Abstract
The dopaminergic neurons of the basal ganglia play critical roles in CNS function and human disease, but specification of dopamine neuron phenotype is poorly understood in vertebrates. We performed an in vivo screen in zebrafish to identify dopaminergic neuron enhancers, in order to facilitate studies on the specification of neuronal identity, connectivity, and function in the basal ganglia. Based primarily on identification of conserved non-coding elements, we tested 54 DNA elements from four species (zebrafish, pufferfish, mouse, and rat), that included 21 genes with known or putative roles in dopaminergic neuron specification or function. Most elements failed to drive CNS expression or did not express specifically in dopaminergic neurons. However, we did isolate a discrete enhancer from the otpb gene that drove specific expression in diencephalic dopaminergic neurons, although it did not share sequence conservation with regulatory regions of otpa or other dopamine-specific genes. For the otpb enhancer, regulation of expression in dopamine neurons requires multiple elements spread across a large genomic area. In addition, we compared our in vivo testing with in silico analysis of genomic regions for genes involved in dopamine neuron function, but failed to find conserved regions that functioned as enhancers. We conclude that regulation of dopaminergic neuron phenotype in vertebrates is regulated by dispersed regulatory elements.
Keywords: dopamine, enhancers, zebrafish, CNS, basal ganglia
Introduction
The basal ganglia and their dopaminergic neurons play critical roles in CNS function with vital roles in initiation and regulation of movement, limbic emotional responses, and reward-mediated aspects of behavior and learning. Several human diseases including Parkinson's disease, attention-deficit hyperactivity disorder, Tourette's syndrome, and addiction behaviors have been linked with pathology of basal ganglia dopaminergic neurons (Nussbaum and Ellis, 2003; Albin and Mink, 2006; Hyman, Malenka, and Nestler, 2006). In mammals, groups of dopaminergic neurons are located in discrete nuclei in the telencephalon, diencephalon, and mesencephalon. The mesodiencephalic dopaminergic (mesDA) neurons constitute the largest fraction of dopaminergic neurons in the brain (roughly 75%; Wallen and Perlmann, 2003) and include three distinct sets of nuclei (Dahlstroem and Fuxe, 1964): the substantia nigra pars compacta (group A9), the retrorubral field (group A8), and the ventral tegmental area (group A10).
The mesDA system is highly complex both in its organization and function. Its neurons integrate pathways of information from the striatum and cortex in which there is somatotopic representation of distinct body parts, and also sorting of parallel functional pathways for different cortical modalities (such as limbic behaviors or motor function (DeLong and Wichmann, 2009). Accordingly, mesDA neurons are heterogeneous with respect to their development (Smidt et al., 2000), projections, intrinsic electrophysiologic properties (Lammel et al., 2008), and neurotransmitter identities.
Specification of mesDA neurons is controlled by a developmental cascade of transcription factors (Abeliovich and Hammond, 1997; Smidt and Burbach, 2007). However, it is not known whether this hierarchical cascade alone is sufficient to specify mesDA neuron identity (including neurotransmitter status and axon projections), or whether additional information is necessary to provide more precise identities for subtypes of mesDA neurons. For example, the orphan nuclear receptor nurr1 is necessary for generation and maintenance of mesDA neurons, as assayed by absent tyrosine hydroxylase (TH) expression in knock-out mice (Zetterstroem et al., 1997; Saucedo-Cardenas et al., 1998; Wallen et al., 1999). However, in nurr1 knock-out mice, some neurons destined to express TH still differentiate partially; nigrostriatal projections still develop normally; and the neurons express other markers (such as cholecystokinin) specific to dopaminergic neurons (Witta et al., 2000). nurr1 is only partially responsible for specifying dopamine neuron-specific gene expression: while vmat2 and dat require nurr1, aadc expression is induced independently (Smits et al., 2003). Therefore, while nurr1 is necessary for terminal differentiation of mesDA neurons including the expression of several dopamine-neuron specific biosynthetic enzymes and receptors, it does not regulate other key elements of mesDA identity such as axon pathfinding, and does not appear to affect development of other dopaminergic neuron groups in the CNS (Baeckman et al., 1999).
Another key unanswered question is how the genes necessary for dopaminergic neuron function are regulated. Multiple genes necessary for dopamine neuron function, survival, and axon pathfinding must be coordinately expressed in the correct subset of neurons. Elegant work from C. elegans has shown that a single cis-regulatory element and associated transcription factor (Ast-1) are necessary and sufficient for establishing dopamine neuron neurotransmitter identity (Flames and Hobert, 2009). Whether such a system is present in vertebrates is unknown. Because of the complexity of dopaminergic neuron development, as well as the involvement of the mesDA neurons in disease processes, identifying a discrete enhancer element specific for mesDA neurons would facilitate studies on the specification of neuronal identity and function in the basal ganglia.
In the present study we have performed an in vivo screen in zebrafish to identify dopaminergic neuron-specific enhancers. In zebrafish, dopaminergic neurons are present in the diencephalon but not the mesencephalon (Holzschuh et al., 2001; Kastenhuber et al., 2009), with projections in the adult to the subpallium (striatum) (JLB, unpublished data; Rink and Wulliman, 2001; Rink and Wulliman, 2002; Kastenhuber et al., 2009). Further, chemical ablation of the diencephalic dopaminergic (diDA) neurons phenotypically mimics loss of mesDA neurons in mammals (Lam et al., 2005; McKinley et al., 2005; Wen et al., 2008). We have identified a minimal 4.5kb enhancer element associated with the otpb gene that is sufficient to drive expression in specific dopaminergic neurons of the diencephalon in zebrafish. However, this enhancer (otpb.A) only drives expression in a subset of CNS dopaminergic neurons, and analysis of other dopaminergic-specific gene regions failed to identify a discrete enhancer with function in CNS neurons. Further, we were unable to detect any conservation between the sequence of the otpb.A enhancer element and the roughly 50 kb genomic neighborhoods (the most likely location of regulatory regions) of other genes specific to dopaminergic neurons. Our analysis of zebrafish dopaminergic gene regulatory regions reveals that conserved DNA elements are widely dispersed over large genomic loci.
Materials and Methods
Fish stocks and embryo raising
Adult fish were bred according to standard methods. Embryos were raised at 28.5°C in E3 embryo medium and staged by time and morphology (Kimmel et al., 1995). For in situ staining, embryos were fixed in 4% paraformaldehyde (PFA) in PBS for 3 hours at room temperature (RT) or overnight (O/N) at 4°C, washed briefly in PBS, dehydrated, and stored in 100% MeOH at -20°C until use.
Transgenic fish lines and alleles used in this paper were the following: Tg(otpb.A:egfp)zc48 (official ZFIN nomenclature Tg(otpb:EGFP)zc48), Tg(fezf2:egfp)zc55, Tg(pitx3:egfp)zc50, Tg(f.TH.A:egfp)zc56, Tg(otpb.A:GAL4)zc57 (official ZFIN nomenclature Tg(otpb:Gal4-VP16)zc57), Tg(otpb.I:GAL4)zc66, and Tg(UAS:GFP) [(Tg(5xUAS:GFP)nkuasgfp1a - kind gift of K. Kawakami]. Lines are available upon request.
Immunohistochemistry and In situ hybridization
Following fixation and dehydration in methanol, embryos were rehydrated, permeabilized using proteinase K [10μg/mL in PBST (PBS with 0.1% Tween-20)] at 28°C for 60′ (8′ for 24hpf; 20′ for 36hpf; and 30′ for 48hpf) without rocking, washed twice in PBST for 5′ then re-fixed for 15′. Embryos were then washed in PBST, blocked in PBST/1% DMSO/2% BSA/5% normal goat serum (NGS), and then incubated O/N in a primary antibody solution diluted in PBST/1%DMSO/2%BSA/2%NGS at 4°C. The next day embryos were washed in PBST/1%DMSO/1%NGS for a minimum of 6 hours, followed by incubation O/N with secondary antibodies, and washing the following day. Antibodies and concentrations used were rabbit polyclonal anti-tyrosine hydroxylase 1:400 (Millipore), mouse monoclonal anti-GFP 1:400 (Millipore), Cy-3 anti-rabbit 1:400, and Alexa 488 anti-mouse 1:400.
Double immunohistochemistry/in situ labeling was performed by permeabilization using 0.1% collagenase in PBST, re-fixation for 10′ with 4% PFA, and then performing anti-GFP antibody staining and detection in PBST using rabbit polyclonal anti-GFP 1:400 (Millipore #11122) followed by anti-rabbit Alexa 488 1:250 (Invitrogen). Following washing in PBST, embryos were fixed for 1 hour, washed with PBST, then re-permeabilized using 0.1% collagenase at RT. Whole-mount in situ labeling for dat (Holzschuh et al., 2001) and isotocin (Blechman et al., 2007) was then performed, followed by plastic sectioning, as previously described (Bonkowsky and Chien, 2005).
Genomic PCR and Enhancer Cloning
PCR primers used to clone genomic fragments are listed in Table S1. PCR and cloning of genomic fragments into pDONR P4-P1R was performed as described (Bonkowsky et al., 2008). The identity of the genomic fragments was confirmed by restriction enzyme digests and partial sequencing. Unless otherwise specified, the minimal promoter used for expression in Gateway constructs was the adenovirus E1b TATA box with the carp β-actin 5′-UTR fragment (Kwan et al., 2007; Bonkowsky et al., 2008). Specific plasmids used for cloning were pME-basEGFP (middle entry clone with EGFP preceded by minimal promoter), pDestTol2pA2, pDestTol2CG2, pDestTol2CR3 (pDestTol2pA3 with cmlc2:TagRFP transgenesis marker); pME-basGal4-VP16413-470 (Koester and Fraser, 2001; Ogura et al., 2009) was used for generation of GAL4 transgenic lines. pME-gata2EGFP is a middle entry clone with EGFP proceeded by the gata2a minimal promoter (Meng et al., 1997; Bessa et al., 2009). pME-cfosEGFP is a middle entry clone with EGFP preceded by the mouse c-fos minimal promoter (Dorsky et al., 2002).
Injection of DNA constructs and raising of stable transgenic lines was performed essentially as described (Fisher et al., 2006; Kwan et al., 2007; Bonkowsky et al., 2008). Patterns of enhancer expression were confirmed by transient injections of each construct (>100 embryos per construct), as well as isolation of two or more independent stable transgenic lines (in cases where stable transgenics were isolated). Plasmids and specific PCR conditions are available upon request.
Microscopy and image analysis
Image acquisition and analysis were performed essentially as described previously (Suli et al., 2006). Images of embryos processed for immunohistochemistry were taken using a confocal microscope; embryos were taken step-wise into a solution of 80%glycerol/20% PBST, then mounted on a glass slide with a #0 coverslip fixed into place over a well made using electrical tape. NIH ImageJ software (W. Rasband, NIMH) was used to merge slices to create maximal intensity z-stack projections.
Comparative genomic analysis
Cross-species non-coding conservation was determined by examination of the zebrafish genome assembly Zv5 at the UCSC genome browser (http://genome.ucsc.edu/) (Siepel et al., 2005). The “DA motif” was defined using the position weight matrix scoring from Flames and Hobert (2009). Sequence comparisons for DA motifs and cross-gene comparisons were done using sequence from Zv7. DA motif searches of DNA fragments were performed using ConSite (http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite) (Sandelin et al., 2004). Genomic DNA comparisons between non-coding regions of genes were performed using Shuffle-LAGAN in rVISTA (Brudno et al., 2004; Frazer et al., 2004). Genomic regions chosen for comparison were centered on each coding regions, encompassing 86.5kb from the ddc locus, 51.3kb from the slc6a3 locus, 76.1 kb from the th1 locus, and 42.2 kb from the slc18a2 locus. Shared synteny was determined from genetic maps from the Ensembl and UCSC genome browsers (Hubbard et al., 2009; Rhead et al., 2009).
Results
Screen for dopaminergic neuron enhancers
We undertook a screen to identify genomic DNA fragments which might serve as enhancers to drive expression in dopaminergic neurons of the zebrafish brain, with minimal expression in other neuron types. The identification of potential enhancer fragments was based on the concept of conserved non-coding sequence serving as potential enhancers (Allende et al., 2006; Gomez-Skarmeta et al., 2006; Pennachio et al., 2006). This assumes that regions of nucleotide conservation (usually of greater than 60-70% identity) between species in regions of conserved syntenic blocks of genes may function as enhancers (or silencers) of transcription.
We cloned genomic fragments using PCR into a Tol2 transposon-based vector (Kawakami, 2004; Kwan et al., 2007; Villefranc et al., 2007). Genomic DNA fragments were chosen based on their location in relative proximity to a target gene (described below), location upstream of the first coding exon (for some targets), and conservation of non-coding sequence (using the UCSC genome web browser http://genome.ucsc.edu/) (Kent et al., 2002; Bejerano et al., 2005). To visualize expression driven by the potential enhancers, the DNA fragments were cloned immediately upstream of a minimal promoter followed by GFP (Kwan et al., 2007; Villefranc et al. 2007). We have previously demonstrated that this minimal promoter is competent to drive expression in diverse CNS cell types without ectopic expression (Bonkowsky et al., 2008). We also tested the gata2a and c-fos minimal promoters using previously characterized enhancers (Dorsky et al., 2002; Bessa et al., 2009), but found higher rates of ectopic expression in non-target tissues (JLB, unpublished data). To test for expression, we injected one-cell stage embryos and looked for GFP expression from 12hpf through 96hpf. The first expression of th in zebrafish is between 16-20hpf (Holzschuh et al., 2001). Transient expression was analyzed in 100-200 embryos; if we observed consistent CNS expression in a region that potentially had overlap with diDA neurons, we raised stable transgenic lines for characterization. Characterization of stable transgenic lines consisted of double immunohistochemistry for GFP and for tyrosine hydroxylase (TH).
We used a comprehensive strategy to identify potential target genes, choosing them from two classes (Table 1). The first class were genes with known or putative roles in dopaminergic neuron specification [including FEZ family zinc finger 2 (fezf2), LIM homeobox transcription factor 1 alpha 2 (lmx1a.2), muscle segment homeobox E (msxE), neurogenin 1 (ngn1), nuclear receptor subfamily 4 group A member 2a (nr4a2a, a homolog of mammalian nurr1), orthopedia homolog a and b (otpa and otpb), and paired-like homeodomain transcription factor 3 (pitx3)] (Abeliovich and Hammond, 2007; Smidt and Burbach, 2007). The second class were genes with roles in dopaminergic neuron function: aromatic acid decarboxylase (ddc, previously known as aadc), dopamine receptor (drd2b), tyrosine hydroxylase (th, to be distinguished from its paralog th2), dopamine transporter (slc6a3, previously dat), and vesicular monoamine transporter 2 (slc18a2, previously vmat2). In some cases we tested for overlap of expression of a particular gene with th by double-labeling for its mRNA by in situ hybridization, and TH by immunohistochemistry (data not shown). In addition to analyzing zebrafish genomic fragments for potential dopaminergic enhancer activity, we also tested elements from pufferfish, mouse, and rat. Genomic DNA fragments were also cloned from pufferfish (Fugu rubripes) because of its compact genome size and the assumption that intergenic regions would be enriched for sequences that could serve as enhancers (Brenner et al., 1994). Enhancers from mouse and rat were chosen because of their known expression in dopaminergic neurons (such as the rat TH promoter) (Schimmel et al., 1999), or expression in the mesodiencephalon (VISTA Enhancer Browser- Visel et al., 2007). Finally, we tested a BAC with GFP inserted at the mouse slc6a3 region (GENSAT1- BX1837) (Gong et al., 2003).
Table 1.
CNS dopaminergic neuron enhancer screen: DNA elements tested.
Table of genomic DNA fragments tested in vivo, grouped according to functional category, gene, and species. Position relative to translation start site (chosen instead of transcriptional start site because of variability in zebrafish genome annotation), and enhancer fragment size are also listed. Primer sequences are in Table S1.
| Gene | Enhancer | Species | Category+ | CNS Expression* | Stable/Allele″ | Positionˆ (kb) | Fragment Size (kb) |
|---|---|---|---|---|---|---|---|
| ddc | Dr | F | - | 6.6 | 3.4 | ||
| slc6a3 | A | Dr | F | - | -0.7 | 1.9 | |
| E | Dr | F | - | -5.1 | 5.1 | ||
| slc6a3 (BAC) | BX1837** | Mm | F | - | |||
| drd2B | A | Dr | F | - | -0.2 | 6.7 | |
| f.slc6a3 | A | Fr | F | - | -0.4 | 4.5 | |
| B | Fr | F | - | 1.7 | 3.8 | ||
| fezl | Dr | T | + | -3.4 | 3.7 | ||
| LB1 | 219 | Mm | - | FOXP2 intragenic | 1.3 | ||
| 298 | Mm | + | SHFM1-DLX5 | 0.7 | |||
| 304 | Mm | + | JMJD2C-PTPRD | 0.6 | |||
| Lmx1a.2 | A | Dr | T | + | + | -0.1 | 5.2 |
| B | Dr | T | + | + | -2.0 | 1.3 | |
| C | Dr | T | + | + | -0.1 | 2.5 | |
| msxE | Dr | T | + | + | -0.1 | 2 | |
| ngn12 | 3.1delLATE | Dr | T | + | + | 0 | 2.4 |
| nurr1 | Dr | T | - | 4.5 | 4.5 | ||
| otpa | Dr | T | - | 1.0 | 2.4 | ||
| f.otpb | A | Fr | T | - | -0.1 | 2.0 | |
| B | Fr | T | - | -2.1 | 1.8 | ||
| otpb | A | Dr | T | + | +; zc48 | -4.8 | 4.4 |
| B | Dr | T | - | -4.8 | 1.8 | ||
| C | Dr | T | + | + | -4.1 | 3.7 | |
| D | Dr | T | - | -7.2 | 2.5 | ||
| E | Dr | T | + | + | -4.1 | 2.7 | |
| F | Dr | T | - | -1.3 | 1.3 | ||
| G | Dr | T | + | + | -4.8 | 3.4 | |
| H | Dr | T | - | -4.1 | 1.1 | ||
| I | Dr | T | + | + | -2.8 | 0.4 | |
| pitx3 | A | Dr | T | + | +; zc50 | -0.3 | 6.7 |
| f.TH | A | Fr | F | + | +; zc56 | -5.4 | 5.1 |
| B | Fr | F | + | + | -2.7 | 2.7 | |
| C | Fr | F | - | - | -5.4 | 3.1 | |
| D | Fr | F | - | - | -8.0 | 2.7 | |
| E | Fr | F | + | + | -2.6 | 0.8 | |
| F | Fr | F | - | - | -1.1 | 1.3 | |
| G | Fr | F | + | + | -7.7 | 7.8 | |
| H | Fr | F | - | - | -6.1 | 6.0 | |
| I | Fr | F | - | - | -6.1 | 3.5 | |
| J | Fr | F | - | - | -2.6 | 0.3 | |
| K | Fr | F | + | + | -2.3 | 0.5 | |
| L | Fr | F | + | + | -2.2 | 0.4 | |
| M | Fr | F | + | + | -2.3 | 0.3 | |
| O | Fr | F | - | - | -1.8 | 0.7 | |
| rat TH3 | Rn | F | - | -0.1 | 4.5 | ||
| th | A | Dr | F | - | -12 | 2.3 | |
| B | Dr | F | - | -4.7 | 4.5 | ||
| C | Dr | F | - | -2.1 | 5.8 | ||
| D | Dr | F | - | 7.3 | 6 | ||
| uchL1 | A | Dr | F | - | -0.8 | 1.1 | |
| slc18a2 | A | Dr | F | - | 0.1 | 0.3 | |
| B | Dr | F | - | -3.1 | 1 | ||
| C | Dr | F | - | -10.6 | 1 | ||
| D | Dr | F | - | 0.1 | 2 | ||
Expression was evaluated from 12hpf through 96hpf.
GENSAT1 BAC clones (Gong et al., 2003).
“Category” refers to the gene type-F: functional (function of dopamine neurons), T: transcription factor involved in specifying dopamine neurons.
Stable indicates that a stable transgenic line was generated and tested. If the line has been maintained this is indicated by an allele number.
“Position” is relative to translation start site (negative is upstream); except for Lawrence-Berkeley clones, which lists flanking or intragenic region.
Abbreviations: LB: Lawrence-Berkeley clones; Fr: Fugu rubripes (pufferfish); Mm: Mus musculus (mouse); Rn: Rattus norvegicus (rat); Dr: Danio rerio (zebrafish).
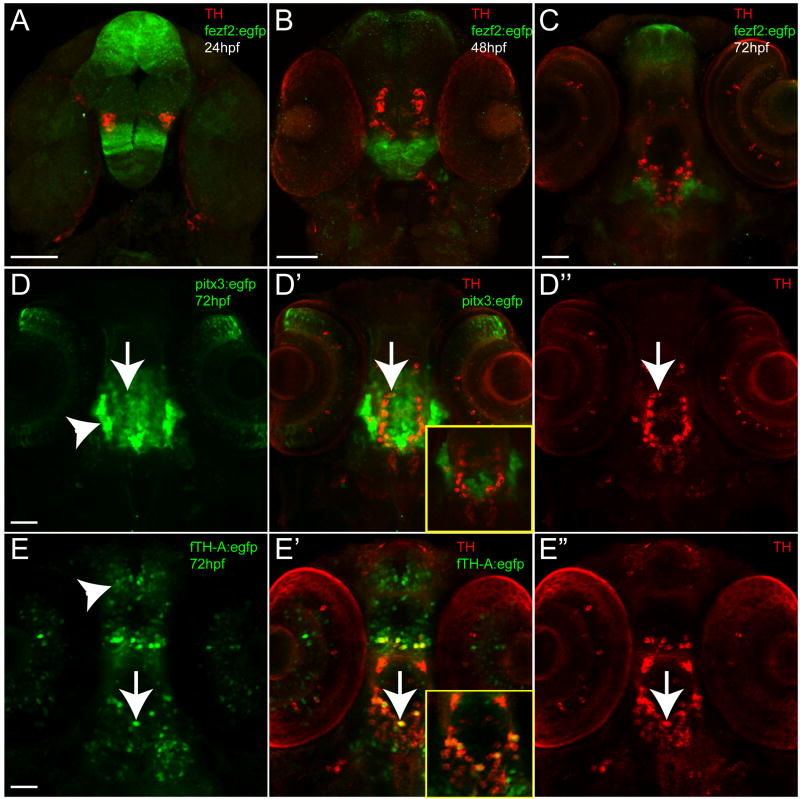
We tested 54 fragments from 21 different genes. In many cases no CNS expression was seen. We found that DNA fragments isolated from regions near genes encoding transcription factors were more likely to function in vivo as enhancers, a phenomenon that has also been noted for other tissue types (Sandelin et al., 2004; Woolfe et al., 2005). For fragments that functioned as CNS enhancers during development, most showed either minimal or no overlap with TH-positive neurons (as for Tg(fezf2:egfp)zc55 and msxE:EGFP, Figure 1A-C and Figure S2E-G) or had widespread expression in non-TH neurons (Tg(pitx3:egfp)zc50, Tg(f.TH.A:egfp)zc56, and Tg(lmx1a.2:egfp) (Figure 1D-E and Figure S2A-D). pitx3 has subsequently been shown not to express in dopaminergic neurons in zebrafish (Filippi et al., 2008). Expression patterns from the transgenic lines Tg(lmx1a.2:egfp), Tg(fezf2:egfp)zc55 and Tg(pitx3:egfp)zc50 appear to match with the known expression domains of lmx1a.2, fezf2 and pitx3, respectively (Figure S2; Blechman et al., 2007; Filippi et al., 2008), indicating that our enhancer lines recapitulate endogenous gene expression at least to some extent.
Figure 1.
Examples of stable transgenic enhancer lines generated; ventral views, anterior to the top, double-immunohistochemistry for GFP and TH (green and red), confocal z-stacks. Scale bars are 50 μm. (A-C) Time-series of Tg(fezf2:egfp)zc55 expression at 24hpf, 48hpf, and 72hpf, shows non-overlap of TH and GFP expression. (D-D″) Tg(pitx3:egfp)zc50 and (E-E″) Tg(f.TH.A:egfp)zc56 at 72hpf, shows partial overlap of enhancer expression and TH (arrows), but also wide spread expression of the enhancer in non-TH neurons (arrowheads). Insets in D′ and E′ show single confocal plane of double-labeling, emphasizing the minimal overlap of TH and GFP labeling.
Since it is possible that some enhancers failed to function in vivo because they might require a specific promoter (Gehrig et al., 2009), we also tested two of our genomic DNA fragments with two additional alternate promoters (gata2a and c-fos). We tested a fragment from the ddc gene, and the optb. A enhancer (described further below). The ddc fragment did not drive expression with any of the three promoters (data not shown). Expression driven by the otpb.A enhancer in transient injections was similar with all three promoters (Supplemental Figure S1), although more ectopic non-CNS expression (compared to the stable transgenic otpb.A line and the endogenous otpb expression pattern) was seen using the c-fos and gata2a minimal promoters. Another possibility is that a gene's endogenous promoter might be necessary for enhancer-driven expression. However, we tested the known endogenous promoter for slc6a3 in one of our constructs, and for many other tested fragments, their size and location immediately upstream of the translation start site makes it very likely that they include the endogenous promoter (including fragments for the genes th, drd2b, Fugu dat, otpa, Fugu otpb, and slc18a2).
We characterized most enhancers that had CNS expression by generation of a stable transgenic line, and have maintained some of these lines (Table 1). The failure of some DNA fragments to function as enhancers despite high cross-species non-coding conservation might be due to function as a repressor or silencer of expression; to the fragment regulating expression at a different (non-embryonic) stage; or to insufficiency of the element in isolation to drive expression.
Analysis of genomic regions of dopaminergic neuron phenotype genes
Because of our difficulty in identifying a discrete enhancer with expression specific to dopaminergic neurons (with one exception, see below), we sought to address how dopaminergic neuron phenotype is regulated in vertebrates. To address whether vertebrate dopaminergic neuron phenotype requires a single, discrete cis-regulatory element, or a dispersed complex code of binding sites, we examined a core group of genes whose expression is relatively specific to dopamine neurons, using both in silico and in vivo approaches. For our core group of dopaminergic phenotype genes we chose genes expressed by all neurons that use dopamine as a neurotransmitter, including the rate-limiting enzyme for dopamine synthesis th, the enzyme for converting L-dopa to dopamine ddc, the pre-synaptic uptake receptor slc6a3, and the cytosol to synaptic vesicle transporter slc18a2.
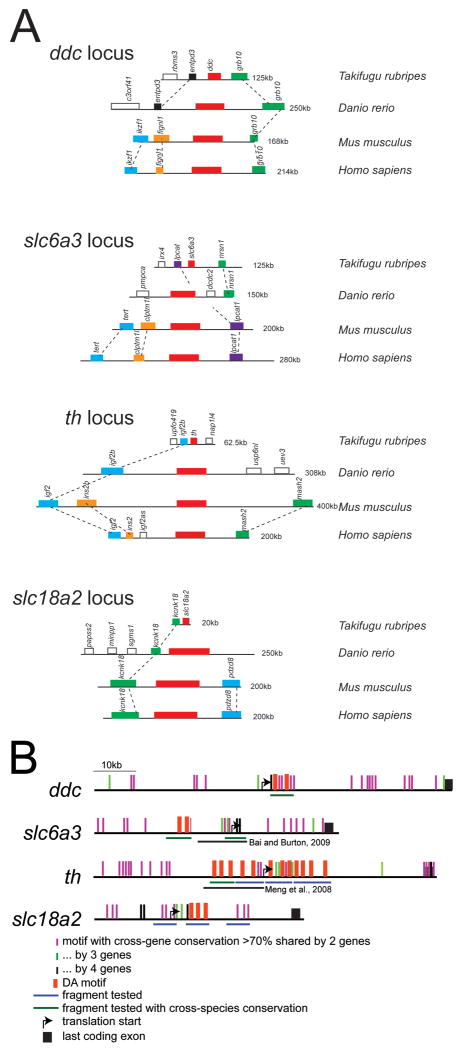
First, we examined whether the genomic neighbors for the dopaminergic phenotype genes are conserved across evolutionary time. For vertebrate genes, conserved synteny of genes in “genomic regulatory blocks” is associated with dispersal of cis-regulatory elements amongst the coding exons of the different genes (reviewed in Kikuta et al, 2007). We compared conserved synteny between human, mouse, zebrafish, and pufferfish regions for the genes ddc, slc6a3, th, and slc18a2 (Figure 2A). In all cases, there was at least partial conservation of synteny between zebrafish and other vertebrates, suggesting that necessary cis-regulatory elements may be present in these regions.
Figure 2.
(A) Gene order and conserved synteny at ddc, slc6a3, th, and slc18a2 loci between pufferfish, zebrafish, mouse, and human; scale is approximate (indicated to right). Zebrafish has partial conservation of synteny with mammalian orthologs. (B) Schematic representation of conserved DA motifs in zebrafish dopamine pathway genes (scale is approximate). DA motifs in tested genomic fragments are represented by red boxes shown above; enhancer fragments tested are shown below. If significant cross-species conservation (from the UCSC genome site) was present the enhancer fragment is labeled in green, otherwise the enhancer fragments are shown in blue. Shared (non-coding) DNA elements between 2 genes are shown as vertical purple lines above the gene locus; elements shared by 3 genes are shown as green vertical lines; and by all 4 genes as black vertical lines. None of these enhancers was sufficient to drive expression in dopaminergic neurons in vivo. Also shown are the genomic fragments tested by Meng et al. (2008) and Bai and Burton (2009), which also failed to recapitulate th and dat expression, respectively.
Next, we looked for conserved cross-species conservation in genomic regions surrounding DA neuron-specific genes, and tested in vivo the ability of different genomic fragments to act as enhancers. Surprisingly, none of the genomic fragments we tested in vivo drove expression in DA neurons, or even in the CNS (Table 1; Figure 2B). Further, this was despite the presence in many of the fragments of multiple copies of the “DA motif” (Flames and Hobert, 2009), as well as high cross-species conservation in some fragments. We found that the presence of the DA motif was no more frequent (and with no higher matrix scores) than in a CNS enhancer not expressed in DA neurons (foxP2-enhancerA, Bonkowsky et al., 2008). We also constructed and tested a DA motif (5′-gcagaggaggaagagtggaga-3″) triplet multimer, fused to a basal promoter and GFP, but did not find specific CNS expression. A separate study has also tried to identify a DA-specific enhancer from the slc6a3 region, and tested an 11-kb fragment encompassing the transcription start and regions upstream (Bai and Burton, 2009). This 11-kb enhancer drove expression in dopaminergic neurons of the pre-tectal region, but not in other dopaminergic neurons, and also had ectopic expression in many CNS cell groups, implying the absence of both necessary enhancer elements as well as of silencing elements. Together, these results show that in vertebrates a single discrete DA motif is not sufficient for expression in DA neurons.
We also performed a detailed expression analysis of potential enhancers from the Fugu th genomic region (Figure S3). The original transgenic line Tg(f.TH.A:egfp)zc56 showed expression in most diDA neurons (Figure 1), but also had expression in many non-DA neurons. Although we tested a large number of fragments and transgenic lines based on this original enhancer (Figure S3), none gave specific expression in DA neurons alone.
While we did not find that the DA motif was sufficient for expression in dopaminergic neurons, an alternative explanation is that a different conserved element is used in vertebrates. To try to identify whether there were other elements in the loci of the dopamine phenotype genes that might specify for DA neuron expression, we did a comparative analysis of the zebrafish genomic loci for th, ddc, slc6a3, and slc18a2, to identify highly conserved (>70%) regions of 50bp or more. We found 42, 53, and 65 regions in the th locus that were highly conserved with regions in the slc6a3, slc18a2, and ddc loci, respectively; and only 1 region that was conserved in all 4 genomic loci (Figure 2B). These regions of conservation were dispersed over the entirety of each genomic locus. Further, our previous in vivo analysis had tested some of these regions which failed to drive CNS expression. Similarly, genomic fragments for the th and slc6a3 regions partially overlapping the fragments we tested had also been tested in vivo by other groups (Meng et al., 2008 and Bai and Burton, 2009, respectively; Figure 2B) and had also failed to drive specific dopaminergic neuron CNS expression.
We used BLAST and CLUSTALW analyses of the most highly conserved regions shared by the different gene loci to look for conserved DNA motifs. No common shared sequence motifs were identified in these highly conserved regions. We conclude that there is not an obvious single candidate cis-regulatory element that controls expression in vertebrate dopamine neurons, and that the core regulatory elements necessary for dopaminergic expression are widely dispersed.
This dispersed pattern of non-coding conservation in the dopamine pathway genes, together with our in vivo testing of specific genomic fragments, strongly argues that regulation of the genes necessary to maintain a dopaminergic phenotype is complex in vertebrates. In contrast to C. elegans, each group of dopaminergic neurons in vertebrates may require a distinct combinatorial code to establish its mature phenotype.
Characterization of otpb enhancer
From the enhancer screen (Table 1) we found a genomic DNA fragment (otpb.A) from the region upstream of the orthopedia-b (otpb) gene that drove expression in TH-positive neurons of the diencephalon with minimal expression in other neurons (Figure 3). otpb encodes a homeobox transcription factor necessary for dopaminergic neuron development in zebrafish and in mouse (Del Giacco et al., 2006; Ryu et al., 2007). Double-labeling for GFP and for TH in the transgenic line Tg(otpb.A:egfp)zc48 of embryos at 72hpf showed co-expression in the diencephalon, primarily in dopaminergic neuron groups 4 and 6 (based on the nomenclature of Rink and Wullimann, 2002) (Figure 3E-E″). Expression of GFP in Tg(otpb.A:egfp)zc48 was first visible between 18-24hpf, and became more widespread between 36-48hpf (Figure 3F-G″).
Figure 3.
Characterization of Tg(otpb.A:egfp)zc48; confocal images of whole mount embryos double-labeled for GFP and for TH immunohistochemistry, ventral views, anterior to the top. Scale bar is 50 μm. (A) Schematic diagram of TH-positive cell groups in the zebrafish brain at 72hpf, based on Rink and Wullimann, 2002. (B) Confocal z-stack projection of TH immunohistochemistry at 72hpf in the zebrafish brain. (C) Confocal z-stack projection of GFP immunohistochemistry in Tg(otpb.A:egfp)zc48. (D-D″) Confocal z-stack projections at different dorsal-ventral levels in the brain of Tg(otpb.A:egfp)zc48 at 72hpf, showing co-expression of TH and GFP in diDA neuron groups 4 and 6, but not groups 1 and 2 (arrow); arrowhead points to the NPO neurons. (E-E″) Magnified views of the region boxed in B″, showing extensive overlap of GFP-positive neurons in the diencephalon with TH expression. (F-F″) Expression at 24hpf (G-G″) Expression at 48hpf.
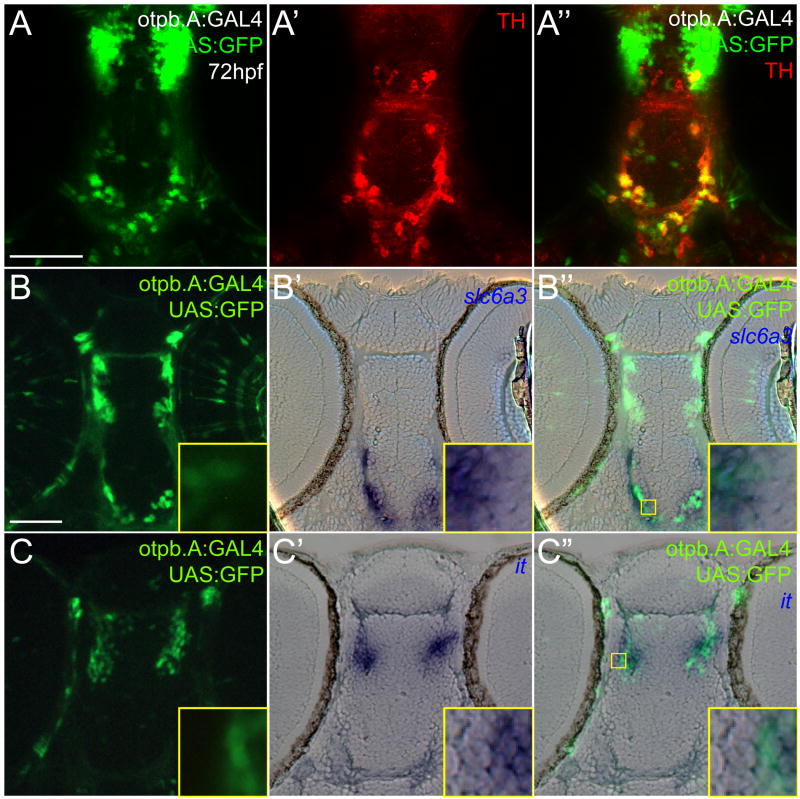
TH labels all catecholaminergic neurons, including adrenergic, noradrenergic, and dopaminergic types. To confirm that the TH-positive neurons labeled by the otpb.A enhancer were in fact dopaminergic, we performed double-labeling for the dopamine transporter (slc6a3) gene. slc6a3 encodes a reuptake transporter of dopamine that is specifically expressed in dopaminergic neurons and not other catecholaminergic neuron types (Nirenberg et al., 1996; Nirenberg et al., 1997; Holzschuh et al. 2001). We used Tg(otpb.A:GAL4)zc57, in which GAL4-VP16 (Koester and Fraser, 2001) drives expression under the control of otpb.A (Figure 4A-A″), to analyze co-expression. Interestingly, when Tg(otpb.A:GAL4)zc57 was crossed to Tg(UAS:GFP), expression of GFP was found in all TH-positive neurons of the diencephalon, in contrast to the original Tg(otpb.A:egfp)zc48 line, in which not all diDA neurons were labeled. This may be due to a position effect of the original Tg(otpb.A:egfp)zc48 line, or to stronger expression due to amplification by the GAL4/UAS system (Koester and Fraser, 2001). We observed that diencephalic neurons expressing GFP in the diencephalon also co-expressed slc6a3 (Figure 4B-B″), confirming that they were dopaminergic. The otpb.A enhancer also drives expression in the rostral diencephalon, in the neurosecretory preoptic (NPO) neurons that require otpb expression for their development (Blechman et al., 2007). Double-labeling in Tg(otpb.A:GAL4)zc57;Tg(UAS:GFP) embryos for GFP and for isotocin (the chief neurohypophysial peptide expressed in the NPO- Unger and Glasgow, 2003) revealed that most of this more rostral group co-expressed both markers (Figure 4C-C″). Other neuroendocrine-specific genes are also co-expressed with the otpb.A reporter in the NPO region (J. Schweitzer, H. Loehr, W. Driever, J.L.B., manuscript in preparation). These results show that the otpb.A enhancer specifically recapitulates otpb gene expression in non-dopaminergic NPO cells and most if not all diDA neurons.
Figure 4.
Characterization of otpb.A expression in dopaminergic and neuroendocrine cells. Embryos at 72hpf, Tg(otpb.A:GAL4)zc57, Tg(UAS:GFP). Scale bar is 50 μm. (A-A″) Confocal z-stack projection of whole-mount embryos double-labeled for GFP and for TH immunohistochemistry, ventral views, anterior to the top. (B) and (C) Plastic horizontal sections of double-labeled Tg(otpb.A:GAL4)zc57; Tg(UAS:GFP) embryos at 72hpf, anterior to the top. Higher magnification inset showing co-localization is indicated by yellow boxed area and is shown at bottom right of each panel. (B-B″) Labeled for GFP antibody (B) and slc6a3 mRNA (B′), GFP-positive neurons in the diencephalon co-express slc6a3, confirming their identity as dopaminergic. (C-C″) Labeled for GFP antibody (C) and isotocin mRNA (C′), rostral diencephalon GFP-positive NPO neurons co-express GFP and isotocin.
Regulation of dopaminergic identity and the otpb enhancer
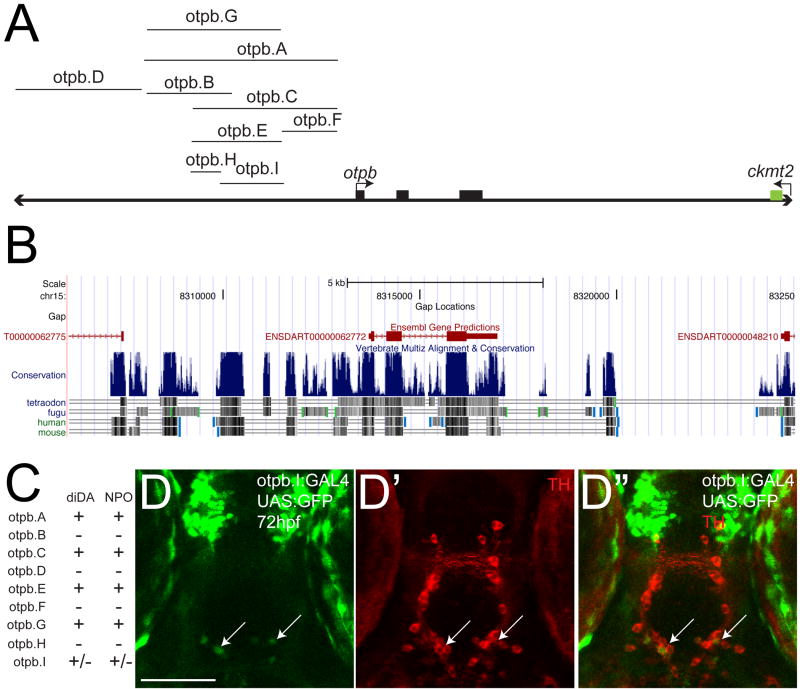
otpb is necessary for development of the diDA neurons and is expressed in all diDA neurons as well as in the NPO cells (Ryu et al., 2007; Loehr et al., 2009). In an effort to identify a minimal sufficient region for diDA expression, we tested multiple genomic fragments in the otpb genomic locus, including a more distal genomic region (otpb.D) (Figure 5A-C). Some of the fragments failed to drive any CNS expression (fragments otpb.B, otpb.D, otpb.F, and otpb.H), while some overlapping fragments drove essentially identical expression in approximately 22-24 diDA neurons (enhancers otpb.C, otpb.E, and otpb.G) (Figure 5C). otpb.I recapitulated only part of the original otpb.A pattern (Figure 5C, D). To demonstrate that this partial expression was in fact due to an absence of necessary cis-binding elements, and not simply low levels of expression, we tested the otpb.I fragment (a 444-bp fragment derived from optb.A) both when driving GFP directly, as well as driving GAL4-VP16 expression in a Tg(UAS:GFP) background. optb.I only drove partial expression in the NPO and diDA neurons, even when using the GAL4/UAS:GFP system (Figure 5D). Thus, otpb.A must contain other important sequences not included in otpb.I, consistent with our hypothesis that expression in diDA neurons is regulated by multiple independent cis-regulatory binding sites spread out over a large genomic region.
Figure 5.
Genomic structure and characterization of otpb genomic locus enhancers. (A) otpb genomic region (not to scale). Coding exons are shown as solid black boxes. DNA fragments tested for enhancer activity are shown. Region pictured is approximately 20kb; 3′-most exon of ckmt2 is shown. (B) Screenshot from the UCSC genome browser (http://genome.ucsc.edu) showing the location of the exons of genes, relative to regions of cross-species conservation (shown as vertical blue lines). Increasing conservation is indicated by increasing height and/or density of the blue lines. Species used to determine the conservation are shown below. (C) Summary table of expression patterns of the different enhancers at 72hpf in the CNS with respect to expression in the diencephalic dopaminergic neurons (diDA) and neurosecretory preoptic nucleus (NPO). (D-D″) Confocal z-stack projection of whole-mount Tg(otpb.I:GAL4)zc57 embryos at 72hpf, shown crossed to Tg(UAS:GFP) and double-labeled for GFP (D) and for TH (D′) immunohistochemistry, ventral views, anterior to the top. Arrows point to the sparse diDA neuron expression.
Since we had identified a relatively small region in otpb.A (4.5 kb) that was sufficient for expression in diDA neurons, we wondered whether this region contained motifs that would be shared with other genes expressed in diDA neurons. We performed a comparative analysis of the otpb.A genomic region (using the sequence obtained by direct sequencing of the cloned fragment), with the regions surrounding the ddc, slc6a3, TH, and slc18a2 genes using rVISTA. In addition, we examined the region upstream of otpa in zebrafish, a paralog of otpb. We failed to observe any significant conservation (>70%) of sequence from the otpb.A region with otpa, or ddc, slc6a3, th, or the slc18a2 gene regions. Therefore, the cis-regulatory sequences, and by extension the transcription factors that bind to these sequences, are probably different for otpb compared to otpa or to the dopaminergic phenotype genes.
Discussion
Through a detailed screen for enhancers that drive expression in CNS dopaminergic neurons, we have identified a single discrete enhancer that functions in diencephalic dopaminergic (diDA) neurons of the zebrafish. This enhancer fragment, otpb.A, drives expression specifically in diDA neurons and in NPO neurons of the hypothalamus. Despite testing a large number of potential enhancers (54 fragments from 21 genes) with conserved non-coding conservation, most of the genomic regions we tested failed to have CNS expression in embryos, with the exception of regions derived from transcription factor genes. Other groups have also noted previously that genomic regions derived from locations near transcription factors are more likely to act as enhancers (Sandelin et al., 2004; Woolfe et al., 2005).
The reason(s) why certain genomic fragments did not work as enhancers are uncertain. A fragment might work as a silencer of expression; it might regulate expression at a non-embryonic stage (for example, Fujimori, 2009); or it might drive expression at very low levels, although we have tested fragments from th, slc6a3, and ddc driving GAL4-VP16 and failed to see expression when injected into UAS:GFP transgenic embryos (data not shown). Another possibility is that the endogenous promoter associated with a gene might be necessary for enhancer-driven expression (Gehrig et al., 2009). However, for slc6a3 we included its known endogenous promoter in one of our constructs, and for th, drd2b, Fugu dat, otpa, Fugu otpb, and slc18a2 we tested large fragments immediately upstream of the translation start site, which were very likely to encompass the endogenous promoter. Furthermore, we have tested two alternative minimal promoters (from c-fos and gata2a), and did not find substantive differences in expression compared to the E1b-based minimal promoter that we used. We suggest a model in which single DNA elements in isolation are insufficient to specify dopaminergic neuron phenotype in vertebrates. This is based on our work examining the otpb.A enhancer in detail, and our analysis (in vivo and in silico) of the genomic regions surrounding the dopaminergic phenotype genes. However, our strategy of using non-coding conservation as a marker of potential enhancers is of limited use in cases where the genomic annotation is incomplete. For example, in zebrafish a second tyrosine hydroxylase paralog has recently been identified (th2) (Candy and Collet, 2005; Chen et al., 2009; Filippi et al., 2010; Yamamoto et al., 2010), but which does not have significant expression until 3-4 dpf. Alternative strategies for identifying enhancers have also recently been described; for example, using tissue-specific ChIP-seq to identify p300 binding sites (Visel et al., 2009). To test more formally our model that multiple combined DNA elements regulate dopaminergic expression, ideally we would like to have a locus-spanning BAC with a recombineered GFP cassette, and compare it to isolated genomic fragments either alone or in combination. However, for the slc6a3 locus for example, there is no available BAC (database searches, JLB, and personal communication, Sanger Institute, zebrafish sequencing group) (presumably in part because of its telomeric location).
Our goal of identifying a vertebrate dopaminergic enhancer was only partially successful, in contrast to work in C. elegans that identified a simple “DA motif” that is necessary for terminal selection of neuron phenotype (Flames and Hobert, 2009). Loss of the DA motif leads to loss of expression in dopaminergic neurons, and ectopic expression of the transcription factor ast-1 (which binds the DA motif) is sufficient to induce a dopaminergic phenotype. Flames and Hobert propose a model in which dopaminergic neurotransmitter status is regulated by a single terminal selector gene and its corresponding cis-motif (the “bar code model”- Spitzer, 2009). The concept of “terminal selector genes” is appealing and several examples have been demonstrated in C. elegans (Hobert, 2008). However, the organization of both gene structure and the nervous system are considerably less complex in C. elegans compared to vertebrates. Most enhancers in C. elegans are located in the 1-2 kb immediately 5′ of the translation start (Okkema and Krause, 2005). The nervous system of C. elegans is considerably simpler than that of vertebrates. For example, C. elegans hermaphrodites have a total of eight dopaminergic neurons, with projections only to the nerve ring and nerve cord (reviewed in Nass and Blakely, 2003). Thus, the regulation of dopaminergic phenotype in C. elegans matches the relative simplicity of its dopaminergic circuits.
Using both bioinformatics and in vivo testing, we were unable to isolate a compact enhancer from the genomic loci of dopaminergic phenotype genes (th, slc6a3, slc18a2, and ddc). Rather, we found multiple conserved motifs dispersed across large genomic regions around these genes. While in some cases we tested these motifs in vivo and did not detect enhancer activity, we did not test all of these motifs, nor did we test them in combination. Further, the otpb.A enhancer, which drives expression in dopaminergic neurons, does not share any detectable motifs with neighboring genomic regions of other dopaminergic phenotype genes, and furthermore cannot be reduced to a compact DNA module that is sufficient for expression in dopamine neurons. The otpb.I subfragment (444bp) of otpb.A only expresses in 3-4 dopamine neurons, compared to 20-30 for the original otpb.A fragment, and this does not appear to be due to low levels of expression. Therefore, otpb.I does not have all the necessary cis-information to regulate expression in dopaminergic neurons. It is still formally possible that both our in vivo and in silico analyses have failed to detect a small, conserved cis-motif in the genomic regions of the dopaminergic phenotype genes or in the otpb.A enhancer.
The otpb.A enhancer drives expression in most of the diencephalic dopamine neurons in zebrafish, as well as in neuroendocrine cells, matching otpb's endogenous expression pattern (Del Giacco et al., 2006; Ryu et al., 2007). Despite extensive efforts, we were unable to isolate a minimal region of the otpb.A enhancer that was sufficient for either dopaminergic or neuroendocrine expression alone. Thus, regulation of the otpb.A enhancer appears to be coordinated across the entire 4.5 kb region. In silico analysis did not identify sequences shared with the genomic regions of otpa or of dopaminergic phenotype genes. The otpb.A enhancer does provide a valuable tool for investigating zebrafish diencephalic dopamine neuron development and function, with the potential for dopamine neuron specific-expression by using combinatorial expression approaches (EF, CBC, and JLB, unpublished data).
We conclude that in vertebrates dopaminergic cell identity regulation is dispersed over large genomic regions, and that a complex regulatory system is necessary for expression of a dopaminergic phenotype. This is consistent with other studies showing that vertebrate gene expression can depend upon widely dispersed cis-elements (Komisarczuk et al., 2009), in “genomic regulatory blocks” (reviewed in Kikuta et al., 2007). These findings suggest that dopaminergic cell identity is regulated by a mosaic of factors that dictate not only the dopaminergic neurotransmitter phenotype, but also other elements of neuronal identity such as synaptic targets and function (Figure 6). Our findings support a model in which distinct groups of dopaminergic neurons use unique solutions to achieve a dopaminergic phenotype.
Figure 6.

Model of dopaminergic neuron phenotype specification in vertebrates. Different groups of dopamine neurons are specified by different combinations of transcription factors, specifying both their dopamine phenotype and other aspects of their identity (such as their synaptic targets).
Supplementary Material
Figure S1. Comparison of different minimal promoters upstream of EGFP, driven by otpb.A enhancer. The transient injections with the E1b minimal promoter demonstrate expression in both the NPO and diDA cell groups, while expression under the control of the gata2a or c-fos minimal promoters is more limited in the desired cell groups. Confocal maximum projections, ventral views, anterior to the top, double-immunohistochemistry for GFP and TH (green and red). Arrows point to NPO cells, arrowheads to diDA neurons. Scale bar is 50 μm. (A) E1b minimal promoter; (B) pgata2 minimal promoter; (C) c-fos minimal promoter.
Figure S2. Characterization of lmx1a.2.A and msxE enhancer construct expression (A-A″ and E-G; maximum-intensity projections of double immunostaining for GFP and TH) and in situ expression (B-D, H-J) in whole-mount embryos. (A-A″) Stable transgenic Tg(lmx1a.2.A:egfp) embryo at 72hpf shows minimal overlap of TH and GFP expression. Ventral views, anterior to the top. (B-D) Whole-mount in situ expression patterns of lmx1a.2 at 24hpf, 36hpf, and 72hpf, ventral view, anterior to the top (except B, lateral view, anterior to the left). Note that in situ expression pattern at 72hpf correlates with enhancer expression pattern in (A). (E-G) Transient expression of msxE:EGFP. Ventral views, anterior to the top, at 24hpf, 36hpf, and 72hpf. (H-J) Whole-mount in situ expression patterns of msxE at 24hpf, 36hpf, and 72hpf, ventral views, anterior to the top (except H, lateral view, anterior to the left, dorsal up). Neither in situ nor transient transgenic expression at 72 hpf labels diDA neurons.
Figure S3. Genomic structure and functional enhancer characterization of Fugu TH genomic fragments. (A) Fugu TH genomic region (not to scale). Coding exons are shown as solid black boxes. DNA fragments tested for enhancer activity are shown. Region pictured is approximately 14kb; upstream 3′-most exon of nap1l4 is shown. (B) Summary table of enhancer CNS expression at 72hpf, categorized as expression in either as diDA neurons or non-dopaminergic cells (non-DA). (C, D) Maximum-intensity projections of whole-mount Tg(f.TH.M:egfp) embryo (C) and Tg(f.TH.O:egfp) embryo (D) at 72hpf, double-labeled for GFP and TH immunostaining. Ventral views, anterior to the top. Neither enhancer's expression overlaps with TH expression (compare extensive co-expression in Figure 1, showing Tg(f.TH.A:egfp)zc56).
Table S1. Primer sequences used for amplification of genomic DNA fragments tested for enhancer activity.
Acknowledgments
We would like to thank H. Otsuna and other members of the Chien lab for their assistance in preparing this work; W. Driever, J. Gomez-Skarmeta, K. Kawakami, G. Levkowitz, L. Pennacchio, and A. Visel for sharing plasmids and fish lines; B. Gaynes for making the pME-gata2aEGFP construct; and R. Dorsky and K. Kwan for helpful discussions. This work was supported by a PCMC Foundation grant to JLB, NIH R01 MH092256 to CBC, and NIH K12 5HD001410 and K08 DA024753 to JLB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich A, Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev Biol. 2007;304:447–54. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Albin RL, Mink JW. Recent advances in Tourette syndrome research. Trends Neurosci. 2006;29:175–82. doi: 10.1016/j.tins.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–6. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–60. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekman C, Perlmann T, Wallen A, Hoffer BJ, Morales M. A selective group of dopaminergic neurons express nurr1 in the adult mouse brain. Brain Res. 1999 Dec 18;851(1-2):125–32. doi: 10.1016/s0006-8993(99)02149-6. [DOI] [PubMed] [Google Scholar]
- Bai Q, Burton EA. Cis-acting elements responsible for dopaminergic neuron-specific expression of zebrafish slc6a3 (dopamine transporter) in vivo are located remote from the transcriptional start site. Neuroscience. 2009;164(3):1138–51. doi: 10.1016/j.neuroscience.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Siepel AC, Kent WJ, Haussler D. Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat Methods. 2005;2:535–545. doi: 10.1038/nmeth0705-535. [DOI] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, Fernández A, Montoliu L, Akalin A, Lenhard B, Casares F, Gómez-Skarmeta JL. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238:2409–17. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, Strähle U. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131:5627–37. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- Blechman J, Borodovsky N, Eisenberg M, Nabel-Rosen H, Grimm J, Levkowitz G. Specification of hypothalamic neurons by dual regulation of the homeodomain protein Orthopedia. Development. 2007;134:4417–4426. doi: 10.1242/dev.011262. [DOI] [PubMed] [Google Scholar]
- Bonkowsky JL, Chien CB. Molecular cloning and developmental expression of foxP2 in zebrafish. Dev Dyn. 2005;234:740–746. doi: 10.1002/dvdy.20504. [DOI] [PubMed] [Google Scholar]
- Bonkowsky JL, Wang X, Fujimoto E, Lee JE, Chien CB, Dorsky RI. Domain-specific regulation of foxP2 CNS expression by lef1. BMC Dev Biol. 2008;8:103. doi: 10.1186/1471-213X-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, et al. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1994;366:265–26. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Brudno M, et al. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003;19 1:i54–62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- Candy J, Collet C. Two tyrosine hydroxylase genes in teleosts. Biochim Biophys Acta. 2005;1727:35–44. doi: 10.1016/j.bbaexp.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Chen L, O'Keefe SL, Hodgetts RB. Control of Dopa decarboxylase gene expression by the Broad-Complex during metamorphosis in Drosophila. Mech Dev. 2002;119:145–56. doi: 10.1016/s0925-4773(02)00346-5. [DOI] [PubMed] [Google Scholar]
- Chen YC, Priyadarshini M, Panula P. Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochem Cell Biol. 2009;132(4):375–81. doi: 10.1007/s00418-009-0619-8. [DOI] [PubMed] [Google Scholar]
- Clark ME, Mellon PL. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995 Nov;15(11):6169–77. doi: 10.1128/mcb.15.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129(11):2639–48. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- Dahlstroem A, Fuxe K. Evidence for the existence of monoamine-containing neurons in the central nervous system. I. Demonstration of monoamines in the cell bodies of brain stem neurons. Acta Physiol Scand Suppl. 1964 232:1–55. [PubMed] [Google Scholar]
- Del Giacco L, Sordino P, Pistocchi A, Andreakis N, Tarallo R, Di Benedetto B, Cotelli F. Differential regulation of the zebrafish orthopedia 1 gene during fate determination of diencephalic neurons. BMC Dev Biol. 2006;6:50. doi: 10.1186/1471-213X-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong M, Wichmann T. Update on models of basal ganglia function and dysfunction. Parkinsonism Relat Disord. 2009 Dec;15 3:S237–40. doi: 10.1016/S1353-8020(09)70822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241(2):229–37. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Filippi A, Dürr K, Ryu S, Willaredt M, Holzschuh J, Driever W. Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development. BMC Dev Biol. 2007;7:135. doi: 10.1186/1471-213X-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi A, Mahler J, Schweitzer J, Driever W. Expression of the paralogous tyrosine hydroxylase encoding genes th1 and th2 reveals the full complement of dopaminergic and noradrenergic neurons in zebrafish larval and juvenile brain. J Comp Neurol. 2010;518(4):423–38. doi: 10.1002/cne.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–9. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server):W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori KE. Characterization of the regulatory region of the dopa decarboxylase gene in Medaka: an in vivo green fluorescent protein reporter assay combined with a simple TA-cloning method. Mol Biotechnol. 2009;41(3):224–35. doi: 10.1007/s12033-008-9120-1. [DOI] [PubMed] [Google Scholar]
- Gehrig J, Reischl M, Kalmár E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Müller F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat Methods. 2009 Dec;6(12):911–6. doi: 10.1038/nmeth.1396. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003 Oct 30;425(6961):917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006 Jan 13;124(1):47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19(23):10348–56. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003 Jan 23;37(2):233–47. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A. 2008 Dec 23;105(51):20067–71. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschuh J, Ryu S, Aberger F, Driever W. Dopamine transporter expression distinguishes dopaminergic neurons from other catecholaminergic neurons in the developing zebrafish embryo. Mech Dev. 2001 Mar;101(1-2):237–43. doi: 10.1016/s0925-4773(01)00287-8. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P. Ensembl 2009. Nucleic Acids Res. 2009 Jan;37(Database):D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- Jeong JY, Einhorn Z, Mercurio S, Lee S, Lau B, Mione M, Wilson SW, Guo S. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):5143–8. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenhuber E, Kratochwil CF, Ryu S, Schweitzer J, Driever W. Genetic dissection of dopaminergic and noradrenergic contributions to catecholaminergic tracts in early larval zebrafish. J Comp Neurol. 2010 Feb 15;518(4):439–58. doi: 10.1002/cne.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003 Jul 1;31(13):3576–9. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H, Fredman D, Rinkwitz S, Lenhard B, Becker TS. Retroviral enhancer detection insertions in zebrafish combined with comparative genomics reveal genomic regulatory blocks - a fundamental feature of vertebrate genomes. Genome Biol. 2007;8 1:S4. doi: 10.1186/gb-2007-8-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koester RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001 May 15;233(2):329–46. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Komisarczuk AZ, Kawakami K, Becker TS. Cis-regulation and chromosomal rearrangement of the fgf8 locus after the teleost/tetrapod split. Dev Biol. 2009 Dec 15;336(2):301–12. doi: 10.1016/j.ydbio.2009.09.029. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lam CS, Korzh V, Strahle U. Zebrafish embryos are susceptible to the dopaminergic neurotoxin MPTP. Eur J Neurosci. 2005 Mar;21(6):1758–62. doi: 10.1111/j.1460-9568.2005.03988.x. [DOI] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Häckel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008 Mar 13;57(5):760–73. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Zeller J, Sirotkin HI, French D, Schilbach S, Hashimoto H, Hibi M, Talbot WS, Rosenthal A. Zinc finger protein too few controls the development of monoaminergic neurons. Nat Neurosci. 2003 Jan;6(1):28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- Lillesaar C, Tannhäuser B, Stigloher C, Kremmer E, Bally-Cuif L. The serotonergic phenotype is acquired by converging genetic mechanisms within the zebrafish central nervous system. Dev Dyn. 2007 Apr;236(4):1072–84. doi: 10.1002/dvdy.21095. [DOI] [PubMed] [Google Scholar]
- Löhr H, Ryu S, Driever W. Zebrafish diencephalic A11-related dopaminergic neurons share a conserved transcriptional network with neuroendocrine cell lineages. Development. 2009 Mar;136(6):1007–17. doi: 10.1242/dev.033878. [DOI] [PubMed] [Google Scholar]
- Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J Physiol. 2006 Sep 1;575(Pt 2):379–87. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley ET, Baranowski TC, Blavo DO, Cato C, Doan TN, Rubinstein AL. Neuroprotection of MPTP-induced toxicity in zebrafish dopaminergic neurons. Brain Res Mol Brain Res. 2005 Nov 30;141(2):128–37. doi: 10.1016/j.molbrainres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Meng A, Tang H, Ong BA, Farrell MJ, Lin S. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc Natl Acad Sci U S A. 1997 Jun 10;94(12):6267–72. doi: 10.1073/pnas.94.12.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S, Ryu S, Zhao B, Zhang DQ, Driever W, McMahon DG. Targeting retinal dopaminergic neurons in tyrosine hydroxylase-driven green fluorescent protein transgenic zebrafish. Mol Vis. 2008;14:2475–83. [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MJ, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. The dopamine transporter is localized to dendritic and axonal plasma membranes of nigrostriatal dopaminergic neurons. J Neurosci. 1996 Jan 15;16(2):436–47. doi: 10.1523/JNEUROSCI.16-02-00436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MJ, Chan J, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. Immunogold localization of the dopamine transporter: an ultrastructural study of the rat ventral tegmental area. J Neurosci. 1997 Jun 1;17(11):4037–44. doi: 10.1523/JNEUROSCI.17-11-04037.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum RL, Ellis CE. Alzheimer's disease and Parkinson's disease. N Engl J Med. 2003;348(14):1356–64. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- Ogura E, Okuda Y, Kondoh H, Kamachi Y. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn. 2009;238(3):641–55. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Krause M. Transcriptional regulation. WormBook. 2005;23:1–40. doi: 10.1895/wormbook.1.45.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita P, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2009 Nov; doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal telencephalon (posterior tuberculum) Brain Res. 2001;889:316–330. doi: 10.1016/s0006-8993(00)03174-7. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain Res Dev Brain Res. 2002;137:89–100. doi: 10.1016/s0165-3806(02)00354-1. [DOI] [PubMed] [Google Scholar]
- Ryu S, Mahler J, Acampora D, Holzschuh J, Erhardt S, Omodei D, Simeone A, Driever W. Orthopedia homeodomain protein is essential for diencephalic dopaminergic neuron development. Curr Biol. 2007;17:873–880. doi: 10.1016/j.cub.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Bailey P, Bruce S, Engström PG, Klos JM, Wasserman WW, Ericson J, Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004 Dec 21;5(1):99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998 Mar 31;95(7):4013–8. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel JJ, Crews L, Roffler-Tarlov S, Chikaraishi DM. 4.5 kb of the rat tyrosine hydroxylase 5′ flanking sequence directs tissue specific expression during development and contains consensus sites for multiple transcription factors. Brain Res Mol Brain Res. 1999 Dec 10;74(1-2):1–14. doi: 10.1016/s0169-328x(99)00234-x. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008 Mar; doi: 10.1002/0471250953.bi0206s21. Chapter 2:Unit 2.6. [DOI] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000 Apr;3(4):337–41. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Spitzer N. A bar code for differentiation. Nature. 2009;458:843–4. doi: 10.1038/458843a. [DOI] [PubMed] [Google Scholar]
- Suli A, Mortimer N, Shepherd I, Chien CB. Netrin/DCC signaling controls contralateral dendrites of octavolateralis efferent neurons. J Neurosci. 2006;26:13328–13337. doi: 10.1523/JNEUROSCI.2858-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger JL, Glasgow E. Expression of isotocin-neurophysin mRNA in developing zebrafish. Gene Expr Patterns. 2003 Mar;3(1):105–8. doi: 10.1016/s1567-133x(02)00064-9. [DOI] [PubMed] [Google Scholar]
- Villefranc JA, Amigo J, Lawson ND. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007 Jan;35(Database):D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009 Feb 12;457(7231):854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallén A, Zetterström RH, Solomin L, Arvidsson M, Olson L, Perlmann T. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999 Dec 15;253(2):737–46. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- Wallén A, Perlmann T. Transcriptional control of dopamine neuron development. Ann N Y Acad Sci. 2003 Jun;991:48–60. doi: 10.1111/j.1749-6632.2003.tb07462.x. [DOI] [PubMed] [Google Scholar]
- Wang VE, Schmidt T, Chen J, Sharp PA, Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol Cell Biol. 2004 Feb;24(3):1022–32. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Turner EE. Expression of dopamine pathway genes in the midbrain is independent of known ETS transcription factor activity. J Neurosci. 2010;30(27):9224–7. doi: 10.1523/JNEUROSCI.1977-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Wei W, Gu W, Huang P, Ren X, Zhang Z, Zhu Z, Lin S, Zhang B. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev Biol. 2008 Feb 1;314(1):84–92. doi: 10.1016/j.ydbio.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Witta J, Baffi JS, Palkovits M, Mezey E, Castillo SO, Nikodem VM. Nigrostriatal innervation is preserved in Nurr1-null mice, although dopaminergic neuron precursors are arrested from terminal differentiation. Brain Res Mol Brain Res. 2000 Dec 8;84(1-2):67–78. doi: 10.1016/s0169-328x(00)00211-4. [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005 Jan;3(1):e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Ruuskanen JO, Wullimann MF, Vernier P. Two tyrosine hydroxylase genes in vertebrates New dopaminergic territories revealed in the zebrafish brain. Mol Cell Neurosci. 2010;43(4):394–402. doi: 10.1016/j.mcn.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Zetterström RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997 Apr 11;276(5310):248–50. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of different minimal promoters upstream of EGFP, driven by otpb.A enhancer. The transient injections with the E1b minimal promoter demonstrate expression in both the NPO and diDA cell groups, while expression under the control of the gata2a or c-fos minimal promoters is more limited in the desired cell groups. Confocal maximum projections, ventral views, anterior to the top, double-immunohistochemistry for GFP and TH (green and red). Arrows point to NPO cells, arrowheads to diDA neurons. Scale bar is 50 μm. (A) E1b minimal promoter; (B) pgata2 minimal promoter; (C) c-fos minimal promoter.
Figure S2. Characterization of lmx1a.2.A and msxE enhancer construct expression (A-A″ and E-G; maximum-intensity projections of double immunostaining for GFP and TH) and in situ expression (B-D, H-J) in whole-mount embryos. (A-A″) Stable transgenic Tg(lmx1a.2.A:egfp) embryo at 72hpf shows minimal overlap of TH and GFP expression. Ventral views, anterior to the top. (B-D) Whole-mount in situ expression patterns of lmx1a.2 at 24hpf, 36hpf, and 72hpf, ventral view, anterior to the top (except B, lateral view, anterior to the left). Note that in situ expression pattern at 72hpf correlates with enhancer expression pattern in (A). (E-G) Transient expression of msxE:EGFP. Ventral views, anterior to the top, at 24hpf, 36hpf, and 72hpf. (H-J) Whole-mount in situ expression patterns of msxE at 24hpf, 36hpf, and 72hpf, ventral views, anterior to the top (except H, lateral view, anterior to the left, dorsal up). Neither in situ nor transient transgenic expression at 72 hpf labels diDA neurons.
Figure S3. Genomic structure and functional enhancer characterization of Fugu TH genomic fragments. (A) Fugu TH genomic region (not to scale). Coding exons are shown as solid black boxes. DNA fragments tested for enhancer activity are shown. Region pictured is approximately 14kb; upstream 3′-most exon of nap1l4 is shown. (B) Summary table of enhancer CNS expression at 72hpf, categorized as expression in either as diDA neurons or non-dopaminergic cells (non-DA). (C, D) Maximum-intensity projections of whole-mount Tg(f.TH.M:egfp) embryo (C) and Tg(f.TH.O:egfp) embryo (D) at 72hpf, double-labeled for GFP and TH immunostaining. Ventral views, anterior to the top. Neither enhancer's expression overlaps with TH expression (compare extensive co-expression in Figure 1, showing Tg(f.TH.A:egfp)zc56).
Table S1. Primer sequences used for amplification of genomic DNA fragments tested for enhancer activity.