Thus, mediastinal gray zone lymphoma cannot be assigned to either classical Hodgkin’s lymphoma or primary mediastinal large B-cell lymphoma, validating the decision to create an intermediate category in the World Health Organization classification.
Keywords: mediastinal gray zone lymphoma, classical Hodgkin’s lymphoma, primary mediastinal large B-cell lymphoma, epigenetics, microdissection
Introduction
The term “gray zone lymphoma” was introduced for the first time in the proceedings of the “Workshop on Hodgkin’s disease and related diseases” in 1998 when it was used to define cases of malignant lymphoma which could not be classified with certainty as either Hodgkin’s lymphoma or B-cell non-Hodgkin’s lymphoma.1 This diagnostic gray zone is of theoretical interest, but also represents a practical problem, as the treatment approaches for Hodgkin’s lymphoma traditionally differ from those of aggressive B-cell lymphomas.
Recent data suggest that the malignant cell of classical Hodgkin’s lymphoma (CHL) is a B cell.2 If CHL is derived from an altered B-lymphocyte, it is not surprising that areas of overlap with B-cell non-Hodgkin’s lymphoma should occur biologically as well as clinically. CHL may be seen as a sequential or composite lymphoma in association with a variety of B-cell lymphomas, including follicular lymphoma, chronic lymphocytic leukemia, and diffuse large B-cell lymphoma (DLBCL).3 However, this occurrence is usually not of clinical concern, as the differential diagnosis between these malignancies is generally straightforward.
A neoplasm of greater clinical concern is mediastinal gray zone lymphoma (MGZL), first described by Traverse-Glehen et al. in 2005.4 MGZL underscores the close relationship between primary mediastinal large B-cell lymphoma (PMLBCL) and CHL, nodular sclerosis (CHLNS) of the mediastinum, and demonstrates transitional features, morphologically and phenotypically, between PMLBCL and CHLNS.5 In 2008, this category of disease was included in the World Health Organization (WHO) classification as ‘B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and CHL’.6
Clinically, PMLBCL and CHLNS share a number of common characteristics. They both present frequently with an anterior mediastinal mass involving the thymus gland and supraclavicular lymph nodes in young women. Instances of composite lymphoma and sequential occurrences of PMLBCL and CHLNS have also been reported.4,7–9 MGZL has a similar clinical presentation, but, interestingly, is more common in males than in females.4,10
From a biological perspective, gene expression profiling studies demonstrated differences between PMLBCL and non-mediastinal DLBCL but similarities to CHLNS.11–13 A number of common genetic aberrations in PMLBCL and CHL further underscore the close relationship of these types of lymphomas. PMLBCL show frequent gains of gene regions on chromosomes 9p and 2p, which have also been described in CHL but are only rarely detected in DLBCL.14–18
The exact molecular mechanisms responsible for the transformation of a B cell to a Hodgkin and Reed-Sternberg cell are not fully understood. Recent studies have suggested that down-regulation of the B-cell program in CHL may be responsible for tumorigenesis and controlled at the epigenetic level.19–22 MGZL represent a unique resource to study this question, since the cells appear to have the capacity to undergo re-programming of their phenotype during the disease course. Cases of CHL have recurred as PMLBCL and the opposite sequence has also been seen.4
The goal of this study was to derive a large scale DNA methylation profile inclusive of high-density and low-density CpG genome space of MGZL in comparison with CHLNS and PMLBCL. The study cohort was further compared with cases of DLBCL and germinal center B cells from reactive tonsils (RTB), and five representative lymphoma cell lines.
Design and Methods
Tissue and cell specimens
The study cohort consisted of CHLNS (10 cases), PMLBCL (10 cases), and 10 cases of MGZL or mediastinal composite/sequential lymphoma with components of CHLNS and PMLBCL, respectively. All cases were retrieved from the National Cancer Institute (NCI) Laboratory of Pathology (Table 1). The study was approved by the NCI Institutional Review Board including a waiver of written consent. Histopathological diagnoses were made by the authors (ESJ or SP) according to WHO criteria.6 MGZL was defined as a mediastinal lymphoma with discordance between morphology and immunopheno-type precluding classification as PMLBCL or CHLNS.4 Four cases morphologically resembled CHL but had a phenotype of PMLBCL (CD20++, CD15−). One case presented as CHLNS and had concurrent PMLBCL. Only the PMLBCL component was available for analysis. Four cases more closely resembled PML-BCL but had a phenotype closer to CHL, with expression of CD30, CD15, or loss of CD20, CD79a. One case of mediastinal composite lymphoma comprised two sharply delineated components of PMLBCL and CHLNS, both of which were separately examined. DLBCL (10 cases) and RTB (3 cases) were included as controls. DLBCL were classified by two of the authors (FCE and SP) as germinal center B-cell-like or activated B-cell-like based on the immunophenotypic profile, as previously validated in our laboratories by comparison with gene expression profiling (Table 1).23,24 All selected cases were preserved as formalin-fixed paraffin-embedded tissue specimens. Sections were stained with anti-CD20 (clone L26, Dako, Denmark) or with anti-CD30 (clone Ber-H2, Dako, Denmark) antibodies for immuno-guided laser-assisted microdissection as described previously (Table 1).25 As a validation of the successful isolation of tumor cells by laser-assisted microdissection, we compared the epigenetic profiles of DNA isolated by this method of microdissection with those of DNA obtained from whole tissue sections using 10 cases of CHLNS as the most challenging lymphoma subtype for microdissection included in this study. The results of this test are outlined in Online Supplementary Figure S1 and confirmed that the epigenetic profiles of the DNA from microdissected Hodgkin and Reed-Sternberg cells differed from those of DNA obtained from whole tissue sections in which varying numbers of tumor cells were present in a reactive background. In this study all data on CpG target methylation of tissue samples were generated using only microdissected tumor cells or microdissected RTB. Tumor cells or RTB yielding 250 ng DNA were microdissected using a Leica LMD 6000 (Leica Microsystems, Wetzlar, Germany) (Online Supplementary Figure S1A). The lymphoma cell lines Farage, K1106, L428, L1236 and U2940 were also studied. Cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum.
Table 1.
Characteristics and immunostaining of samples analyzed for DNA methylation.
DNA methylation profiling using universal BeadArrays
Microdissected tumor cells, microdissected RTB or cultured cells were lysed and DNA was extracted. In brief, microdissected cells were incubated in 40 μL ATL buffer (Qiagen, CA, USA), 5 μL Proteinase K (Qiagen, CA, USA) and 4 μL Dako Target Retrieval solution High pH 10x Concentrate (Dako, Glostrup, Denmark) at 65°C for 16 h. DNA was extracted from the cell lines using a QIAamp DNA Micro Kit (Qiagen, CA, USA), bisulfite-modified using an EZ DNA Methylation-Gold Kit (Zymo Research, CA, USA) and then used for the Illumina GoldenGate Methylation assay (Illumina, San Diego, CA; USA). As described previously, methylation of 1505 CpG sites selected from over 800 genes was detected using the GoldenGate Methylation Cancer Panel I (Illumina, San Diego, CA, USA).26 This panel includes tumor suppressor genes, oncogenes, and genes involved in DNA repair, cell cycle control, cell differentiation, and apoptosis.
Images were processed and intensity data extracted with Illumina-supplied equipment. Each methylation data point is represented by fluorescent signals from the unmethylated (U) and methylated (M) alleles. The methylation status of the investigated CpG site is calculated as the ratio of the fluorescent signal from one allele relative to the sum of the signals from both methylated and unmethylated alleles (β-value):
The β-value provides a continuous measurement of levels of DNA methylation at a CpG site and ranges from 0 for completely unmethylated sites to 1 for completely methylated sites.
Validation
Pyrosequencing was performed using bisulfite-modified DNA of tissue samples and lymphoma cell lines according to a previously published protocol with minor modifications.27 Quantitative methylation values at four CpG sites corresponding to four GoldenGate probes from three genes (CDH1, FAT, and SLIT2) were analyzed as described previously.28
Statistical analysis
Since chromosome X is methylated in women, all 84 CpG on chromosome X were excluded before data analysis to avoid a gender-specific bias. Therefore, a total of 1421 CpG entered further statistical analysis. First, CpG targets were categorized as hypomethylated (β≤0.25), intermediate (0.25<β<0.75) or hypermethylated (β≥0.75). Selection of hypo- and hypermethylated CpG in tumor samples was based on: (i) this definition, and (ii) a P value of less than 0.05 as determined by one way ANOVA with Benjamini-Hochberg multiple testing correction. Secondly, differential methylation was computed and defined as follows: (i) a methylation level difference (delta beta) of at least 0.30 (Δβ≥ 0.30), and (ii) a P value of less than 0.05 as determined by one way ANOVA with Benjamini-Hochberg multiple testing correction. De novo hypomethylation was defined as a difference of at least 0.30 between β values for RTB and tumor cell samples (βRTB–βtumor ≥ 0.30), whereas de novo hypermethylation was defined as a difference of at least 0.30 between β values for tumor cell samples and RTB (βtumor–βRTB ≥ 0.30).
Fisher’s exact test (GraphPad Prism 5.0) was used to calculate whether target genes of the polycomb repressor complex 2 (PRC2) as defined by Lee et al.29 were enriched among genes with de novo hypomethylation or de novo hypermethylation in MGZL, CHLNS, PMLBCL, and DLBCL.
Data analysis, including principal component analysis, Venn diagrams and cluster analysis, was performed using GeneSpring GX 10.0. Detailed information on the statistical analysis used to build predictive models and identify a gene set for class prediction is provided in the Online Supplementary Methods.
Results
Mediastinal gray zone lymphoma appears to have a distinct epigenetic profile intermediate between those of classical Hodgkin’s lymphoma, nodular sclerosis and primary mediastinal large B-cell lymphoma, and clearly different from that of diffuse large B-cell lymphoma
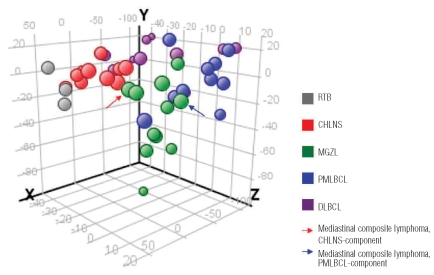
As shown in Figure 1, MGZL appears to have a distinct epigenetic profile intermediate between those of CHLNS and PMLBCL. The principal component analysis demonstrated a clustering of CHLNS cases with a distribution distinct from that of PMLBCL. A clear association of MGZL with either CHLNS or PMLBCL was not detected. One of the lymphomas studied was a composite lymphoma, comprising two distinct components of CHLNS and PMLBCL in the same biopsy with distinctive morphologies and immunophenotypes (Figure 2).4 The two different portions of the tumor were microdissected separately. Notably, the epigenetic profile of the Hodgkin and Reed-Sternberg cells dissected from the CHLNS component clustered with CHLNS, whereas the tumor cells selected from the PMLBCL areas of the same tumor showed an epigenetic profile closer to that of PMLBCL samples. The epigenetic profile of MGZL was clearly different from that of DLBCL and, not unexpectedly, greater heterogeneity was observed in cases of DLBCL which included germinal center B-cell-like and activated B-cell-like types of DLBCL (Figure 1, Table 1).23
Figure 1.
Distinct epigenetic profile of MGZL as assessed by principal component analysis. The methylation data for 1421 CpG targets from all studied tissue samples were subjected to principal component analysis and projected onto the first three principal components. MGZL appears to have a distinct epigenetic profile intermediate between CHLNS and PMLBCL, but clearly different from that of DLBCL. One of the lymphomas studied was a composite lymphoma, comprising two distinct components of CHLNS and PMLBCL in the same biopsy which were microdissected separately. Both elements of the composite lymphoma clustered with cases of MGZL, but the two components also demonstrated a particularly close association with cases of CHLNS or PMLB-CL, respectively.
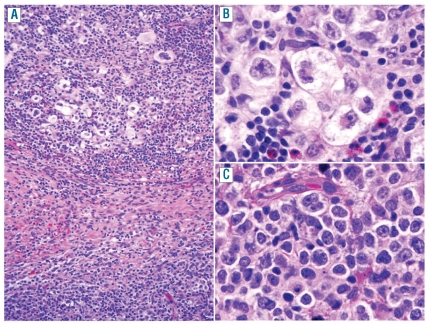
Figure 2.
Histological features of mediastinal composite lymphoma showing two different histological components in the same biopsy: one characteristic of CHLNS and another of PMLBCL. (A) CHLNS-component with Hodgkin and Reed-Sternberg cells in an inflammatory background with numerous eosinophils (top) separated by fibrous collagen bands from the PMLBCL-component composed of tumor cells with abundant pale cytoplasm (bottom) (original magnification: x10). (B) High magnification of the CHLNS-component showing characteristic lacunar cells (original magnification: x40). (C) High magnification of the PMLBCL-component showing sheets of relatively monomorphic large cells (original magnification: x40).
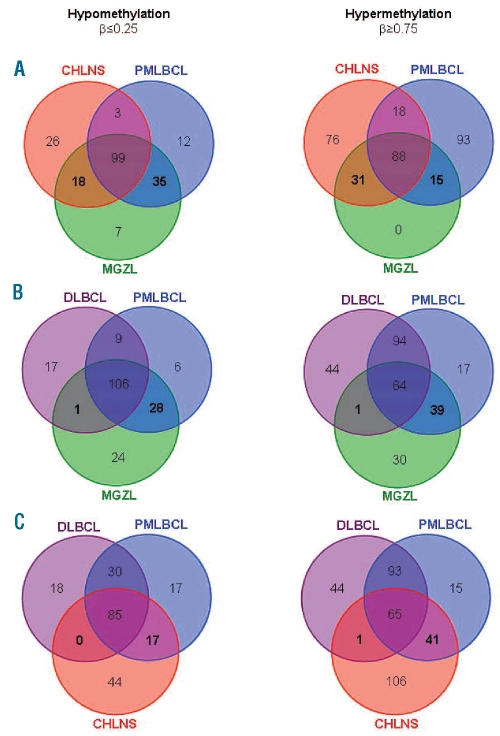
Next, we compared hypo- and hypermethylated CpG targets of the different lymphoma types (Figure 3). The total number of hypomethylated sites was similar in MGZL, CHLNS, PMLBCL and DLBCL. In contrast, MGZL showed a clearly lower total number of hypermethylated sites (n=134) compared to the other lymphoma entities (n=213 in CHLNS, 214 in PMLBCL and 203 in DLBCL). We then compared common hypomethylated or hypermethylated targets in MGZL, CHLNS and PMLBCL, which were only shared by MGZL and CHLNS or MGZL and PMLBCL (Figure 3A). Interestingly, MGZL shared twice as many common hypomethylated targets with PMLBCL (n=35) compared to CHLNS (n=18). The opposite phenomenon was found when comparing common hypermethylated targets (31 shared between MGZL and CHLNS, 15 between MGZL and PMLBCL). Subsequently we compared common hypomethylated or hypermethylated CpG between MGZL, PMLBCL and DLBCL (Figure 3B). As expected, we found a closer relationship of MGZL with PMLBCL than with DLBCL. While MGZL shared 28 common hypomethylated and 39 common hypermethylated targets with PMLBCL that were not shared by DLBCL, only one common hypomethylated and one common hypermethylated target was shared by MGZL and DLBCL. Further analysis demonstrated that the epigenetic profile of CHLNS is closer to that of PMLBCL than to that of DLBCL (Figure 3C). Although we did not observe any targets that were exclusively hypermethylated in our study cohort of MGZLs, our analysis revealed seven CpG exclusively hypomethylated in MGZL. These hypomethylated CpG were located on the genes ACVR1C, ERN1, HOXA5, ISL1, PTGS2, TMEFF2 and ZNF215. Of note, all seven identified CpG were located within CpG islands.
Figure 3.
CpG target hypomethylation and hypermethylation in MGZL, CHLNS, PMLBCL and DLBCL. The Venn diagrams show the numbers of common hypomethylated (β≤0.25) targets (left hand panels) or common hypermethylated (β≥0.75) targets (right hand panels) in the indicated lymphoma entities. Comparison of MGZL with CHLNS and PMLBCL (A) or DLBCL and PMLBCL (B) and comparison of CHLNS with DLBCL and PMLBCL (C). (A) MGZL shares a number of unique common hypo- and hypermethylated targets with CHLNS and PMLBCL (numbers in bold). (B) In contrast, only one hypomethylated and one hypermethylated target are shared between MGZL and DLBCL, exclusive of those shared by PMLBCL. Several hypomethylated and hypermethylated targets are shared in common by MGZL and PMLBCL but not shared by DLBCL, 28 and 39 respectively (numbers in bold). (C) Some overlap of target methylation is present between CHLNS and PMLBCL (17 hypomethylated CpG, 41 hypermethylated CpG) but almost none between CHLNS and DLBCL (numbers in bold).
Hodgkin’s lymphoma is characterized by a lack of de novo hypomethylation within or outside CpG islands
To assess a lymphoma-associated distribution pattern we then compared the methylation status of promoter regions and their relationship to CpG islands. We therefore compared methylation of promoters within CpG islands and outside CpG islands of tumor samples with methylation in normal controls (RTB) (Online Supplementary Table S1). In MGZL, 58% of the hypomethylation occurred outside CpG islands and 42% within CpG islands, while hypermethylation was mainly present within CpG islands (99%). In PMLBCL and DLBCL de novo hypomethylation was mainly detected outside CpG islands (68% and 81%, respectively), whereas de novo hypermethylation was located exclusively within CpG islands. Interestingly, de novo hypomethylation was not found in CHLNS, either within or outside CpG islands. Moreover, de novo hypermethylation in CHLNS occurred not only in CpG islands, but also in a fraction of promoters located outside CpG islands (14%) (Online Supplementary Table S1).
De novo DNA methylation in the lymphomas studied seems to be mediated by members of the polycomb complex in a large proportion of genes
Analysis of the association between PRC2 targets and genes differentially methylated in MGZL, CHLNS, PMLB-CL and DLBCL as compared to in normal controls showed that between 45% and 50% of the genes experiencing de novo hypermethylation are PRC2 targets.29 This enrichment was statistically significant in all four lymphoma groups when compared with the 21% of PRC2 targets in all genes studied (all P values ≤0.0014). In contrast, only 3% to 11% of genes with de novo hypomethylation were PRC2 targets in embryonic stem cells.
Validation
Excellent intra- and inter-array correlations between replicate DNA samples have already been demonstrated in multiple studies using the Illumina GoldenGate Methylation Cancer Panel I.28,30 Furthermore, we have recently demonstrated that matched formalin-fixed paraffin-embedded and frozen surgical pathology replicates showed the complete preservation of the cancer methylome,31 and that immunohistochemistry studies of formalin-fixed paraffin-embedded tissue specimens do not alter the DNA methylation profile.25 Here, we performed pyrosequencing for four different CpG sites from three different genes (CDH1, FAT, SLIT2) and demonstrated a high accuracy of the methylation array (Online Supplementary Table S2).
Identification of differentially methylated targets in mediastinal gray zone lymphoma, classical Hodgkin’s lymphoma, nodular sclerosis and primary mediastinal large B-cell lymphoma
After investigating common hypo- and hypermethylated targets to elucidate similarities of MGZL with CHLNS and PMLBCL, computation of differential methylation was performed for identification of tumor-specific changes in methylation. MGZL and CHLNS showed 20 differentially methylated targets, whereas 35 targets were found to be differentially methylated between MGZL and PMLBCL (Online Supplementary Table S3). Analysis of differential methylation between CHLNS and PMLBCL revealed a total of 180 differentially methylated targets (data not shown). Having identified these 235 differentially methylated targets between MGZL, CHLNS and PMLB-CL, we next sought to use a biomathematic approach for predicting the diagnosis of MGZL, CHLNS or PMLBCL based on the epigenetic profile.
Prediction models identify 22 differentially methylated targets that can distinguish between mediastinal gray zone lymphoma, classical Hodgkin’s lymphoma, nodular sclerosis and primary mediastinal large B-cell lymphoma
Using 23 samples (7 MGZL, 8 CHLNS, and 8 PMLBCL), we performed multiple exclusive linear discriminant analysis implemented in Partek Genomics Suite 6.5 to build a total of 13 predictive models that could best separate MGZL, CHLNS, and PMLBCL (see Online Supplementary Methods for details). The predictive models were then validated by applying the models to eight independent samples (2 MGZL, the 2 components of one composite lymphoma, 2 CHLNS, and 2 PMLBCL) which were not part of the model building process, using a simple majority voting scheme. As shown in Online Supplementary Table S4, the final combined prediction for these samples was 100%. Of note, both components of the mediastinal composite lymphoma were predicted as MGZL.
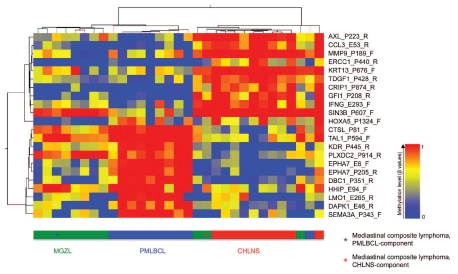
This model building process identified 100 predictive methylation loci, 22 of which met the criteria for differential methylation (P<0.01, one-way ANOVA; with Δβ≥ 0.30 between any two sample conditions). These 22 differentially methylated targets represent a selection of the 235 CpG targets that were identified as differentially methylated targets between MGZL, CHLNS and PMLBCL. Hierarchical cluster analysis of β-values from these 22 CpG loci from tissue samples of MGZL, PMLBCL and CHLNS separated the samples into three distinct groups with only two outliers of MGZL and one outlier of PML-BCL (Figure 4). Interestingly, the outlier PMLBCL case that clustered with CHLNS was a lymphoma with occasional CD30+ tumor cells in which a differential diagnosis of CHLNS was considered histologically. However, based on clinical, morphological and immunophenotypic features, the diagnosis of PMLBCL was favored. One of the two MGZL that clustered with CHLNS was a case that was morphologically more reminiscent of PMLBCL. Immunophenotypically, the tumor cells were positive for both CD20 and CD79a, but interestingly also positive for CD30 and CD15. The other MGZL contained some cells morphologically resembling Reed-Sternberg cells with variable expression of CD30 but strongly positive for CD20 and PAX-5. CD79a and CD15 were both negative in this lymphoma. The two components of the single composite lymphoma were analyzed separately. Whereas the PMLBCL-component was closer to the profile of MGZL, the CHLNS-component clustered with cases of CHLNS. As shown in Figure 4, HOXA5 hypomethylation was typically observed in MGZL, while MMP9 hypermethylation was characteristic of CHLNS. Hypermethylation of EPHA7 and DAPK1 was a hallmark of PMLBCL.
Figure 4.
Hierarchical cluster analysis of 22 differentially methylated targets in MGZL, CHLNS and PMLBCL. Columns represent individual cases of microdissected tumor samples, rows represent the indicated CpG sites (selected targets of prediction models; P<0.01, one-way ANOVA; Δβ≥ 0.30 between any two sample conditions; see Design and Methods section for details). The color represents the methylation level (β-values) from 0 (0% methylation, blue) to 1 (100% methylation, red) as given in the vertical color bar. The horizontal color bar indicates the tumor samples. The two components of a single mediastinal composite lymphoma were analyzed separately. Whereas the PMLBCL-component was closest to the profile of MGZL, the CHLNS-component clustered with the cases of CHLNS. PMLB-CL and CHLNS are readily distinguished, while MGZL shows overlap with both groups.
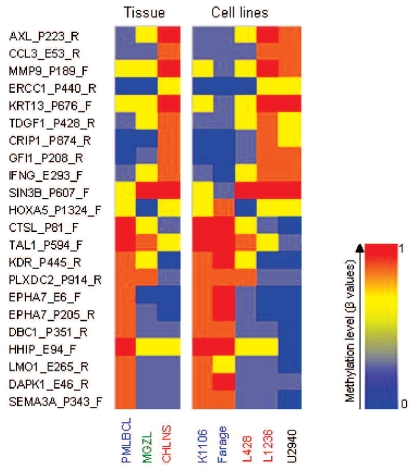
We also compared the methylation status of MGZL, PMLBCL and CHLNS tissue samples to that of representative lymphoma cell lines (Figure 5). The methylation profile of the PMLBCL-cell lines K1106 and Farage showed great similarity to the profile obtained with PMLBCL tissue DNA. The profile of the Hodgkin-cell line L428 closely approximated that of the corresponding tissue, whereas the results for the Hodgkin-cell line L1236 were identical to those obtained with CHLNS tissue samples. Interestingly, the methylation profile of the cell line U2940, which was derived from a patient with DLBCL that developed sequentially to CHL, was most similar to the tissue methylation profile of CHLNS (Figure 5).
Figure 5.
Epigenetic characterization of lymphoma cell lines using the identified 22 differentially methylated targets of the prediction models. Columns represent the indicated lymphoma tissue samples or the indicated lymphoma cell lines. Rows represent the 22 selected CpGs based on Figure 4 (selected targets of prediction models; see Design and Methods section for details). The color represents the methylation level (mean of β-values of tissue samples and β-values of individual cell lines) as given in the vertical color bar (0, 0% methylation, blue; 1, 100% methylation, red). The analysis of the 22 selected CpG loci reveals great similarity of the methylation pattern of the PMLBCL-cell lines (blue) and the Hodgkin-cell lines (red) to their tissue counterparts. The pattern of U2940, a cell line derived from a patient with a DLBCL sequential to CHL, was most similar to the tissue methylation profile of CHLNS.
Discussion
In 2005, MGZL was described for the first time as a lymphoma demonstrating transitional morphological and immunophenotypic features intermediate between CHLNS and PMLBCL.4 Three years later, this category was included in the WHO classification as a separate entity.6 Despite its inclusion in the WHO classification, its delineation to date has been based solely on histological and immunohistochemical features and data on the genetic or epigenetic features of these lymphomas are lacking. However, there is an urgent need to explore the biology of MGZL, since this entity presents a challenge both to the pathologist and the clinician. Comprehensive diagnostic criteria are still missing and the optimal therapy for MGZL is as yet undetermined. Clinical experience shows that these tumors fail to respond to therapeutic regimens effective in either CHL or PMLBCL. In a first clinical NCI trial of this rare entity, 11 patients with MGZL and 35 patients with PMLBCL were treated with a DA-EPOCH-R regimen.32 Preliminary data from this study confirm prior reports outside the setting of a clinical trial, indicating that MGZL generally have a more aggressive clinical course than PMLBCL or CHL. In the NCI trial radiation therapy was required to obtain sustained failure-free survival. The present study is a first step in the exploration of the biological basis for the poor clinical responsiveness of this tumor in order to develop new and more effective therapies. Moreover, having new biological tools to diagnose MGZL will aid the accurate identification of those patients requiring different management.
If MGZL, CHLNS and PMLBCL are derived from a common precursor, such as thymic B cells, the genetic or epigenetic events that alter the morphology and immunophenotype of the neoplastic cells would be of special interest. Because the morphological and immunophenotypic changes in MGZL appear reversible, it is likely that epigenetic rather than genetic alterations are responsible for these changes.4,19 The best-established epigenetic marker is DNA methylation, which is an important event during carcinogenesis.33 Accordingly, studies of DNA methylation help to better define factors influencing tumor behavior and can be beneficial not only for clinical diagnostics but also for tumor therapy.34 Promising results have been achieved with histone deacetylase and DNA methyltransferase inhibitors for the treatment of hematologic malignancies and lymphoproliferative disorders.35,36 Thus, unraveling the epigenetic characteristics of lymphomas may help to establish new therapeutic approaches with demethylating agents, especially in cases of MGZL that respond poorly to polychemotherapy.
In the present study we investigated DNA methylation of 1,044 CpG sites located within CpG islands and 461 CpG sites located outside CpG islands. As described previously for other types of lymphoma,31,37 our results confirm that de novo hypomethylation is mainly detected outside CpG islands, whereas de novo hypermethylation is typically located within CpG islands. Of note, we did not observe any de novo hypomethylation in CHLNS, clearly distinguishing this lymphoma subtype from MGZL, PML-BCL and DLBCL. This finding indicates that the development of Hodgkin and Reed-Sternberg cells may be driven by specific mechanisms inducing gene silencing due to DNA methylation. In contrast, de novo hypomethylation was present in MGZL, PMLBCL and DLBCL. Thus, mechanisms influencing epigenetic modifications may distinguish Hodgkin and Reed-Sternberg cell pathogenesis from tumor cell development in MGZL, PMLBCL and DLBCL.
The pathogenic role of tumor-specific aberrant hypermethylation in CHLNS is further underscored by the fact that hypermethylation in this lymphoma also occurs in a fraction of promoters located outside CpG islands. Our results indicate that DNA methylation may be an important step for directing the B-cell program towards a Hodgkin and Reed-Sternberg cell. This theory is in line with the findings of previous studies suggesting a central role of DNA methylation in the pathogenesis of Hodgkin’s lymphoma.20–22 Further analysis of de novo DNA methylation showed that a high proportion of genes de-novo-methylated in MGZL, CHLNS, PMLBCL and DLBCL were repressed by PRC2 in embryonic stem cells. This finding is consistent with previous descriptions of the same phenomenon for solid tumors and hematologic neoplasms such as DLBCL.37,38
Our results further reveal that while the epigenetic profile of MGZL shows many similarities to the profiles of CHLNS and PMLBCL, the profile of MGZL has distinctive features, indicating that this lymphoma cannot be assigned to either one of the parent entities. The epigenetic data obtained from microdissected tumor cells allowed accurate prediction of the three lymphoma types: MGZL, CHLNS or PMLBCL. Interestingly, both the CHLNS and PMLBCL components of the composite lymphoma were predicted as MGZL and not as CHLNS or PMLBCL. Thus, the prediction model identified the plasticity of the neoplastic cells, leading to the two different morphological and immunophenotypic expressions, suggesting a close relationship to MGZL. In the principal component analysis both elements of the composite lymphoma also clustered with cases of MGZL, but individually the two components shifted, falling closer to cases of CHLNS and PMLBCL, respectively (Figure 1).
The prediction model building process also identified 100 predictive methylation loci, 22 of which were also differentially methylated. Among these 22 CpG we found genes encoding proteins implicated in cell proliferation, differentiation or death, catabolic enzymes, receptor tyrosine kinases, transcription factors and zinc finger proteins, some of which are of particular interest. Hypomethylation of HOXA5 was observed in MGZL suggesting a unique role of this gene in MGZL tumorigenesis. Interestingly, HOXA5 has been reported to promote leukemic transformation in concert with certain chromosomal translocations.39 EPHA7 and DAPK1 were exclusively hypermethylated in PMLB-CL. These results are in line with data from earlier studies, describing silencing of EPHA7 and DAPK1 in related B-cell lymphomas.40,41 MMP9 was predominantly hypermethylated in CHLNS and may reflect the sclerotic morphology of this lymphoma suggesting a low expression of matrix metalloproteinases. Hierarchical cluster analysis of these 22 CpG demonstrated that PMLBCL and CHLNS are readily distinguished, while MGZL shows overlap with both groups. Only two cases of MGZL and one PMLBCL did not cluster with their respective lymphoma groups. Of note, both outliers of MGZL clustered with cases of CHLNS. One such case was morphologically more reminiscent of CHLNS, while the other had morphological features closer to PMLBCL. As with most MGZL, the immunophenoptypic features of both cases were ambiguous.
Analysis of PMLBCL and CHL cell lines for these 22 CpG showed great similarity to the results obtained for the corresponding tissue samples. Interestingly, the cell line U2940, derived from a DLBCL arising in a patient with a history of CHL, had a DNA methylation profile most similar to that of CHLNS, although the cell line itself had features of a mature B-cell malignancy.42 Thus, the cell line retained the methylation signature of the originally diagnosed malignancy.
Although the number of study samples was restricted due to the rarity of MGZL and the labor-intensive approach of microdissection, the confirmation of our tissue data by the independent cell line data and the possibility of developing a prediction model with a final combined prediction of 100% underscore the strength of the data obtained from microdissected tumor cells. Similar studies employing statistical analysis of microdissected tumor cells were able to draw significant conclusions, despite analysis of even fewer cases than in the present study.43
We also performed pathway analyses focusing on JAK/STAT signaling, NFκB signaling and B-cell-associated genes using Ingenuity Pathway Analysis (Ingenuity Systems). Among these pathways, we found that genes with differential methylation between tumor samples and reactive tonsils and relation to the NFκB pathway were significantly enriched (P=0.0156, data not shown). These findings suggest that the NFκB pathway may play an important role not only in the pathogenesis of CHLNS and PMLBCL,11,12 but also in the pathogenesis of MGZL. The methylation array used in this study is currently relatively unique in its ability to perform methylation profiling of DNA extracted from formalin-fixed paraffin-embedded tissue. Therefore pathway analysis is limited to the available gene panel of this platform. Future studies should attempt to validate these findings further with platforms containing a more comprehensive selection of genes.
It has been uncertain whether MGZL represents a variant of PMLBCL, CHLNS, or an independent entity. We show a close epigenetic relationship between MGZL, CHLNS and PMLBCL but also demonstrate a unique epigenetic signature for MGZL, validating its WHO classification as a separate disease. The precise pathobiology of MGZL does, however, remain to be explored in greater detail.
Acknowledgments
We thank Kris Ylaya, Robert Walker, Daniel Edelman, Lixin Rui, George Wright, Shelley Hoover, Nicole Grant, Theresa Davies-Hill, and Silke Williams for their excellent support.
Footnotes
The online version of this article has a supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
Funding: this work was supported by the intramural research program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
References
- 1.Rudiger T, Jaffe ES, Delsol G, deWolf-Peeters C, Gascoyne RD, Georgii A, et al. Workshop report on Hodgkin’s disease and related diseases (‘grey zone’ lymphoma) Ann Oncol. 1998;9 (Suppl 5):S31–8. doi: 10.1093/annonc/9.suppl_5.s31. [DOI] [PubMed] [Google Scholar]
- 2.Kuppers R. The biology of Hodgkin’s lymphoma. Nature Rev. 2009;9(1):15–27. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- 3.Jaffe ES, Wilson WH. Gray zone, synchronous, and metachronous lymphomas: diseases at the interface of non-Hodgkin’s lymphomas and Hodgkin’s Lymphoma. In: Mauch PM, Armitage JO, Coiffier B, Dalla-Favera R, Harris NL, editors. Non-Hodgkin’s Lymphoma. Philadelphia, PA: Lippincott, Williams, and Wilkins; 2004. pp. 69–80. [Google Scholar]
- 4.Traverse-Glehen A, Pittaluga S, Gaulard P, Sorbara L, Alonso MA, Raffeld M, et al. Mediastinal gray zone lymphoma: the missing link between classic Hodgkin’s lymphoma and mediastinal large B-cell lymphoma. Am J Surg Pathol. 2005;29(11):1411–21. doi: 10.1097/01.pas.0000180856.74572.73. [DOI] [PubMed] [Google Scholar]
- 5.Eberle FC, Mani H, Jaffe ES. Histopathology of Hodgkin’s lymphoma. Cancer J. 2009;15(2):129–37. doi: 10.1097/PPO.0b013e31819e31cf. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed . Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 7.Gonzalez CL, Medeiros LJ, Jaffe ES. Composite lymphoma. A clinicopathologic analysis of nine patients with Hodgkin’s disease and B-cell non-Hodgkin’s lymphoma. Am J Clin Pathol. 1991;96(1):81–9. doi: 10.1093/ajcp/96.1.81. [DOI] [PubMed] [Google Scholar]
- 8.Perrone T, Frizzera G, Rosai J. Mediastinal diffuse large-cell lymphoma with sclerosis. A clinicopathologic study of 60 cases. Am J Surg Pathol. 1986;10(3):176–91. doi: 10.1097/00000478-198603000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Zarate-Osorno A, Medeiros LJ, Longo DL, Jaffe ES. Non-Hodgkin’s lymphomas arising in patients successfully treated for Hodgkin’s disease. A clinical, histologic, and immunophenotypic study of 14 cases. Am J Surg Pathol. 1992;16(9):885–95. doi: 10.1097/00000478-199209000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Garcia JF, Mollejo M, Fraga M, Forteza J, Muniesa JA, Perez-Guillermo M, et al. Large B-cell lymphoma with Hodgkin’s features. Histopathology. 2005;47(1):101–10. doi: 10.1111/j.1365-2559.2005.02175.x. [DOI] [PubMed] [Google Scholar]
- 11.Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102(12):3871–9. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- 12.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable sub-group of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198(6):851–62. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calvo KR, Traverse-Glehen A, Pittaluga S, Jaffe ES. Molecular profiling provides evidence of primary mediastinal large B-cell lymphoma as a distinct entity related to classic Hodgkin lymphoma: implications for mediastinal gray zone lymphomas as an intermediate form of B-cell lymphoma. Adv Anat Pathol. 2004;11(5):227–38. doi: 10.1097/01.pap.0000138144.11635.f8. [DOI] [PubMed] [Google Scholar]
- 14.Joos S, Otano-Joos MI, Ziegler S, Bruderlein S, du Manoir S, Bentz M, et al. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 1996;87(4):1571–8. [PubMed] [Google Scholar]
- 15.Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, et al. Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res. 2000;60 (3):549–52. [PubMed] [Google Scholar]
- 16.Bentz M, Barth TF, Bruderlein S, Bock D, Schwerer MJ, Baudis M, et al. Gain of chromosome arm 9p is characteristic of primary mediastinal B-cell lymphoma (MBL): comprehensive molecular cytogenetic analysis and presentation of a novel MBL cell line. Genes Chromosomes Cancer. 2001;30(4):393–401. doi: 10.1002/1098-2264(2001)9999:9999<::aid-gcc1105>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 17.Wessendorf S, Barth TF, Viardot A, Mueller A, Kestler HA, Kohlhammer H, et al. Further delineation of chromosomal consensus regions in primary mediastinal B-cell lymphomas: an analysis of 37 tumor samples using high-resolution genomic profiling (array-CGH) Leukemia. 2007;21(12):2463–9. doi: 10.1038/sj.leu.2404919. [DOI] [PubMed] [Google Scholar]
- 18.Oschlies I, Burkhardt B, Salaverria I, Rosenwald A, ES DA, Szczepanowski M, et al. Clinical, pathological and genetic features of primary mediastinal large B-cell lymphomas and mediastinal gray zone lymphomas in children. Haematologica. 2011;96(2):262–8. doi: 10.3324/haematol.2010.030809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertel CB, Zhou XG, Hamilton-Dutoit SJ, Junker S. Loss of B cell identity correlates with loss of B cell-specific transcription factors in Hodgkin/Reed-Sternberg cells of classical Hodgkin lymphoma. Oncogene. 2002;21(32):4908–20. doi: 10.1038/sj.onc.1205629. [DOI] [PubMed] [Google Scholar]
- 20.Ushmorov A, Ritz O, Hummel M, Leithauser F, Moller P, Stein H, et al. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood. 2004;104(10):3326–34. doi: 10.1182/blood-2003-04-1197. [DOI] [PubMed] [Google Scholar]
- 21.Doerr JR, Malone CS, Fike FM, Gordon MS, Soghomonian SV, Thomas RK, et al. Patterned CpG methylation of silenced B cell gene promoters in classical Hodgkin lymphoma-derived and primary effusion lymphoma cell lines. J Mol Biol. 2005;350 (4):631–40. doi: 10.1016/j.jmb.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Ehlers A, Oker E, Bentink S, Lenze D, Stein H, Hummel M. Histone acetylation and DNA demethylation of B cells result in a Hodgkin-like phenotype. Leukemia. 2008;22(4):835–41. doi: 10.1038/leu.2008.12. [DOI] [PubMed] [Google Scholar]
- 23.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 24.Dunleavy K, Pittaluga S, Czuczman MS, Dave SS, Wright G, Grant N, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113(24):6069–76. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberle FC, Hanson JC, Killian JK, Wei L, Ylaya K, Hewitt SM, et al. Immunoguided laser assisted microdissection techniques for DNA methylation analysis of archival tissue specimens. J Mol Diagn. 2010;12(4):394–401. doi: 10.2353/jmoldx.2010.090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006 Mar 16;3 :383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2(9):2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 28.O’Riain C, O’Shea DM, Yang Y, Le Dieu R, Gribben JG, Summers K, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;23(10):1858–66. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Subero JI, Kreuz M, Bibikova M, Bentink S, Ammerpohl O, Wickham-Garcia E, et al. New insights into the biology and origin of mature aggressive B-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113(11):2488–97. doi: 10.1182/blood-2008-04-152900. [DOI] [PubMed] [Google Scholar]
- 31.Killian JK, Bilke S, Davis S, Walker RL, Killian MS, Jaeger EB, et al. Large-scale profiling of archival lymph nodes reveals pervasive remodeling of the follicular lymphoma methylome. Cancer Res. 2009;69 (3):758–64. doi: 10.1158/0008-5472.CAN-08-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunleavy K, Pittaluga S, Tay K, Grant N, Chen CC, Shovlin M, et al. Comparative clinical and biological features of primary mediastinal B-cell lymphoma (PMBL) and mediastinal grey zone lymphoma (MGZL) Blood (ASH Annual Meeting Abstracts) 2009;114:106. [Google Scholar]
- 33.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 34.Kondo Y, Issa JP. DNA methylation profiling in cancer. Expert Rev Mol Med. 12:e23. doi: 10.1017/S1462399410001559. [DOI] [PubMed] [Google Scholar]
- 35.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol. 2008;68(1):1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Issa JP. Epigenetic changes in the myelodys-plastic syndrome. Hematol Oncol Clin North Am. 2010;24(2):317–30. doi: 10.1016/j.hoc.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Subero JI, Ammerpohl O, Bibikova M, Wickham-Garcia E, Agirre X, Alvarez S, et al. A comprehensive microarray-based DNA methylation study of 367 hematological neoplasms. PLoS One. 2009;4(9):e6986. doi: 10.1371/journal.pone.0006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 39.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006;8(9):1017–24. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson DW, Hong JS, Shen RR, French SW, Troke JJ, Wu YZ, et al. Global DNA methylation profiling reveals silencing of a secreted form of Epha7 in mouse and human germinal center B-cell lymphomas. Oncogene. 2007;26(29):4243–52. doi: 10.1038/sj.onc.1210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Korbi S. Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol. 2008;19(10):1774–86. doi: 10.1093/annonc/mdn374. [DOI] [PubMed] [Google Scholar]
- 42.Sambade C, Berglund M, Lagercrantz S, Sallstrom J, Reis RM, Enblad G, et al. U-2940, a human B-cell line derived from a diffuse large cell lymphoma sequential to Hodgkin lymphoma. Int J Cancer. 2006;118(3):555–63. doi: 10.1002/ijc.21417. [DOI] [PubMed] [Google Scholar]
- 43.Eckerle S, Brune V, Doring C, Tiacci E, Bohle V, Sundstrom C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23(11):2129–38. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]