Abstract
Eukaryotes engage in intimate interactions with microbes that range in age and type of association. Although many conspicuous examples of ancient insect associates are studied (e.g., Buchnera aphidicola), fewer examples of younger associations are known. Here, we further characterize a recently evolved bacterial endosymbiont of the leafhopper Euscelidius variegatus (Hemiptera, Cicadellidae), called BEV. We found that BEV, continuously maintained in E. variegatus hosts at UC Berkeley since 1984, is vertically transmitted with high fidelity. Unlike many vertically transmitted, ancient endosymbioses, the BEV–E. variegatus association is not obligate for either partner, and BEV can be cultivated axenically. Sufficient BEV colonies were grown and harvested to estimate its genome size and provide a partial survey of the genome sequence. The BEV chromosome is about 3.8 Mbp, and there is evidence for an extrachromosomal element roughly 53 kb in size (e.g., prophage or plasmid). We sequenced 438 kb of unique short-insert clones, representing about 12% of the BEV genome. Nearly half of the gene fragments were similar to mobile DNA, including 15 distinct types of insertion sequences (IS). Analyses revealed that BEV not only shares virulence genes with plant pathogens, but also is closely related to the plant pathogenic genera Dickeya, Pectobacterium, and Brenneria. However, the slightly reduced genome size, abundance of mobile DNA, fastidious growth in culture, and efficient vertical transmission suggest that symbiosis with E. variegatus has had a significant impact on genome evolution in BEV.
Electronic supplementary material
The online version of this article (doi:10.1007/s00284-011-9893-5) contains supplementary material, which is available to authorized users.
Introduction
Bacterial interactions with insect hosts have pronounced consequences for bacterial lifestyles and genome evolution. After millions of years of co-evolution, ancient insect endosymbionts have experienced irreversible changes in genome size and content, resulting in an inability to survive outside of hosts [24]. However, transitions from free-living to symbiotic lifestyles are likely ongoing in bacterial lineages [1, 12, 18]. Recently evolved facultative symbionts provide the opportunity to examine the initial processes that affected the genome evolution of long-term, obligate endosymbionts. Such a symbiont was identified and cultivated from the leafhopper Euscelidius variegatus (Hemiptera, Cicadellidae) [33], and its effects on hosts have been studied for over two decades.
The bacterium of E. variegatus (BEV) is transovarially transmitted [34], a factor that in some cases indicates long-term stability or mutualistic tendencies; however, BEV reduces the fitness of its host and is highly pathogenic when injected into other leafhopper species [6, 33, 34]. When tested in 1987, infected E. variegatus displayed reduced fecundity, longevity, and increased development time [34]; since then, the degree of pathogenicity may have declined, under laboratory conditions. BEV can be horizontally acquired by naïve hosts when feeding on plants in the company of infected individuals, but it does not move, replicate, or cause disease within plants [35]. Microscopy of E. variegatus indicates that BEV cells are present in the hemolymph and invade the ovaries and midgut epithelium leading to tissue degeneration in the latter [5, 6]. BEV’s ability to penetrate into different host tissues and avoid the insect immune system may be due in part to a tolerance for acidic conditions. Even at a pH of 5.2, BEV exhibited cell growth over a period of 24 h (Alexander Purcell, personal communication). Unlike obligate endosymbionts of insects, BEV is not essential for its hosts’ survival or reproduction [23]. Additionally, BEV remains one of the few endosymbionts that can be cultivated axenically [32].
Here, we evaluated the efficiency of BEV vertical transmission in E. variegatus and calculated its growth rate in culture. Sequencing endosymbiont genomes generally requires complex isolation and enrichment techniques to acquire DNA for sequencing. However, we have taken advantage of the ability to grow BEV in culture to make a preliminary survey of its genome size, content, and phylogenetic origins.
Materials and Methods
History of the BEV Colony
BEV was discovered by Alexander Purcell in laboratory colonies of the leafhopper E. variegatus in Pont-de-la-Maye, France and has been maintained at UC Berkeley since 1984 [33]. Unfortunately, original material (insect or bacterial) from 1984 is not available. Infected leafhoppers are reared on a mix of barley, rye, and wheat grasses that are changed every 1 to 2 weeks. For the following experiments, BEV was axenically cultured from leafhoppers on Difco purple broth (PB) with 1.5–2.0% agar, acidified to pH 6.3 with 0.1 N HCl and incubated at 28°C in the dark [33].
Quantifying Vertical Transmission Efficiency of BEV
Ten cages were prepared each containing four rye grass plants and twenty BEV-infected E. variegatus adults. The adults were allowed to oviposit for 7 days then removed and frozen. After 5 days of maturation, 5–7 eggs per cage were collected and surface sterilized. DNA was extracted from the eggs and screened with BEV specific 16S ribosomal RNA (rRNA) PCR primers (see Supplemental Methods). Amplicons were run on a 1% agarose gel and visualized with ethidium bromide (EtBr) and several were randomly sequenced for confirmation.
Estimation of BEV Growth Rate
A triple-cloned isolate (re-plated using a single colony, three times) was plated as a lawn on PB agar, collected and used to inoculate two replicate 1 ml liquid PB cultures. Two sets of ten culture tubes were then inoculated from each of the replicates, wrapped in foil, and incubated at 28°C and shaken at 180 rpm. Culture tubes from each replicate were removed serially starting at day zero, plated and colony-forming units per milliliter (CFU/ml) were counted.
Genome Size Determination
Briefly, BEV DNA was purified for pulsed-field gel electrophoresis (PFGE) by first scraping colonies from PB agar plates and spinning down the cells. Then, the pellet was resuspended in PBS, mixed with an equal volume of 1.5% w/v pulsed-field gel agarose (BioRad) in TE (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0) and solidified in plastic plug molds. The plugs, containing intact cells, were then lysed, washed, and digested with the homing endonuclease I-CeuI (NEB). Plugs and appropriate size standards were separated on a PFGE rig (BioRad) using 0.5× TBE buffer, 1% w/v pulsed-field agarose gels and run conditions listed in the Supplemental Methods. Gels were visualized with EtBr and fragment sizes were estimated manually by measuring fragment migration of the size standard and plotting it by size on a semi-log plot.
BEV Genome Library Construction and Annotation
Total genomic DNA was extracted from a single triple-cloned BEV isolate grown on PB agar using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). This required two extractions to obtain a 20 μg pooled DNA sample (as determined by UV spectrophotometery). This sample was used to generate a blunt-end, short-insert pUC19 library according to published protocols [36] (Supplemental Methods). Transformants bearing plasmids were purified using a boiling mini prep protocol [36], quantified, and Sanger sequenced with M13F primer on an ABI3730xl (Life Technologies). Individual BEV reads were trimmed, dereplicated, and annotated following standard protocols [11]. Reads containing identical transposase gene fragments were manually clustered then assembled into complete insertion sequences. Raw sequence reads have been submitted to the GenBank Trace Archive (2292004866–2292005569).
Reconstruction of BEV Phylogeny
Identified BEV CDSs were screened for orthologs of 203 single-copy, vertically inherited, gammaproteobacterial core genes identified by Lerat et al. [22]. Orthologs from BEV and enterobacterial genomes in GenBank were identified (as in [22]), aligned and manually edited to remove uncertain amino acids (Xs) and gap containing columns (Table S1). Single gene alignments were concatenated and analyzed using maximum likelihood methods with a gamma model of rate heterogeneity estimated from the data and WAG or JTT amino acid substitution model in RAxML [38] and PhyML [16], respectively. Sequences from BEV matching multilocus sequence type (MLST) loci from Pectobacterium spp. Dickeya spp. and Brenneria spp. (EF550652–EF550704, EF550758–EF550810) were aligned and analyzed as above.
Results
BEV Transmission and Growth
Transmission of BEV occurs both vertically and horizontally in laboratory-reared E. variegatus [35]. In this experiment, however, we explicitly tested the vertical transmission efficiency of BEV. We randomly selected eggs laid by 200 adults, and surface sterilized and screened them by PCR for BEV. All of the E. variegatus eggs were positive for BEV infection (64/64). Adult leafhoppers were not screened for BEV infections, as all of the eggs were positive for BEV. Although BEV does not propagate within plant tissues, it can be grown axenically under microaerophilic conditions (5–18% O2) at room temperature (22–28°C) in the dark [33]. We measured its growth rate in PB media and estimated that BEV cells doubled every 7.0 h during exponential growth. The lag phase for BEV was about 24 h, the exponential growth phase lasted 36 h, and was followed by a very short stationary phase, after which the numbers of colony-forming units quickly declined (Fig. S1).
Genome Size Estimate
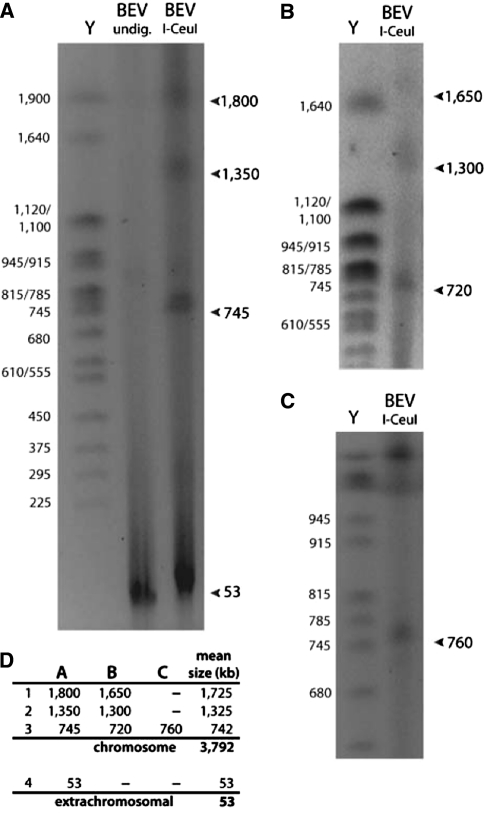
BEV cells were embedded in agarose, lysed, and enzymatically digested, permitting the estimation of its genome size through PFGE. The homing endonuclease I-CeuI recognizes and cuts the 23S rRNA gene. Therefore, the number of bands recovered from a circular chromosome is equivalent to the number of rRNA operons present. Multiple pulsed-field gels indicate that BEV possesses a 3.8 Mbp chromosome (fragment 1 = 1.73, 2 = 1.33, and 3 = 0.74 Mbp) and one ~53 kb extrachromosomal element (Fig. 1). We note that the BEV DNA seemed susceptible to degradation, which required the altering the pH of the recovery media (pH 6.5 vs. 7.4) and keeping the lysis and digestion times to a minimum.
Fig. 1.
Genome size estimation of BEV. The BEV genome size was estimated using pulsed-field gel electrophoresis (PFGE) and the S. cerevisiae chromosomal ladder (Y) was used as a size standard. a BEV DNA was run undigested and digested with I-CeuI to resolve a broad range of fragment sizes (50–1,500 kb). Three chromosomal fragments and one extrachromosomal fragment are apparent. Narrower range conditions were used to better resolve the upper b and middle bands c. d Size estimates for each BEV fragment were determined manually by plotting the migration of the size standard on a semi-log plot and mean fragment sizes are presented
BEV Genome Survey
To assess the genome content of BEV, we uni-directionally sequenced 854 clones from a random, short-insert library. Of these, 704 reads representing 626 unique clones and 438,406 bp were assigned to the BEV genome and annotated (Table 1, Table S2) the remaining reads were vector or poor sequence. The genome-wide G + C content of the reads is 52%, which is consistent with previous estimates from the 16S rRNA gene [33]. Despite size selecting for 1.5–2.0 kb inserts, many clones had inserts <400 bp. Therefore, most CDSs are partial gene fragments, which limits our ability to reconstruct the metabolism of BEV and to identify inactivated genes (pseudogenes). However, the reads still provided insight into the genome content and evolution of the BEV facultative endosymbiont.
Table 1.
Functional distribution of CDSs identified in BEV
| No. of unique partial CDSs | % of total CDSs | |
|---|---|---|
| Core genome | ||
| Cell processes | 10 | (1.4) |
| Cell structure | 6 | (0.8) |
| Information transfer | 40 | (5.6) |
| Metabolism | 124 | (17.2) |
| Regulation | 4 | (0.6) |
| Transport | 40 | (5.5) |
| Putative | 83 | (11.5) |
| Unknown | 72 | (10) |
| Mobile DNA | ||
| Prophage | 65 | (9) |
| Transposase | 276 | (38.3) |
| Plasmid | 1 | (0.1) |
The BEV reads encode a number of CDSs involved in pathways related to BEV being a microaerophilic heterotroph (glycolysis, TCA cycle, NADH dehydrogenase I, and ATP synthase, etc.). BEV also contains several specific genes involved in anaerobic respiration including dimethyl sulfoxide (DMSO) reductase (dmsABC), nitrate reductase A (narIJ), and the biosynthesis of its cofactor molybdenum (moaAB). Partial CDSs were detected for the biosynthesis of nine of the amino acids essential for animals. Genes involved in various essential processes such as DNA replication, transcription, translation, and RNA, DNA and protein modification were also identified (Table S2). Unlike obligate mutualists, BEV has also retained a number of genes involved in lipopolysaccharide (LPS) biosynthesis, bacteriophage, as well as genes from type three secretion systems (T3SS).
Destroying Genomes one Transposition at a Time
Overall, 38% of the CDSs are homologous to transposases (276/721) (Table 1). All of the reads can be assembled into 15 distinct insertion sequence (IS) elements, nine of which have readily identifiable boundaries (inverted repeats) (Table 2). Several reads exhibited transposition of IS elements within other elements (e.g., ISBEV08, ISBEV09). Other reads were identified with transposases adjacent to a bacterial CDS (n = 44), 18 of which clearly disrupt the CDS. If the library represents a random sampling of the BEV genome, then the read coverage of each IS element reflects its overall abundance and consequently its transpositional activity.
Table 2.
Characteristics of consensus insertion sequence (IS) elements in BEV
| Transposase | IS family | Total No. of readsa | Length | G + C% | Inverted repeats (IRs) | IR length (bp) |
|---|---|---|---|---|---|---|
| ISBEV01 | IS1 | 19 | 768 | 51.0 | – | – |
| ISBEV02 | IS1650, IS4, IS427 | 18 | 920 | 49.8 | + | 15 |
| ISBEV03 | IS4 | 62 | 1,464 | 55.3 | + | 16 |
| ISBEV04 | IS2 | 52 | 1,336 | 54.1 | + | 11 |
| ISBEV05 | ISSod13 (integrase catalytic subunit) | 50 | 1,201 | 48.9 | + | 8 |
| ISBEV06 | IS911 | 77 | 1,250 | 56.4 | + | 8 |
| ISBEV07 | – | 13 | 2,269 | 55.1 | + | 5 |
| ISBEV08 | IS100, IS110, IS1328, IS902 | 14 | 1,194 | 46.8 | – | – |
| ISBEV09 | IS116, IS110, IS902 | 7 | 932 | 54.6 | – | – |
| ISBEV10 | IS4, IS903 | 20 | 1,046 | 49.8 | + | 15 |
| ISBEV11 | IS1414, IS285 (mutator type) | 13 | 1,314 | 53.6 | + | 8 |
| ISBEV12 | IS630 | 20 | 1,242 | 51.0 | + | 8 |
| ISBEV13 | IS100, IS110, IS1328, IS902 | 4 | 1,063 | 48.1 | – | – |
| ISBEV14 | IS630 | 2 | 551 | 47.4 | – | – |
| ISBEV15 | IS204, IS1001, IS1096, IS1165 | 2 | 1,335 | 53.4 | – | – |
a68 reads have 2–3 separate transposase fragments
Origins of BEV
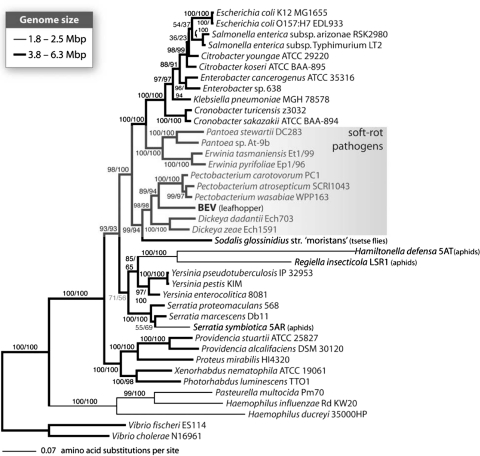
Previous phylogenetic analyses of BEV using 16S rRNA sequences found that it was a member of the Enterobacteriaceae and closely related to a bacterial endosymbiont in bedbugs (Cimex lectularius [Hemiptera, Cimicidae]) [4, 17]. Given that protein phylogenies provide a significantly better resolution of the enteric genera than those generated with 16S rRNA [26], we capitalized on the presence of numerous proteins to estimate a well-resolved phylogeny. Using 14 of 203 proteins conserved in most Gammaproteobacteria (Cca, DapA, GyrA, HslU, IspH, MiaA, MurG, PepA, Pth, PurH, RibA, RplQ, RpsC, SucB) we reconstructed a maximum likelihood phylogeny of the Enterobacteriaceae (Fig. 2). BEV was placed with high support within a clade containing the soft-rot plant pathogens Dickeya and Pectobacterium. A similar placement was found using BEV sequences for 2 MLST loci and a broader sample of taxa representing 49 strains of Dickeya, Pectobacterium, and Brenneria (Fig. S2). However, the delineation between BEV, Pectobacterium spp., Dickeya spp., and Brenneria spp. is unclear due to poor bootstrap support. Consistent with the findings of Naum et al. [26], we found that phylogenies and estimates of pairwise divergence using 16S rRNA were not useful in resolving the relationships of BEV and related enterics (unpublished results).
Fig. 2.
Concatenated protein phylogeny of BEV and enterobacterial relatives. The BEV-symbiont lineage falls firmly among the plant pathogenic genera Pectobacterium and Dickeya, making BEV distinct from other facultative insect endosymbionts from tsetse flies and aphids (in black text). The BEV lineage is less divergent relative to most other facultative endosymbionts (shorter branch length). As for Sodalis the genome has not undergone as radical a reduction in size. Thick branches designate bacteria with large genomes (3.8–6.3 Mbp) and thin branches those bacteria with smaller genomes (1.8–2.5 Mbp). Support values for each node were estimated from 100 non-parametric bootstrap replicates in RAxML (first) and PhyML (second) and those less than 75 are shown in gray
Discussion
Defining Endosymbiotic Origins
It is notoriously difficult to determine the evolutionary origins of long-term intracellular endosymbionts of insects due to their divergence from free-living bacteria and attendant changes in nucleotide composition [19]. However, recently formed insect endosymbioses provide the opportunity to compare the symbiont with closely related free-living bacteria and examine early changes that are involved in the establishment of the association (e.g., [7, 8]).
Previous work suggested that BEV is indeed a recently established endosymbiont [4]. Our current phylogenetic analyses clearly place BEV among the plant pathogenic genera Dickeya, Pectobacterium, and Brenneria (Fig. 2, Fig. S2). This relationship is perhaps unsurprising as (i) its host E. variegatus feeds on the phloem sap of a variety of grasses and other plants, (ii) BEV is horizontally acquired during feeding, and (iii) plant and animal pathogens rely on common mechanisms to invade tissues, cause disease, and evade immune surveillance [35, 37]. These same molecular mechanisms are in fact crucial for some facultative and obligate endosymbioses [7, 8].
Curiously, when comparing 16S rRNA genes, the most closely related bacterium to BEV is a symbiont of the unrelated blood-feeding bedbug (C. lectularius, ~99% identical) [17]. This same pattern of close relationship between symbionts of blood-feeding and plant-feeding insects has been observed in three other facultative endosymbiont lineages, Sodalis, Arsenophonus, and Rickettsia [28–30]. These represent independent horizontal transmission events between unrelated plant-feeding and blood-feeding insects. BEV, Sodalis, and Arsenophonus are also among the few insect endosymbionts that can be cultivated in sterile media [32]. They are all microaerophilic, slow growing, and require rich media and moderate temperatures (25–30°C) [32]. The ability of these facultative endosymbionts to persist outside of a host possibly facilitates successful horizontal transmission.
Vertical Transmission
BEV is efficiently transmitted by transovarial transmission at a level sufficient for retaining it within a host population. Perfect vertical transmission, a hallmark of obligate endosymbionts, is also common among insect reproductive parasites (e.g., Wolbachia, Cardinium) [20, 31]. We did not test the possibility that E. variegatus nymphs lose the infection as they mature. If this occurs, horizontal transmission may play a role in BEV reinfection or exchange within an E. variegatus colony, potentially allowing the bacterium to maintain its mildly pathogenic qualities [6, 34].
Other leafhopper species are known for vertically transmitting bacterial plant pathogens, supporting the possibility of BEV transitioning from a plant pathogen to an insect endosymbiont. For example, rickettsia-like organisms vertically transmitted by leafhoppers are implicated in clover club leaf, rugose leafcurl, and Papaya bunchy top diseases [2, 10, 15]. Transitions may occur in the opposite direction as well, with insect-bacterial symbionts becoming plant pathogens, as may have occurred in the genus Arsenophonus [3].
Genomic Insights
Given the genome size estimate of 3.8 Mbp, we have recovered only a fraction of the entire genome (~12%). However, the partial genome survey reveals a number of clues regarding the metabolic potential of BEV. For example, we identified several partial gene sequences involved in plant pathogenicity consistent with its phylogenetic affinity with the soft-rot plant pathogens: pectinesterase A (b510), polygalacturonase (b159), polysaccharide deacetylase (b303), exopolysaccharide biosynthesis (b704), and CDSs for two T3SS (b48, b445, b674, b709) [14]. The pectinesterase A locus appears to have been disrupted by an IS element. Inactivation of this and possibly other plant pathogenicity loci due to changes in population dynamics and/or selective regimes likely underlies the inability of BEV to persist or cause damage to host plants [35].
Overall, the BEV genome data share several hallmarks common among young facultative endosymbionts including larger genome sizes (3.5–4.1 Mbp), evidence of diverse metabolic capabilities tempered by gene inactivations and the persistence of virulence-associated loci and mobile DNA [9, 39]. These features contrast sharply with the extremely streamlined genomes of obligate mutualists [24]. The large fraction of CDSs associated with mobile DNA in BEV (276/721) suggests that these elements are active, creating new pseudogenes and possibly leading to repeat-induced genome rearrangements and large-scale deletions [21, 27].
Genomes of bacteria are seldom inundated with such large numbers of mobile DNA elements; however, these elements accumulate in facultative endosymbionts and some pathogens in which small population sizes and changes in selective pressures result in an inability to purge such selfish DNA [11, 21, 25]. Coexistence of distinct endosymbionts within individual hosts may facilitate horizontal transfer of novel mobile elements.
Concluding Remarks
BEV represents the longest-studied, cultivatable insect endosymbiont [33], and studies continue to explore its interactions and impact on its host E. variegatus (e.g., [13]). Therefore, BEV provides important insights into the evolutionary transition from free-living bacteria to host-associated symbiont and raises pertinent questions. Considering that BEV can penetrate multiple host organs and is not maintained within a bacteriome, how does it interact with E. variegatus obligate endosymbionts? How does it escape the host immune system? Does it provide a benefit for the host under certain conditions? In light of our added genome data, BEV will continue to be an important model for the exploration of insect-bacterial symbiosis.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1 BEV Growth in Purple Broth Liquid Medium. BEV was grown in liquid purple broth medium and then dilution-plated each day from Day 0-–9 onto solid PB at dilutions of 1/1, 1/100, and 1/10,000. Colony-forming units per milliliter (CFU/mL) of BEV were counted at the dilution, which resulted in 20–1000 colonies. The results from two replicates are shown here with Standard Deviations. The lag phase for BEV is about 24 h, the exponential growth phase is 36 h, and the stationary phase is very short, after which the numbers of colony-forming units quickly decline. Fig. S2 MLST phylogeny of BEV and related plant pathogens. The maximum likelihood phylogeny was estimated with RAxML from the aligned and concatenated nucleotide sequences for glyceraldehyde-3-phosphate dehydrogenase A (gapA) and malate dehydrogenase (mdh). BEV falls with moderate support within the clade associated with Dickeya and Brenneria species. This phylogeny generated from only two genes is generally consistent with original analysis with seven genes in Ma et al. 2008. Numbers adjacent to nodes indicate bootstrap values from 100 non-parametric replicates and values greater than 50 are shown. (DOCX 374 kb)
Acknowledgments
We thank Sandy Purcell for helpful comments and suggestions throughout this research and an anonymous reviewer for comments on the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
P. H. Degnan and L. S. Bittleston contributed equally to this work.
References
- 1.Aksoy S, Chen X, Hypsa V. Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera: Glossinidae) Insect Mol Biol. 1997;6:183–190. doi: 10.1111/j.1365-2583.1997.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 2.Black LM. A plant virus that multiplies in its insect vector. Nature. 1950;166:852–853. doi: 10.1038/166852a0. [DOI] [PubMed] [Google Scholar]
- 3.Bressan A, Sémétey O, Arneodo J, et al. Vector transmission of a plant-pathogenic bacterium in the Arsenophonus clade sharing ecological traits with facultative insect endosymbionts. Phytopathology. 2009;99:1289–1296. doi: 10.1094/PHYTO-99-11-1289. [DOI] [PubMed] [Google Scholar]
- 4.Campbell BC, Purcell AH. Phylogenetic affiliation of BEV, a bacterial parasite of the leafhopper Euscelidius variegatus, on the basis of 16S rDNA sequences. Curr Microbiol. 1993;26:37–41. doi: 10.1007/BF01577240. [DOI] [PubMed] [Google Scholar]
- 5.Cheung WWK, Purcell AH. Ultrastructure of the digestive system of the leafhopper Euscelidius variegatus Kirshbaum (Homoptera: Cicadellidae) with and without congenital bacterial infections. Int J Insect Morphol Embryol. 1993;22:49–61. doi: 10.1016/0020-7322(93)90033-W. [DOI] [Google Scholar]
- 6.Cheung WWK, Purcell AH. Invasion of bacteroids and BEV bacterium into oocytes of the leafhopper Euscelidius variegatus Kirschbaum (Homoptera: Cicadellidae): an electron microscopic study. Zool Stud. 1999;38:69–75. [Google Scholar]
- 7.Dale C, Young SA, Haydon DT, et al. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci USA. 2001;98:1883–1888. doi: 10.1073/pnas.021450998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale C, Plague GR, Wang B, et al. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA. 2002;99:12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby AC, Choi J-H, Wilkes T, et al. Characteristics of the genome of Arsenophonusnasoniae, son-killer bacterium of the wasp Nasonia. Insect Mol Biol. 2010;19:75–89. doi: 10.1111/j.1365-2583.2009.00950.x. [DOI] [PubMed] [Google Scholar]
- 10.Davis MJ, Ying Z, Brunner BR, et al. Rickettsial relative associated with papaya bunchy top disease. Curr Microbiol. 1998;36:80–84. doi: 10.1007/s002849900283. [DOI] [PubMed] [Google Scholar]
- 11.Degnan PH, Yu Y, Sisneros N, et al. Hamiltonella defensa, genome evolution of a protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estes AM, Hearn DJ, Bronstein JL, et al. The olive fly endosymbiont, “Candidatus Erwinia dacicola”, switches from an intracellular existence to an extracellular existence during host insect development. Appl Environ Microbiol. 2009;75:7097–7106. doi: 10.1128/AEM.00778-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galetto L, Nardi M, Saracco P, et al. Variation in vector competency depends on chrysanthemum yellows phytoplasma distribution within Euscelidius variegatus. Entomol Exp Appl. 2009;131:200–207. doi: 10.1111/j.1570-7458.2009.00845.x. [DOI] [Google Scholar]
- 14.Glasner JD, Marquez-Villavicencio M, Kim HS, et al. Niche-specificity and the variable fraction of the Pectobacterium pan-Genome. Mol Plant Microbe Interact. 2008;21:1549–1560. doi: 10.1094/MPMI-21-12-1549. [DOI] [PubMed] [Google Scholar]
- 15.Grylls N. Rugose leaf curl- a new virus disease transovarially transmitted by the leafhopper Austroagallia torrida. Aust J Biol Sci. 1954;7:47–58. [PubMed] [Google Scholar]
- 16.Guindon S, Gascuel O. A simple, fast accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 17.Hypsa V, Aksoy S. Phylogenetic characterization of two transovarially transmitted endosymbionts of the bedbug Cimex lectularius (Heteroptera: Cimicidae) Insect Mol Biol. 1997;6:301–304. doi: 10.1046/j.1365-2583.1997.00178.x. [DOI] [PubMed] [Google Scholar]
- 18.Heddi A, Charles H, Khatchadourian C, et al. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J Mol Evol. 1998;47:52–61. doi: 10.1007/PL00006362. [DOI] [PubMed] [Google Scholar]
- 19.Herbeck JT, Degnan PH, Wernegreen JJ. Non-homogeneous model of sequence evolution indicates independent origins of primary endosymbionts within the Enterobacteriales (γ-Proteobacteria) Mol Biol Evol. 2005;22:520–532. doi: 10.1093/molbev/msi036. [DOI] [PubMed] [Google Scholar]
- 20.Jiggins FM, Randerson JP, Hurst GD, et al. How can sex ratio distorters reach extreme prevalences? Male-killing Wolbachia are not suppressed and have near-perfect vertical transmission efficiency in Acraea encedon. Evolution. 2002;56:2290–2295. doi: 10.1111/j.0014-3820.2002.tb00152.x. [DOI] [PubMed] [Google Scholar]
- 21.Klasson L, Westberg J, Sapountzis P, et al. The mosaic genome structure of the WolbachiawRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-proteobacteria. PLoS Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 25.Moran NA, Plague GR. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 2004;14:627–633. doi: 10.1016/j.gde.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Naum M, Mason-Gamer RJ, Brown EW. Is 16S rDNA a reliable phylogenetic marker to characterize relationships below the family level in the Enterobacteriaceae? J Mol Evol. 2008;66:630–642. doi: 10.1007/s00239-008-9115-3. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson AI, Koskiniemi S, Eriksson S, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 2005;102:12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nováková E, Hypsa V. A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett. 2007;269:131–135. doi: 10.1111/j.1574-6968.2006.00620.x. [DOI] [PubMed] [Google Scholar]
- 29.Nováková E, Hypša V, Moran NA. Arsenophonus, an emerging clade of intracellular symbionts with a broad host distribution. BMC Microbiol. 2009;9:143. doi: 10.1186/1471-2180-9-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perlman SJ, Hunter MS, Zchori-Fein E. The emerging diversity of Rickettsia. Proc R Soc B. 2006;273:2097–2106. doi: 10.1098/rspb.2006.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perlman SJ, Kelly SE, Hunter MS. Population biology of cytoplasmic incompatibility: maintenance and spread of Cardinium symbionts in a parasitic wasp. Genetics. 2008;178:1003–1011. doi: 10.1534/genetics.107.083071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontes MH, Dale C. Culture and manipulation of insect facultative symbionts. Trends Microbiol. 2006;14:406–412. doi: 10.1016/j.tim.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Purcell AH, Steiner T, Mégraud F, et al. In vitro isolation of a transovarially transmitted bacterium from the leafhopper Euscelidius variegatus (Hemiptera: Cicadellidae) J Invert Pathol. 1986;48:66–73. doi: 10.1016/0022-2011(86)90144-8. [DOI] [Google Scholar]
- 34.Purcell AH, Suslow KG. Pathogenicity and effects on transmission of a mycoplasmalike organism of a transovarially infective bacterium on the leafhopper Euscelidius variegatus (Homoptera: Cicadellidae) J Invert Pathol. 1987;50:285–290. doi: 10.1016/0022-2011(87)90094-2. [DOI] [Google Scholar]
- 35.Purcell AH, Suslow KG, Klein M. Transmission via plants of an insect pathogenic bacterium that does not multiply or move in plants. Microb Ecol. 1994;27:19–26. doi: 10.1007/BF00170111. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, MacCallum P, Russell D (2006) The condensed protocols from molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- 37.Staskawicz BJ, Mudgett MB, Dangl JL, et al. Common and contrasting themes of plant and animal diseases. Science. 2001;292:2285–2289. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 38.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 39.Toh H, Weiss BL, Perkin SAH, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 BEV Growth in Purple Broth Liquid Medium. BEV was grown in liquid purple broth medium and then dilution-plated each day from Day 0-–9 onto solid PB at dilutions of 1/1, 1/100, and 1/10,000. Colony-forming units per milliliter (CFU/mL) of BEV were counted at the dilution, which resulted in 20–1000 colonies. The results from two replicates are shown here with Standard Deviations. The lag phase for BEV is about 24 h, the exponential growth phase is 36 h, and the stationary phase is very short, after which the numbers of colony-forming units quickly decline. Fig. S2 MLST phylogeny of BEV and related plant pathogens. The maximum likelihood phylogeny was estimated with RAxML from the aligned and concatenated nucleotide sequences for glyceraldehyde-3-phosphate dehydrogenase A (gapA) and malate dehydrogenase (mdh). BEV falls with moderate support within the clade associated with Dickeya and Brenneria species. This phylogeny generated from only two genes is generally consistent with original analysis with seven genes in Ma et al. 2008. Numbers adjacent to nodes indicate bootstrap values from 100 non-parametric replicates and values greater than 50 are shown. (DOCX 374 kb)