Abstract
Class I phosphoinositide 3-kinases exert important cellular effects through their two primary lipid products, phosphatidylinositol 3,4,5-trisphosphate and phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2). As few molecular targets for PtdIns(3,4)P2 have yet been identified, a screen for PI 3-kinase-responsive proteins that is selective for these is described. This features a tertiary approach incorporating a unique, primary recruitment of target proteins in intact cells to membranes selectively enriched in PtdIns(3,4)P2. A secondary purification of these proteins, optimized using tandem pleckstrin homology domain containing protein-1 (TAPP-1), an established PtdIns(3,4)P2 selective ligand, yields a fraction enriched in proteins of potentially similar lipid binding character that are identified by liquid chromatography-tandem MS. Thirdly, this approach is coupled to stable isotope labeling with amino acids in cell culture using differential isotope labeling of cells stimulated in the absence and presence of the PI 3-kinase inhibitor wortmannin. This provides a ratio-metric readout that distinguishes authentically responsive components from copurifying background proteins. Enriched fractions thus obtained from astrocytoma cells revealed a subset of proteins that exhibited ratios indicative of their initial, cellular responsiveness to PI 3-kinase activation. The inclusion among these of tandem pleckstrin homology domain containing protein-1, three isoforms of Akt, switch associated protein-70, early endosome antigen-1 and of additional proteins expressing recognized lipid binding domains demonstrates the utility of this strategy and lends credibility to the novel candidate proteins identified. The latter encompass a broad set of proteins that include the gene product of TBC1D2A, a putative Rab guanine nucleotide triphosphatase activating protein (GAP) and IQ motif containing GAP1, a potential tumor promoter. A sequence comparison of the former protein indicates the presence of a pleckstrin homology domain whose lipid binding character remains to be established. IQ motif containing GAP1 lacks known lipid interacting components and a preliminary analysis here indicates that this may exemplify a novel class of atypical phosphoinositide (aPI) binding domain.
The cell-surface receptor-regulated, class I phosphoinositide (PI)1 3-kinases play a central role in the intracellular mechanisms that regulate important biological processes including cell division, growth, motility, and metabolism (1, 2). This is emphasized by the association between defects in PI 3-kinase signaling and several disease states (3, 4). Class I PI 3-kinases exert their effects by the unique 3-phosphorylation of their phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) lipid substrate, to produce phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) (5). This lipid acts either to localize and/or modulate the activity of specific effector proteins expressing appropriate PI recognition domains (1, 2, 5, 6). The PtdIns(3,4,5)P3 signal is degraded by both the 3-phosphatase PTEN (phosphatase and tensin homolog deleted on chromosome ten) (7) and by 5-phosphatases including the Src homology 2 domain containing inositol phosphatases 1 and 2 (SHIP1 and SHIP2) (8). However, although these alternative routes both serve to remove PtdIns(3,4,5)P3, they are clearly distinct functionally as impairment of the activity of each has discrete consequences, a feature that may reflect their separate lipid metabolites (9). Thus, whereas PTEN acts simply to reverse PI 3-kinase action by regenerating PtdIns(4,5)P2, 5-phosphatase activity allows the additional synthesis of phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) and, through subsequent 4-phosphatase activity, of PtdIns3P. The latter lipid is also synthesized directly from phosphatidylinositol by class III PI 3-kinases and many of its functions are well established (10).
By contrast, the biological functions of PtdIns(3,4)P2 remain obscure although recent evidence suggests that the specific enzymes responsible for both the synthesis and removal of this lipid fulfill important roles. Hence, the synthesis of PtdIns(3,4)P2 from PtdIns(3,4,5)P3 by the widely distributed 5-phosphatase, Src homology 2 domain containing inositol phosphatase 2 (SHIP2) appears to be tightly regulated (11) and the loss of this enzyme has important consequences (9, 12) that may reflect its influence on cellular PtdIns(3,4)P2 concentrations as well as on those of PtdIns(3,4,5)P3. Similarly, the importance of PtdIns(3,4)P2 catabolism by the types I and II inositol polyphosphate 4-phosphatases (13) is emphasized by the loss of either enzyme. The type I enzyme regulates cell growth downstream of the GATA transcription factor (14) whereas its deficit results in the neuronal loss characteristic of Weeble mice (15) and vulnerability to excitotoxic neuronal death (16). The type II 4-phosphatase may act as a tumor suppressor and recent work implies that this enzyme regulates Akt-dependent cell proliferation (17), a feature consistent with the known ability of PtdIns(3,4)P2 to bind and to regulate the activity of this pivotal serine/threonine kinase (18–22). However, PtdIns(3,4)P2 may also exert effects through other target proteins such as TAPP-1 (tandem pleckstrin homology domain containing protein-1) (23) and lamellipodin (24), which bind this lipid with high selectivity, or DAPP-1 (dual adapter for phosphotyrosine and 3-phosphoinositides) (25) and Irgm-1 (immunity-related GTPase) (26) that interact preferentially with both PtdIns(3,4)P2 and PtdIns(3,4,5)P3. Indeed, it seems likely that it is by acting through these and other similar effectors that PtdIns(3,4)P2 can act independently of, or co-ordinately with, PtdIns(3,4,5)P3 in some circumstances (16, 27). In this context, the activities of PTEN, SHIP2, and the 4-phosphatases can be viewed collectively as a means of tuning the balance of prevailing signal outputs from PI 3-kinase (28).
The number of defined molecular targets for PtdIns(3,4)P2 however, remains limited by comparison with the established repertoire of PtdIns(3,4,5)P3 effector proteins. In the current study, we have sought to redress this balance by developing a screen for additional, selective PtdIns(3,4)P2 binding proteins. To do this we have adopted a novel tertiary approach. This combines a primary, selective, cellular recruitment and later recovery of candidates with a secondary, lipid-affinity and other purification of these proteins. This is coupled thirdly, with a ratio-metric, isotope labeling technique that, in conjunction with mass spectrometric analysis, allows quantitative discrimination of background proteins from those authentically responsive to PI 3-kinase/PtdIns(3,4)P2 at the primary step. By introducing multiple tiers of selectivity, including an initial cellular screen that allows the presentation of PtdIns(3,4)P2 in an authentic membrane bilayer to binding partners present in the cellular milieu, this approach aims both to complement and extend earlier, lipid-affinity based efforts to isolate PI binding proteins (29–33).
The success of this strategy is demonstrated by our identification of several established 3-PI effectors in addition to several proteins whose potential regulation directly or otherwise by 3-PIs has not been recognized previously. The former demonstrate the feasibility of this approach and lend credibility to the view that the latter include novel, authentic targets whose description here will facilitate a deeper understanding of the subtleties of PI 3-kinase signaling.
EXPERIMENTAL PROCEDURES
Materials
1321N1 Astrocytoma cells were from the European Collection of Animal Cell Cultures. Antibodies against TAPP-1 were as described previously (34) and, together with the GST-tagged PH domains of GRP1, TAPP-1, and PLCδ, were obtained from the Division of Signal Transduction Therapy, University of Dundee. Potassium bisperoxo(1,10-phenanthroline)oxovanadate (V) {bpV(phen)} was from Merck and platelet-derived growth factor was from Sigma. Synthetic inositol phosphates and phosphoinositides (dioctanoyl form) were from Cell Signals (Columbus, OH). Heavy isotope substituted (13C) amino acids were from C. K. Gas Products (Cambridge, U.K.). Neutravidin beads were from Pierce. HPLC pure water was from J. T. Baker and trypsin gold was from Promega (Madison, WI). Streptavidin-dextrose (SA) sensor chips for surface plasmon resonance and chromatography media were from G.E. Healthcare. Other reagents were from the sources indicated or as defined previously (11, 35, 36).
Cell Culture
1321N1 Cells were cultured as described previously (36). Stable isotope labeling with amino acids in cell culture (SILAC) was achieved with medium comprising Dulbecco's modified Eagle medium initially lacking arginine and lysine but supplemented freshly with both amino acids without (light) or with (heavy) 13C isotope substitution and with 5% (w/v) dialyzed fetal bovine serum and penicillin/streptomycin. Prior to experimental use, cells were allowed to progress through a minimum of seven doublings in SILAC medium without or with (13C) amino acids to permit adequate labeling of the cellular protein pool.
Preparation of Cell Extracts
Confluent 1321N1 cells (10–15 × 15 cm dishes for SILAC experiments) were washed twice with 2–10 ml of serum-free Dulbecco's modified Eagle medium or modified Krebs-Henseleit buffer (36) and allowed to equilibrate in a similar volume of the same medium or buffer at 37 °C for 60 min. If required, wortmannin or vehicle was added for the latter half of this period. Cells were then incubated in the absence or presence of PI 3-kinase stimuli as indicated to facilitate the membrane recruitment of appropriate 3-PI binding proteins. The incubation medium was then aspirated rapidly and replaced with a suitable volume (2–10 ml) of ice-cold, digitonin permeabilization buffer (25 mm Hepes, pH 7.2, 100 μg/ml digitonin, 75 mm NaCl, 50 mm NaF, 0.1 mm EDTA, 0.1 mm EGTA supplemented freshly with 0.1% (v/v) 2-mercaptoethanol, 0.1 mm phenylmethylsulphonylfluoride (PMSF), 0.1 mm benzamidine and 1 mm sodium vanadate) and the cells transferred on to ice and maintained on a rocking platform at 4 °C for 5 min before collection of the resulting supernatant as the soluble protein (cytosol) fraction. The remaining adherent layer of insoluble cell debris was washed briefly with permeabilization buffer to remove residual, soluble material. Proteins bound to 3-PIs synthesized in response to prior cell stimulation, were then either recovered selectively from the remaining, insoluble cell material (membrane) by inositol phosphate-specific elution in buffer A (10 mm Hepes, pH 7.2, 0.1 mm EDTA, 0.1 mm EGTA) as indicated or more crudely by extraction into a suitable volume (0.5–5.0 ml) of ice-cold lysis buffer (50 mm Tris, pH 7.5, 50 mm NaF, 1 mm EDTA, 1 mm EGTA, 5 mm sodium pyrophosphate, 10 mm glycerol 2-phosphate, 0.5% (w/v) Triton X-100, supplemented freshly with 0.1% (v/v) 2-mercaptoethanol, 0.1 mm PMSF, 0.1 mm benzamidine and 1 mm sodium vanadate). These extracts were then analyzed by SDS-PAGE and mass spectrometry either directly or following further chromatographic enrichment. For SILAC experiments, the debris postpermeabilization and washing was collected on ice and extracts from control and stimulated cells were pooled immediately to allow the coprocessing of these samples through all subsequent steps.
Ion Exchange and Affinity Chromatography
Membrane extracts containing 3-PI binding proteins were purified further by affinity chromatography on neutravidin beads precoupled to biotinylated PtdIns(3,4)P2 either directly or following prior concentration by ion exchange chromatography. For larger scale SILAC experiments, this served both to reduce the sample complexity and, for some fractions, to minimize carryover of eluent inositol 1,3,4-triphosphate (Ins(1,3,4)P3) from the preceding step. In preliminary experiments (see Fig. 3) samples were applied to 0.8 × 2.0 cm columns of Q-Sepharose and these washed with 10 bed volumes of sample application buffer before elution with 1–2 volumes of the same buffer containing 300 mm NaCl to recover a fraction enriched specifically with respect to TAPP-1. To achieve a wider catchment of potential 3-PI binding proteins, this procedure was modified for SILAC experiments such that in addition to retaining a 600 mm NaCl fraction, the initial flow-through from the anion exchange column was retained and applied separately to a similar column of S-Sepharose. This was eluted in the same manner and a flow-through fraction also collected, so that for SILAC experiments subsequent PtdIns(3,4)P2 affinity recovery was performed on three separate, anion, cation, and flow-through sample fractions.
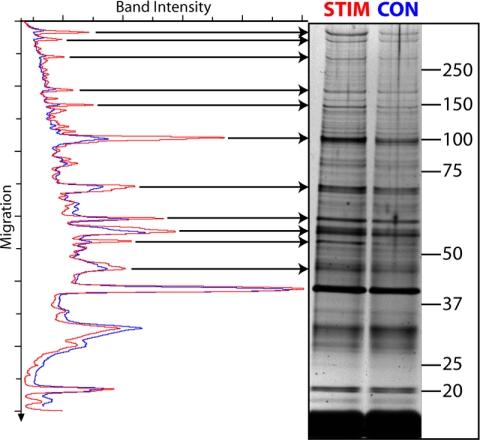
Fig. 3.
A three step enrichment of TAPP-1 reveals multiple, copurifying PI 3-kinase-responsive proteins. Membrane fractions prepared from cells incubated as control (CON, blue) or stimulated for 30 min with bpV(phen) (0.1 mm) (STIM, red) were eluted with Ins(1,3,4)P3 (100 μm), concentrated by anion exchange chromatography, and affinity precipitated with immobilized PtdIns(3,4)P2. The proteins present were analyzed by SDS-PAGE and silver stained. The results represent individual samples from one experiment representative of several (see text) and show a silver stained gel (values indicate molecular mass) and a densitometric scan of the control and stimulated sample lanes to highlight the PI 3-kinase-responsive components.
Biotinylated PtdIns(3,4)P2 was precoupled to neutravidin beads using a sufficient molar excess of lipid that, given the very high established affinity of the biotin-neutravidin interaction and the supplier's stated bead capacity, an estimated ratio of ∼120–240nmol of bound lipid:1.0 ml of packed beads was achieved by mixing an aqueous solution of lipid in 2–4 bead volumes of buffer A or lysis buffer at 4 °C for a minimum of 60 min. Extracts, precleared similarly with an equivalent volume of beads alone for all but SILAC experiments, were mixed at 4 °C for 60 min with a volume of lipid-coupled beads sufficient to give a final concentration of PtdIns(3,4)P2 (25–50 μm) that maximized recovery of TAPP-1. The beads were then collected using a 10 μm polypropylene cup filter (Vectraspin Micro, Whatman) coupled to a reservoir and vacuum filtration system to allow rapid recovery and washing with 2–10 bead volumes of ice-cold buffer A or lysis buffer. Bound proteins were recovered by elution at 70 °C for 5 min with either one bead volume of 2.2% (w/v) sodium dodecyl sulfate (SDS) in 70 mm Tris buffer, pH 6.8 or, for larger bead volumes, a similar volume of buffer diluted fourfold to allow for subsequent vacuum concentration by the same factor. Attempts to recover proteins rapidly by selective elution using Ins(1,3,4)P3 gave poor yields of TAPP-1, of which <30% was eluted by a 10-fold molar excess of InsP3 over PtdIns(3,4)P2.
SDS-PAGE and Immunoblotting
Samples were adjusted to final concentrations of 62.5 mm Tris, pH 6.8, 14.5 mm 2-mercaptoethanol, 2% (w/v) SDS, 0.01% (w/v) bromphenol blue, and 10% (v/v) glycerol and analyzed by SDS-PAGE. Alternatively, if required for subsequent mass spectrometric analysis, samples were first reduced (using 10 mm dithiothreitol at 70 °C for 5 min) and alkylated (using 50 mm iodoacetamide at room temperature for 30 min) prior to dilution and SDS-PAGE analysis. Samples were analyzed on Novex 4–12% (w/v) polyacrylamide gels (Invitrogen) developed in Bis-Tris buffers according to the protocol supplied. For routine analysis and subsequent immunoblotting ∼25 μg of sample protein was loaded per lane. For SILAC samples, the protein load was increased to 50–100 μg of protein per lane and gels were developed only to ∼50% of their full length in order to maximize peptide recovery during subsequent in-gel tryptic digestion. Protein samples were stained using Colloidal Blue (Invitrogen) or Silver Snap (Pierce) according to the supplier's recommendations. Alternatively, proteins were transferred to polyvinylidene difluoride membranes for subsequent immunoblotting as described previously (11, 35). In the latter instance, primary antibodies were applied overnight at 4 °C at 1.0 μg/ml (TAPP-1) and proteins then visualized using appropriate horseradish peroxidase-coupled secondary antibodies with Immobilon Western (Millipore) chemiluminescent substrate. Images were captured on a CCD camera under conditions of linear light accumulation, allowing later quantification using Advanced Image Data Analysis software. Alternatively, individual bands or, for SILAC experiments, complete lanes cut from Colloidal Blue stained gels, were diced, then de-stained and subjected to in-gel tryptic digestion and the resulting peptides recovered for mass spectrometric analysis.
Mass Spectrometry
The tryptic digests were reconstituted in 1% (v/v) formic acid and any debris pelleted by brief centrifugation at 13,000 × g prior to analysis on an LTQ-Orbitrap XL mass spectrometer system (Thermo Fisher Scientific) coupled to a U3000 nano-LC system (Dionex). The samples were loaded at 20 μl/min onto an LC-Packings PepMap C18 trap column (0.3 × 5 mm) equilibrated in 2% (v/v) acetonitrile and 0.1% (v/v) formic acid. The trap column was then switched in-line with an LC-Packings PepMap C18 column (0.075 × 150 mm) equilibrated similarly. The peptides were separated and MS spectra generated as described previously (37). Raw files were processed with MaxQuant (Version 1.0.13.13) (38). Peak lists generated by Quant.exe were searched against the International Protein Index human database version 3.37 containing 69,290 proteins run on an in-house server using the Mascot search engine v.2.2 (Matrix Science). Mass tolerance was set to 7 ppm and 0.5 Da allowing a maximum of three missed cleavages with trypsin. Carboxyamido-methylation of cysteine was set as a fixed modification, oxidation and acetylation of methionine as variable modifications, heavy SILAC labels were 13C(6) arginine (R) and 13C(6) lysine (K).
Subsequent identification and re-quantification of the peptides was performed by Identify.exe (38, 39) with protein ratios compiled from the contribution of individual peptide ratios at a false discovery rate of 1%, achieved on the basis that there were 100-fold more hits in the forward than reverse database. Identification also required peptides to be of at least six amino acids in length and at least one to be unique. The parameters selected for quantification within Identify.exe were; site quantification by least modified peptides, allowing the variable modifications referred to above and a minimum ratio count of two peptides. However, further contributions from razor peptides were permitted. The options for retaining low scoring peptides and for filtering labeled amino acids were deselected as described previously (38, 39).
Molecular Cloning of IQGAP1
The C-terminal region of IQGAP1 (IQGAP1718–1657) was amplified from the cDNA clone MGC: 134938 IMAGE: 40073370 (obtained from the I.M.A.G.E. consortium) by PCR using KOD polymerase (Calbiochem) and the following primers: (5′-3′) sense, GTCTATGTCGACCTATGCAGCTTTCTCGGGAGG and antisense, TATGCGGCCGCTTACTTCCCGTAGAACTTTTTGTTGAG. The PCR product was gel purified on a 1% (w/v) agarose gel, ligated into pGEX-6P-1 for bacterial expression and the sequence confirmed.
Expression of Recombinant Protein
The pGEX-6P-1 construct encoding GST-IQGAP718–1657 was transformed into BL21 E. coli and cultures grown at 37 °C in 500 ml Luria Bertani broth supplemented with 100 μm ampicillin. Expression was achieved by addition of isopropyl β-d-thiogalactoside (50 μm) at 30 °C for 16 h. The cells were collected by centrifugation at 6000 × g at 4 °C for 30 min and the pellets lysed by a single round of freeze-thawing followed by sonication in 25 ml ice cold lysis buffer. Following centrifugation as previously, the supernatant was loaded onto a 1 ml glutathione-Sepharose column pre-equilibrated with lysis buffer at 4 °C. The column was washed with 10 ml of buffer B (50 mm Tris/HCl, 150 mm NaCl, pH 7.2) supplemented with an additional 350 mm NaCl then with 10 ml of buffer B alone. The protein was eluted with 5 ml of buffer B supplemented 15 mm glutathione and the eluate dialyzed against 5 liters of buffer B supplemented with 1 mm dithriothreitol and 20% (v/v) glycerol at 4 °C for 12 h. Aliquots (100 μl) of the protein were frozen in liquid nitrogen and stored at −80 °C.
Measurement of Protein-Lipid Binding
Protein-lipid interaction was measured at 25 °C by surface plasmon resonance (SPR) using a Biacore 3000 biosensor equipped with SA sensor chips coated with biotinylated lipids. This four cell system allowed protein binding to a reference surface (no lipid) to be compared simultaneously with that to test surfaces loaded with PtdIns(3,4)P2, PtdIns(4,5P)2, or PtdIns(3,4,5)P3. The lipids (100 μm), diluted in running buffer (10 mm Hepes, pH 7.2, 250 mm NaCl, 0.1 mm EDTA, 0.1 mm EGTA,) were loaded at a flow rate of 5 μl/min for 5 min or to achieve a comparable loading of each lipid corresponding to ∼400–600 response units. Surfaces were then blocked by washing at the same flow rate with running buffer supplemented with essentially fatty acid-free bovine serum albumin (BSA) (0.2 mg/ml) for a minimum of 15 min before application under the same conditions of the test proteins at the successively increasing concentrations indicated. Surfaces were regenerated between the application of separate proteins using a 30 s pulse of 5 mm NaOH followed by washing with supplemented running buffer as previously described. Sensor chips were discarded following two rounds of regeneration owing to diminished protein binding capacity. Additionally, protein-lipid interaction was assessed using protein-lipid overlay assays performed as described previously (23, 25) using the GST-tagged PH domains of GRP1, TAPP1 and phospholipase (PLC) δ, and GST-tagged test protein (see “Results”) each at 0.2 μg/ml.
RESULTS
A Three-step Enrichment of Potential PtdIns(3,4)P2 Binding Partners
Signaling downstream of class IA PI 3-kinases is mediated by PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (5, 28) but few specific effectors for the latter lipid have been reported. We established a screen for these by optimizing conditions for the PI 3-kinase-dependent recruitment to intact cell membranes of the known PtdIns(3,4)P2 selective protein, TAPP-1. The subsequent enrichment of this fraction allowed the identification of a pool of likewise responsive cellular proteins with potentially similar binding characteristics. To do this we exploited the ability of vanadate analogs to induce in PTEN null, 1321N1 astrocytoma cells a dramatic but selective increase in the cellular concentration of PtdIns(3,4)P2 over that of other 3-PIs mediated by the co-incident activation of class I PI 3-kinase and SHIP2 and the inhibition of PtdIns(3,4)P2 4-phosphatases (11).
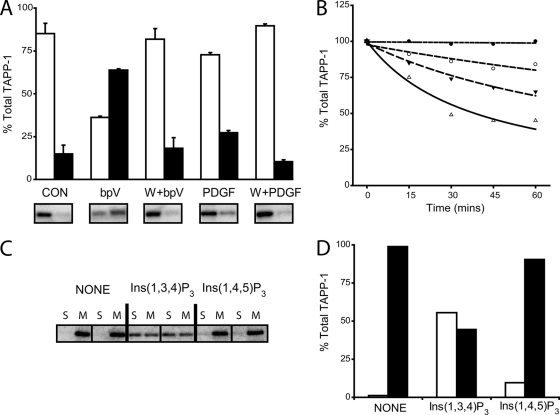
Fig. 1A thus shows that a large fraction of the cellular pool of TAPP-1 protein is recruited PI 3-kinase-dependently from the soluble to the membrane fraction of 1321N1 cells in response to stimulation with the vanadate analog, bpV(phen). In control cells, the soluble fraction released post cell permeabilization (cytosol) contained almost all the detectable TAPP-1, as anticipated for this cytosolic protein (34). The small proportion of TAPP-1 remaining following digitonin treatment (membrane fraction) probably represents trapped protein as this was not diminished by pretreatment of cells with the PI 3-kinase inhibitor, wortmannin (data not shown). Most pertinently however, following stimulation with bpV(phen), the proportions of TAPP-1 protein associated with the cytosol and membrane fractions were reversed such that 60–70% of the cellular total was now bound to the latter. Fig. 1A also demonstrates that stimulation with platelet-derived growth factor (PDGF), which induces a smaller cellular accumulation of PtdIns(3,4)P2 in 1321N1 cells (11), promoted much more limited membrane translocation of TAPP-1. These data further show that the translocation of TAPP-1 induced by either bpV(phen) or PDGF was strictly dependent on stimulated PI 3-kinase activity, because it was prevented fully by wortmannin, though relatively high concentrations (1 μm) of this inhibitor are required to block the ≥1000-fold increase in cellular PtdIns(3,4)P2 concentrations induced by bpV(phen). Nevertheless, the profound effects of this stimulus are an advantage in the current context because they allow the efficient, primary recruitment of cellular TAPP-1 protein and would be expected to facilitate the similar membrane enrichment of other PtdIns(3,4)P2 interacting proteins, perhaps including those of lower binding affinity. Figs. 1B further illustrates that the increased cellular PtdIns(3,4)P2 concentrations (≥250 pmol/mg protein, globally ∼50 μm) achieved by stimulation with bpV(phen) permitted both a large and sustained membrane recruitment of TAPP-1 that persisted without detectable loss for up to 60 min following cell permeabilization. This persistent membrane association of TAPP-1 was, nevertheless, reversible because the addition to digitonin permeabilized cells of Ins(1,3,4)P3, the water-soluble head-group of PtdIns(3,4)P2, at concentrations estimated to give a molar excess of ∼100–1000-fold over cellular PtdIns(3,4)P2, concentration-dependently displaced up to ∼60% of the protein recruited previously. However, whereas addition of Ins(1,3,4)P3 reversed significantly the prior membrane association of TAPP-1, Figs. 1C and 1D shows that the similar addition of Ins(1,4,5)P3 was not effective. These data thus indicate that the membrane association of TAPP-1 is mediated selectively and reversibly by PtdIns(3,4)P2. The features of this system therefore provide the basis for a dual phase enrichment of PtdIns(3,4)P2 binding partners by coupling (i) an efficient, PI 3-kinase-dependent, recruitment of proteins to whole cell membranes enriched in PtdIns(3,4)P2 to (ii) a subsequent, isomer-specific inositol phosphate-effected recovery of these proteins from the same cell membranes post-permeabilization and extensive washing.
Fig. 1.
The use of TAPP-1 as a marker to obtain a fraction enriched in potential PtdIns(3,4)P2 binding partners. A, TAPP-1 is recruited efficiently by membranes enriched in PtdIns(3,4)P2: Cells were incubated for 30min in the presence of 1 μm wortmannin (W) or DMSO vehicle (0.1% v/v) then further incubated as control (Con) or stimulated for 5 min or 30 min with PDGF (50 ng/ml) or bpV(phen) (0.1 mm) (bpV) respectively, as indicated. Cells were then permeabilized and cytosol (open bars) and membrane (closed bars) fractions prepared and immunoblotted for TAPP-1. The results show the proportionate distribution of total, cellular TAPP-1 and represent the mean ± the range of duplicate measurements from one experiment representative of several. Examples of the immunoblots represented by the quantitation are shown below. B, TAPP-1 is eluted concentration-dependently from PtdIns(3,4)P2- enriched membranes by Ins(1,3,4)P3. Membrane fractions prepared from cells prestimulated for 30 min with bpV(phen) (0.1 mm), were incubated at 4 °C as indicated without (closed circles) or with Ins(1,3,4)P3 at 1 μm (open circles), 10 μm (inverted triangles) or 100 μm (triangles) and the TAPP-1 protein remaining measured by immunoblotting. The results show the percentage of membrane associated TAPP-1 remaining as a proportion of that recruited initially and are from one experiment representative of at least three that gave similar results. C and D, The InsP3-mediated recovery of TAPP-1 from membranes is isomer selective. Membrane fractions prepared from cells prestimulated for 30 min with bpV(phen) (0.1 mm), were incubated in duplicate at 4 °C for 60 min in the absence (none) or presence of either 100 μm Ins(1,3,4)P3 or Ins(1,4,5)P3 as indicated and the TAPP-1 remaining membrane associated (M) or recovered in the supernatant (S) measured by immunoblotting. Similar results were obtained on at least one further occasion. Panel D shows the quantitation of the results presented in panel C.
Fig. 2 shows that this enriched TAPP-1 fraction could be further purified using an affinity matrix similar to that employed previously (29–31, 33). For this purpose we used neutravidin beads pre-coupled to biotinylated PtdIns(3,4)P2. To assess the utility of this step, we initially (Fig. 2) used Triton X-100 detergent extracts of digitonin permeabilized cells rather than Ins(1,3,4)P3 eluates from similar samples to avoid the anticipated competition between PtdIns(3,4)P2 and InsP3 for target proteins. In later experiments (Fig. 3 and later) however, Ins(1,3,4)P3-eluted membrane extracts were also purified successfully and we observed that up to a 10-fold molar excess of Ins(1,3,4)P3 over PtdIns(3,4)P2 had only limited effect on the recovery of TAPP-1. Fig. 2A shows that TAPP-1 protein was captured by bead-immobilized PtdIns(3,4)P2 in a concentration-dependent manner, with maximal protein capture occurring with 25–50 μm lipid. Fig. 2B further shows that, at the maximally effective concentration of PtdIns(3,4)P2, ∼75% of TAPP-1 was depleted from detergent extracts of cellular membranes and could then be efficiently recovered by appropriate elution of the affinity matrix. Fig. 2C illustrates that these Triton X-100-derived, TAPP-1 enriched samples, though lacking the benefit of prior Ins(1,3,4)P3-selective extraction, were purified sufficiently to reveal novel, copurified, potential 3-PI binding partners on analysis by SDS-PAGE. Thus, Fig. 2C reveals several protein bands that are either present, or whose concentration is increased markedly, only as a specific consequence of prior cellular activation of PI 3-kinase because the same proteins are absent or less abundant in samples derived from control cells or cells stimulated in the presence of wortmannin. Subsequent analysis of similar protein bands from larger scale cell extracts (see Fig. 2D) identified in the bands indicated at ∼70 kDa and ∼105 kDa respectively, the pleckstrin homology domain containing proteins SWAP-70, a 3-PI-regulated guanine nucleotide exchange factor for Rac GTPase (33, 40) and PARIS-1 (also referred to as TBC1 domain family member 2A), a sparsely characterized protein (41) not recognized before as a possible phosphoinositide effector.
Fig. 2.
The affinity purification of a fraction enriched in TAPP-1 reveals other PI 3-kinase responsive proteins. A, The concentration-dependent recovery of TAPP-1 by bead-immobilized PtdIns(3,4)P2. The Triton X-100 soluble fraction from 1321N1 cells was incubated at 4 °C for 60 min with streptavidin beads precoupled to biotinylated PtdIns(3,4)P2 sufficient to give the final lipid concentrations indicated. The beads were collected and washed and the proportion of total cellular TAPP-1 protein associated was measured by immunoblotting. The results represent the mean of duplicate determinations in one experiment representative of three that gave similar results. B, Cellular recruitment and affinity-capture in combination allow efficient recovery and concentration of TAPP-1. Membrane fractions prepared from cells incubated for 30min without (Con) or with (Stim) bpV(phen) (0.1 mm) were solubilized in lysis buffer and aliquots of the lysate and beads (PD) immunoblotted for TAPP-1 protein before (Pre) and following (Post) affinity precipitation of TAPP-1 with a maximally effective concentration (10–25 μm) of bead-immobilized PtdIns(3,4)P2. The results indicate the proportion of TAPP-1 protein in the lysate (Pre and Post) or that pulled down (PD) normalized to the amount present in the membrane extract from stimulated cells and represent the mean ± S.D. of three experiments performed in duplicate. C, The TAPP-1 fraction reveals other, copurified PI 3-kinase-responsive proteins. Membrane fractions were prepared from cells incubated for 30min with 1 μm wortmannin (W) or DMSO vehicle (0.1% v/v) then further incubated as control (Con) or stimulated for 30 min with bpV(phen) (0.1 mm) (bpV). These fractions were solubilized in lysis buffer and affinity precipitated with immobilized PtdIns(3,4)P2. The precipitated proteins were analyzed by SDS-PAGE and silver stained. The arrows indicate bands that are not apparent in samples from control cells or following wortmannin treatment. The results show duplicate sample lanes. Similar data were obtained on several further occasions (see text). D, Peptide mass fingerprinting of PARIS-1 and SWAP-70. Protein bands were excised and identified as indicated. The threshold for significance of identity by Mascot was set at the commonly accepted value p < 0.05.
Fig. 3 reveals that by incorporating an additional Ins(1,3,4)P3-selective membrane elution step into the enrichment procedure a far greater range of stimulus-responsive proteins became apparent. This shows a silver stained gel comparing the densitometry profiles of samples purified sequentially from whole cells by a combination of (i) a stimulated recruitment to PtdIns(3,4)P2 enriched membranes, (ii) Ins(1,3,4)P3-selective elution, and (iii) PtdIns(3,4)P2-affinity precipitation. The comparative traces clearly reveal multiple (>10) protein bands, the intensity of which is increased as a consequence of cellular PI 3-kinase activation. These results are important because they indicate the variety of potential, cellular PtdIns(3,4)P2 binding partners yet to be recognized and highlight the success of this strategy for identifying candidate proteins. However, owing to the relatively low abundance of the proteins shown and the somewhat variable pattern of individual, PI 3-kinase-responsive protein bands between experiments, it proved difficult to obtain a sufficient amount of these from comparatively small scale experiments for identification by mass spectrometry. The latter feature also precluded pooling material from multiple experiments whereas preliminary attempts to scale-up to yield Colloidal Blue detectable proteins were thwarted by the lower sensitivity of this stain to discriminate stimulus-dependent proteins above background. To avoid these issues and yet retain the advantages of our simple, but effective strategy for the enrichment of TAPP-1 copurifying proteins, we therefore coupled this protocol to SILAC methodology.
Coupling of the Three Phase Affinity Enrichment to SILAC Identifies Multiple Candidate Proteins
SILAC (42, 43) employs the differential, metabolic labeling of control- and test-cell populations, for example with normal (12C) and heavy (13C) isotope substituted amino acids (arginine and lysine) respectively. The subsequent post-challenge pooling and processing of extracts derived from these cells allows the identification of test-responsive proteins via their increased ratio of heavy:light isotope-labeling of discriminating peptides. This elegant process suitably complements our purification strategy as it eliminates the errors associated with the independent processing of separate extracts from control and stimulated cells and permits a quantitative proteomic analysis that better resolves stimulus-responsive proteins from the background.
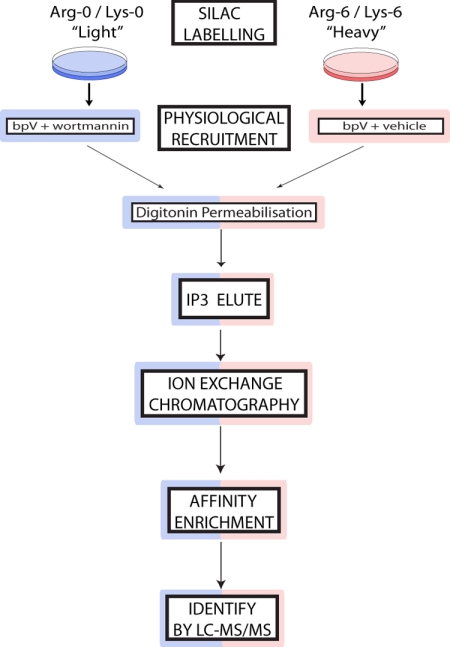
Fig. 4 shows how the SILAC approach was incorporated into our scheme. Thus, control and stimulated cells were prelabeled separately with (12C) and (13C) amino acids respectively. Following cell stimulation, the membrane fractions obtained from both were combined and an Ins(1,3,4)P3 eluate of the pooled membranes was further purified by sequential ion exchange chromatography to obtain three samples, anion, cation, and flow-through, for subsequent affinity enrichment, SDS-PAGE, and MS analysis. Thus, any enhancement in the ratio of heavy:light isotope labeling for individual proteins reflects exclusively that induced by the activation of PI 3-kinase under conditions favoring cellular PtdIns(3,4)P2 accumulation.
Fig. 4.
Incorporating SILAC into the three phase affinity enrichment protocol. A schematic representation is shown of the steps used to label separately control (bpV + wortmannin, blue) and stimulated (bpV + vehicle, red) cell populations respectively with light and heavy isotope-labeled amino acids and, post-stimulation, to then jointly purify PI 3-kinase-responsive proteins from the pooled cell extracts and identify these proteins by their increased heavy isotope labeling using LC-MS/MS.
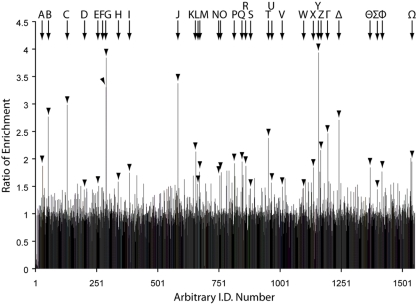
Fig. 5 shows that a total of ∼1500 proteins were identified and assigned a SILAC ratio in the PtdIns(3,4)P2-affinity purified samples obtained from the fractions described. Fig. 5 also demonstrates clearly that although the vast majority (∼95%) of these reflect background proteins with a normalized isotope labeling ratio of 1:1, a significant minority (∼5%, see below) of the proteins that copurify through the protocol for TAPP-1 enrichment also express an isotopic ratio that well exceeds the background, indicating their initial, PI 3-kinase-dependent recruitment. Indeed, the identification of TAPP-1 (Table 1, Y) itself with a heavy:light isotope labeling ratio of close to 4:1 provides strong proof of principle for this process. Equally, the presence of all three known isoforms of Akt (Table 1, F, G, and K) and of other established 3-PI binding proteins, including SWAP-70, centaurin delta (ARAP1) and EEA-1 (Table 1, Z, T, and Γ respectively), among those proteins showing a markedly increased isotope ratio provides powerful support for the practicality of this approach. In all, Fig. 5 shows that ∼80–85 of the proteins identified expressed an increase in their isotopic labeling ratio of at least ∼1:1.5. Above this ratio, the heavy isotope enrichment relative to background was assigned a high degree of significance for all proteins by MaxQuant and is thus likely to reflect their initial PI 3-kinase-dependent membrane recruitment. Strong support for this assertion is provided independently by the positive phosphoinositide binding data reported below for IQGAP1 whose isotope labeling ratio falls just outside this threshold. A full list of all the proteins identified together with other appropriate data relevant to peptide identification, labeling ratio and the significance of the latter relative to that for background proteins as ascribed by MaxQuant (38, 39) is provided in the supplementary Tables S1, S2, and S3). Table 1 however, summarizes some of these features for the proteins that exhibited the most pronounced, stimulus-dependent increase in their 13C:12C isotope ratio. Table 1 also includes other proteins, some with lower ratios but deemed of particular interest either because of the large number of constituent peptides identified (e.g. IQGAP1, Serpin H1) and/or because of established biological features (e.g. localization or association with other signaling components, e.g. IQGAP1, PX domain-containing protein kinase-like protein) that might indicate a plausible connection to the wider phosphoinositide signaling network. This diverse group of proteins were all identified with a high degree of confidence, each with a minimum of two unique peptides. Many, though not all, express potential lipid binding domains consistent with the view that several of those not recognized previously as such may be subject to regulation by 3-PIs, albeit not necessarily by PtdIns(3,4)P2 directly or exclusively. Similar, apparently disparate sets of proteins have been identified previously using lipid-affinity based approaches for the purification of putative 3-PI interacting proteins (29–33). However, we believe that the results presented here represent the first quantitative analysis of potential PI 3-kinase/PtdIns(3,4)P2 effector proteins based uniquely on a combination of their primary, selective cellular recruitment and secondary, preferential enrichment.
Fig. 5.
Coupling SILAC to a three step enrichment protocol identifies multiple 3-PI-responsive proteins. Following purification, the pooled extracts from control and stimulated cells revealed ∼1500 proteins, which were identifiable by discriminating peptides for which there were SILAC pairs. These proteins are presented as an arbitrary identification number plotted against the corresponding heavy:light isotope ratio. Details for the proteins highlighted A-Ω are given in Table I. These proteins include those exhibiting the highest isotope labeling ratio in addition to other candidates (e.g. IQGAP1, see text) whose inclusion is prompted by their established signaling roles. The results are from a single experiment but similar data were obtained on one further occasion.
Table 1. Candidate PtdIns(3,4)P2 and other 3-PI responsive protein partners.
The proteins described are those highlighted in Fig. 5 as key candidates emerging from the SILAC-coupled screen. All the proteins shown were identified with a high degree of confidence (indicated by the posterior error probability, PEP, value) and exhibited markedly elevated ratios of heavy:light (H/L) isotope labelling.
The known lipid-interacting or additional domains/motifs expressed by other proteins are also indicated, excluding those inherent in protein names, together with the other parameters indicated including the significance assigned to the SILAC ratio (Ratio Sig.).
| Protein Names | Gene Name | UniProt. | Domains | Mw [kDa] | Peptides | PEP | Ratio H/L | Ratio Sig. | |
|---|---|---|---|---|---|---|---|---|---|
| A | Fermitin family homolog 2 | FERMT2 | A8K6S3 | FERM, PH | 78.7 | 8 | 5.09E-14 | 1.87 | 2.82E-04 |
| B | Epsin-1 | EPN1 | Q9Y6I3–2 | ENTH | 69.0 | 3 | 1.37E-27 | 2.77 | 3.81E-09 |
| C | Forkhead box protein K2 | FOXK2 | Q01167–1 | FHA, Fork head | 69.1 | 3 | 2.58E-34 | 2.98 | 2.99E-10 |
| D | Ras GTPase-activating-like protein IQGAP1 | IQGAP1 | P46940 | CH, IQ, RasGAP, WW | 189.3 | 163 | 0 | 1.44 | 2.43E-02 |
| E | Peroxiredoxin-4 | PRDX4 | Q13162 | 30.5 | 16 | 9.06E-130 | 1.51 | 1.29E-02 | |
| F | Akt-1; Protein kinase B | AKT1 | P31749 | PH | 55.7 | 55 | 2.41E-294 | 3.30 | 1.09E-11 |
| G | Akt-2; Protein kinase B β | AKT2 | P31751 | PH | 55.8 | 36 | 5.96E-147 | 3.84 | 2.19E-14 |
| H | SPARC | SPARC | P09486 | 34.6 | 6 | 9.65E-15 | 1.58 | 5.53E-03 | |
| I | Protein SOLO | SOLO | Q8TER5–1 | PH | 164.6 | 9 | 5.15E-46 | 1.74 | 1.08E-03 |
| J | Eukaryotic translation initiation factor 4E | EIF4E | P06730 | 28.8 | 13 | 5.84E-29 | 3.38 | 1.42E-11 | |
| K | Akt-3; Protein kinase B γ | AKT3 | Q9Y243–1 | PH | 58.7 | 25 | 4.41E-226 | 2.13 | 1.31E-05 |
| L | Serpin H1 | SERPINH1 | P50454 | 46.4 | 104 | 0 | 1.54 | 8.91E-03 | |
| M | Oxysterol-binding protein-related protein 11 | OSBPL11 | Q9BXB4 | PH | 83.6 | 6 | 9.71E-45 | 1.76 | 5.28E-04 |
| N | KDEL motif-containing protein 2 | KDELC2 | Q7Z4H8–1 | 58.6 | 33 | 1.47E-251 | 1.70 | 1.85E-03 | |
| O | Rho GTPase-activating protein 29 | ARHGAP29 | Q52LW3–1 | C1, RhoGAP | 142.1 | 2 | 1.13E-08 | 1.76 | 5.77E-04 |
| P | ELKS/RAB6-interacting/CAST family member 1 | ERC1 | Q8IUD2–1 | FIP | 128.1 | 3 | 3.03E-09 | 1.92 | 9.46E-05 |
| Q | FK506-binding protein 9 | FKBP9 | O95302 | EF hand | 63.1 | 12 | 3.05E-55 | 1.95 | 1.10E-04 |
| R | La-related protein 1 | LARP1 | Q6PKG0–1 | La domain | 123.5 | 14 | 1.01E-21 | 1.87 | 2.80E-04 |
| S | PICALM | PICALM | A8K5U9 | ANTH | 71.7 | 77 | 0 | 1.46 | 1.99E-02 |
| T | ARAP1 (Centaurin-Δ-2) | ARAP1 | Q96P48–6 | PH, RhoGAP, SAM | 162.2 | 9 | 1.99E-12 | 2.38 | 1.00E-06 |
| U | Annexin A6 | ANXA6 | P08133 | 75.9 | 46 | 7.83E-197 | 1.57 | 7.16E-03 | |
| V | Vinculin | VCL | P18206–1 | 123.8 | 63 | 0 | 1.49 | 1.50E-02 | |
| W | Talin-1 | TLN1 | Q9Y490 | FERM | 269.8 | 103 | 0 | 1.47 | 1.84E-02 |
| X | FK506-binding protein 10 | FKBP10 | Q96AY3 | 64.2 | 30 | 3.95E-138 | 1.84 | 3.70E-04 | |
| Y | TAPP-1 | PLEKHA1 | Q9HB21 | PH | 45.6 | 4 | 1.09E-16 | 3.94 | 5.34E-15 |
| Z | Switch-associated protein 70 | SWAP70 | Q9UH65 | PH | 69.0 | 209 | 0 | 2.16 | 9.68E-06 |
| Γ | Early endosome antigen 1 | EEA1 | Q15075 | FYVE | 162.5 | 14 | 1.27E-69 | 2.47 | 1.43E-07 |
| Δ | Eukaryotic translation initiation factor 4 γ1 | EIF4G1 | Q04637–1 | 175.53 | 50 | 1.55E-139 | 2.71 | 1.33E-08 | |
| ϴ | Endoplasmic reticulum aminopeptidase 2 | ERAP2 | Q6P179–1 | 110.5 | 6 | 2.45E-19 | 1.84 | 2.17E-04 | |
| Σ | PX domain-containing protein kinase-like protein | PXK | Q7Z7A4–1 | PX | 64.9 | 15 | 9.49E-86 | 1.44 | 2.36E-02 |
| φ | Protein canopy homolog 3 | CNPY3 | Q9BT09–1 | 30.7 | 5 | 1.51E-51 | 1.77 | 8.40E-04 | |
| Ω | Ankyrin repeat and FYVE domain-containing protein 1 | ANKFY1 | Q9P2R3–2 | Ankyrin repeat, FYVE | 128.5 | 29 | 1.80E-139 | 1.95 | 1.19E-04 |
The Identification of IQGAP1 as a Potential 3-PI Regulated Protein may Reveal a Novel Category of Atypical Phosphoinositide (aPI) Binding Domain
The 3-PI binding characteristics and the functional consequences of lipid interaction for the candidate proteins identified in Tables 1 and supplemental Table S1 are currently being evaluated systematically by our laboratory. Among these, we placed early focus on IQGAP1 for two reasons. First, this large, multicomponent protein is responsive to inputs from a nexus of signaling pathways and its localization and primary roles in the regulation of cytoskeletal organization and, in particular, of the activity of the small GTPase Rac1 (44, 45), suggest its potential functional integration with the actions of 3-phosphoinositides. Second, the isotopic labeling ratio of ∼1:1.44 obtained for IQGAP1, though arising from a large number (163) of identified peptides, lies around the ∼5% threshold required to achieve the significance value assigned by MaxQuant (38, 39) to distinguish probable candidates and background proteins. Thus, IQGAP1 provides an appropriate test of principle with which to validate the view that proteins expressing isotope ratios greater than this are likely, authentically PI 3-kinase responsive. Fig. 6 shows that a GST-tagged fusion protein comprising the C terminus (amino-acids 718–1657) of IQGAP1 (GST-IQGAP718–1657), incorporating several established protein-protein interaction domains, bound selectively and concentration-dependently to PtdIns(3,4,5)P3 but not to PtdIns(3,4)P2 or PtdIns(4,5)P2. Moreover, Fig. 6 shows that the binding of IQGAP1 to PtdIns(3,4,5)P3 was comparable in character to that displayed by similar concentrations of other well-established phosphoinositide binding domains (6, 46) for their cognate lipid targets under similar conditions. Thus, Figs. 6A–6C illustrate the selective binding of the GST-tagged PH domain, of GRP1 to PtdIns(3,4,5)P3, of TAPP-1 to PtdIns(3,4)P2, and of PLCδ to PtdIns(4,5)P2, measured by surface plasmon resonance (SPR) analysis using immobilized, biotinylated lipids. Fig. 6D demonstrates the binding of the GST-IQGAP718–1657 fusion protein to PtdIns(3,4,5)P3, revealing that this was qualitatively similar to that for GST-GRP1, suggesting that the interaction of IQGAP1 with PtdIns(3,4,5)P3 is authentic and potentially important. Fig. 6E shows that the binding of GST-IQGAP718–1657 to PtdIns(3,4,5)P3 was dependent on protein concentration, with a half-maximal response apparent at ≥30 nm, a value within the affinity range displayed by other 3-PI binding proteins (46, 47). A similar analysis of these interactions using protein-lipid overlay assays routinely revealed the same, anticipated phosphoinositide specificities of the established lipid binding domains and confirmed the ability of the GST-IQGAP718–1657 fusion protein to bind polyphosphoinositides but indicated a lesser apparent selectivity for PtdIns(3,4,5)P3 (data not shown). We would emphasize however, that although the latter overlay approach is used widely to assess the potential for proteins to bind lipids, the identity of the preferred lipid ligand is often determined more reliably by complementary techniques like SPR that allow a more controlled presentation of the lipid (47). Consequently, whereas the data shown reflect a preliminary rather than a definitive characterization of the interaction between IQGAP1 and 3-PIs, they nevertheless argue persuasively that IQGAP1 reflects a plausible and intriguing target for PtdIns(3,4,5)P3 or perhaps, for the Ins(1,3,4,5)P4 head-group of this lipid. Most importantly however, these results emphasize the utility of the screen and highlight the potential value of the wider data set presented.
Fig. 6.
GST-IQGAP718–1657 binds selectively to PtdIns(3,4,5)P3. A–D, The phosphoinositide binding selectivity of GST-IQGAP718–1657 is similar to that of GRP-1: The binding of the GST-tagged PH domains of GRP-1 (A), TAPP-1 (B), or PLC-δ (C) each at 50 nm to streptavidin chips preloaded with equal amounts of PtdIns(3,4,5)P3 (green), PtdIns(3,4)P2 (red), or PtdIns(4,5)P2 (black) was compared with that for 50 nm GST-IQGAP718–1657 (D) by surface plasmon resonance. E, The concentration-dependence of GST-IQGAP718–1657 binding to PtdIns(3,4,5)P3. The binding of GST-IQGAP1718–1657 (0–100 nm, as indicated) to streptavidin chips preloaded with PtdIns(3,4,5)P3 was examined by surface plasmon resonance by sequential application of the protein from low to high concentration. The traces shown are representative of those obtained on at least two separate occasions.
DISCUSSION
The recognition of specific protein domains that selectively bind particular lipids has been responsible for dramatic conceptual and practical advances in the understanding and study of cellular signaling mechanisms (48). Indeed, the coupling of class I PI 3-kinases to the regulation of diverse cellular processes is currently accounted for primarily by the localization and/or activation of an array of effector proteins expressing appropriate PtdIns(3,4,5)P3 and PtdIns(3,4)P2 binding domains (1, 2, 5, 6). However, as few molecular targets selective for the latter lipid have yet been described, the current study has established a screen for these.
Earlier informatics- (23, 25, 49–51) and affinity chromatography- (29–33) based approaches have both been successful in identifying individual protein binding partners for particular PIs. The former approach however, is limited by the bias inherent in the selection of the criteria assumed to govern binding. Alternatively, affinity chromatography, though particularly useful for the identification of protein sets with the potential to interact with specific PI species (29, 30, 32), provides little basis alone on which to distinguish authentic candidates from contaminating proteins. Here we have developed an unbiased strategy that couples the advantages of established binding specificity to affinity enrichment and other, novel steps that avoids these limitations.
In the first step, proteins were recruited to the membranes of intact cells stimulated under conditions that favor dramatically the selective, but not exclusive, accumulation of PtdIns(3,4)P2 (11). Importantly, this allowed a primary enrichment of proteins under conditions in which the target lipid was presented in the appropriate physicochemical context of an authentic membrane bilayer to potential binding partners present at physiologically relevant concentrations. These factors are especially difficult to mimic in vitro but are crucial, cellular determinants of protein-lipid interactions (47). The practical utility of this approach and the effectiveness of secondary purification steps to yield a fraction enriched in likewise responsive proteins of similar lipid binding character was verified by tracking endogenous TAPP-1, a recognized PtdIns(3,4)P2 binding protein (23). The potential of these combined strategies is demonstrated by the identification of SWAP-70 and PARIS-1 among a wider set of apparently PI 3-kinase-responsive proteins. SWAP-70 has been identified previously as a 3-PI-regulated GEF for Rac GTPase, capable of binding PtdIns(3,4,5)P3 (33) or PtdIns(3,4)P2 (40). By contrast, PARIS-1 has only been identified previously as a potential marker for prostate cancer (41) but Pfam analysis of the available amino acid sequence indicates a TBC domain characteristic of Rab GTPase activating proteins and a PH (pleckstrin homology) domain whose role and lipid binding properties are being characterized currently by our laboratory. Finally, to facilitate the broader identification of proteins within this larger group and to evaluate the initial PI 3-kinase responsiveness of individual candidate proteins, this approach was coupled to the ratio-metric power of SILAC analysis.
This strategy allowed the unambiguous, mass spectrometric identification of a diverse group of proteins shown to be responsive to the primary, cellular stimulation of PI 3-kinase and distinguishable from copurifying, background proteins by their increased isotope-labeling ratio determined in the absence and presence of wortmannin. The identification of TAPP-1 as one of those proteins exhibiting the highest ratio confirms that this approach can not only reveal PI 3-kinase-responsive proteins but is also most selective for those that bind PtdIns(3,4)P2 preferentially. Similarly, the high ratio observed for all three isoforms of Akt supports this notion because data obtained from both cellular (18–21) and in vitro (22) studies indicates that Akt interacts with both PtdIns(3,4,5)P3 and PtdIns(3,4)P2. The prominence of TAPP-1 and Akt thus provides both elegant proof of principle for this as an approach to identify other proteins with similar binding characteristics and lends credibility to the novel candidates identified. The expression of PH and/or other recognized lipid binding domains, including FYVE, ENTH, C1 and FERM domains, by several of those proteins revealing the highest labeling ratios, similarly emphasizes this view. Thus, our results introduce a set of proteins whose authentic PI 3-kinase responsiveness and plausible PtdIns(3,4)P2 selectivity are defined and graded on the basis of their cellular susceptibility to wortmannin-sensitive recruitment by membranes enriched in this lipid. The confirmation of any candidates as direct 3-PI effectors proteins will require rigorous characterization of both their interaction with, and/or functional regulation by, appropriate lipids and is our current priority.
This is not to anticipate however, either that our candidates reflect the complete cellular complement of potential PtdIns(3,4)P2 effector proteins or that all of the proteins observed with an elevated isotope ratio necessarily bind PtdIns(3,4)P2 preferentially or directly. Indeed, the absence from our candidate list of other PtdIns(3,4)P2 binding proteins such as lamellipodin (24) and DAPP-1 (25) highlights the first limitation and possibly reflects either the relatively low abundance of these proteins in our samples or a failure of these to yield discriminating peptides sufficient for their ultimate identification. Equally, it is plausible that some proteins though PI 3-kinase responsive, do not bind 3-PIs directly but are recruited via appropriate adaptors such as TAPP-1. Furthermore, although each stage of our screen is optimized to favor the selective isolation of proteins most like TAPP-1 in their lipid binding character, the primary factor responsible for a positive isotope ratio is the activation of PI 3-kinase. Consequently, it can be expected that proteins with other 3-PI binding preferences will also emerge and this may account for the ratios observed for proteins such as SWAP-70 (33, 52) and centaurin delta 2 (ARAP1) (53, 54) or EEA-1 (10, 48) that are thought to interact with PtdIns(3,4,5)P3 or PtdIns3P respectively. Alternatively, as SWAP-70 may also bind PtdIns(3,4)P2 (40) and the in vitro Arf GAP activity of ARAP1, though regulated preferentially by PtdIns(3,4,5)P3, is also sensitive to PtdIns(3,4)P2 (54), our results may indicate that these proteins, like Akt, are effectors for both lipids.
The wider utility of our screen lies in its capacity to identify candidates that may function as components of the broader network of PI 3-kinase signaling proteins. This is best exemplified by our identification of IQGAP1 as a novel, potential 3-PI-regulated protein. IQGAP1 is a ∼190 kDa protein whose numerous protein-interaction domains mediate inputs from diverse, upstream signaling pathways and allow it to execute a multitude of actions at the interface between intracellular signaling and cytoskeletal organization (44, 45, 55, 56). The predominant molecular function of IQGAP1 remains unclear but its involvement in various cellular processes on which PI 3-kinase also exerts a prominent influence, implies a likely close integration of these components. The localization of IQGAP1 at the leading edge of migrating cells and its ability to interact with Rac1 (44, 57), features that are common to PtdIns(3,4,5)P3 (58, 59), emphasize this point and our results indicating that the C-terminal half of IQGAP1 can bind PtdIns(3,4,5)P3 may forge a direct functional link between IQGAP1 and PI 3-kinase signaling. Furthermore, as IQGAP1 lacks a hitherto defined lipid recognition domain, these results also suggest that this protein may exemplify a new category of atypical phosphoinositide (aPI) binding component. This exciting prospect introduces a fresh perspective on the coupling of this putative tumor promoter to upstream signaling events and poses intriguing questions with respect to the potential lipid binding character of this and other proteins of the IQGAP family.
In conclusion, we have described a screen for proteins that may act as novel effectors for PtdIns(3,4)P2 and other 3-PIs. This uses a unique approach shown successfully to isolate and identify known targets for these lipids and to highlight likely authentic, new candidate proteins. It is anticipated that the future, detailed characterization of candidates identified here will extend significantly the current repertoire of 3-PI effector proteins and establish new connections to the PI 3-kinase signaling network.
Acknowledgments
We are grateful to the Division of Signal Transduction Therapy, University of Dundee, for the generous provision of antibodies and proteins and to The Fingerprints Proteomics Facility, University of Dundee for the analysis of SILAC samples.
Footnotes
* Financial support from Medical Research Council, U.K. (Programme Grant No.G 0801865) is also gratefully acknowledged. MJD is supported by the BBSRC. MA is supported by the Wellcome Trust (Grant No. 083481).
 This article contains supplemental Tables S1, S2, S3.
This article contains supplemental Tables S1, S2, S3.
1 The abbreviations used are:
- PI
- phosphoinositide
- PtdIns
- phosphatidylinositol
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PtdIns3P
- phosphatidylinositol 3-phosphate
- Ins(1,4,5)P3
- inositol 1,4,5-trisphosphate
- Ins(1,3,4)P3
- inositol 1,3,4-trisphosphate
- PTEN
- phosphatase and tensin homologue deleted on chromosome ten
- SHIP1 and SHIP2
- Src homology 2 domain containing inositol polyphosphate 5-phosphatases 1 and 2
- TAPP-1
- tandem pleckstrin homology domain containing protein-1
- DAPP-1
- dual adapter for phosphotyrosine and 3-phosphoinositides
- Irgm-1
- immunity-related GTPase
- GST
- glutathione transferase
- PH
- pleckstrin homology
- GRP-1
- general receptor for phosphoinositides
- PLCδ
- phospholipase C delta
- bpV(phen)
- potassium bisperoxo(1,10-phenanthroline)oxovanadate (V)
- SILAC
- stable isotope labelling with amino acids in cell culture
- PDGF
- platelet-derived growth factor
- SWAP-70
- switch-associated protein-70
- PARIS-1
- prostate antigen recognized and identified by SEREX 1
- ARAP1
- ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
- EEA1
- early endosome antigen 1
- IQGAP1
- IQ motif containing GTPase activating protein 1
- GAP
- GTPase activating protein
- GEF
- guanine nucleotide exchange factor
- aPI binding domain
- atypical phosphoinositide binding domain.
REFERENCES
- 1. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 3. Kok K., Geering B., Vanhaesebroeck B. (2009) Regulation of phosphoinositide 3-kinase expression in health and disease. Trends Biochem. Sci. 34, 115–127 [DOI] [PubMed] [Google Scholar]
- 4. Marone R., Cmiljanovic V., Giese B., Wymann M. P. (2008) Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim. Biophys. Acta 1784, 159–185 [DOI] [PubMed] [Google Scholar]
- 5. Hawkins P. T., Anderson K. E., Davidson K., Stephens L. R. (2006) Signalling through Class I PI3Ks in mammalian cells. Biochem. Soc. Trans. 34, 647–662 [DOI] [PubMed] [Google Scholar]
- 6. Lemmon M. A. (2003) Phosphoinositide recognition domains. Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 7. Leslie N. R., Downes C. P. (2002) PTEN: The down side of PI 3-kinase signalling. Cell. Signal. 14, 285–295 [DOI] [PubMed] [Google Scholar]
- 8. Backers K., Blero D., Paternotte N., Zhang J., Erneux C. (2003) The termination of PI3K signalling by SHIP1 and SHIP2 inositol 5-phosphatases. Adv. Enzyme Regul. 43, 15–28 [DOI] [PubMed] [Google Scholar]
- 9. Lazar D. F., Saltiel A. R. (2006) Lipid phosphatases as drug discovery targets for type 2 diabetes. Nat. Rev. Drug Discov. 5, 333–342 [DOI] [PubMed] [Google Scholar]
- 10. Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. (2001) The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13, 485–492 [DOI] [PubMed] [Google Scholar]
- 11. Batty I. H., van der Kaay J., Gray A., Telfer J. F., Dixon M. J., Downes C. P. (2007) The control of phosphatidylinositol 3,4-bisphosphate concentrations by activation of the Src homology 2 domain containing inositol polyphosphate 5-phosphatase 2, SHIP2. Biochem. J. 407, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sleeman M. W., Wortley K. E., Lai K. M., Gowen L. C., Kintner J., Kline W. O., Garcia K., Stitt T. N., Yancopoulos G. D., Wiegand S. J., Glass D. J. (2005) Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 11, 199–205 [DOI] [PubMed] [Google Scholar]
- 13. Majerus P. W., Kisseleva M. V., Norris F. A. (1999) The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 274, 10669–10672 [DOI] [PubMed] [Google Scholar]
- 14. Vyas P., Norris F. A., Joseph R., Majerus P. W., Orkin S. H. (2000) Inositol polyphosphate 4-phosphatase type I regulates cell growth downstream of transcription factor GATA-1. Proc. Natl. Acad. Sci. U.S.A. 97, 13696–13701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nystuen A., Legare M. E., Shultz L. D., Frankel W. N. (2001) A null mutation in inositol polyphosphate 4-phosphatase type I causes selective neuronal loss in weeble mutant mice. Neuron 32, 203–212 [DOI] [PubMed] [Google Scholar]
- 16. Sasaki J., Kofuji S., Itoh R., Momiyama T., Takayama K., Murakami H., Chida S., Tsuya Y., Takasuga S., Eguchi S., Asanuma K., Horie Y., Miura K., Davies E. M., Mitchell C., Yamazaki M., Hirai H., Takenawa T., Suzuki A., Sasaki T. (2010) The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 465, 497–501 [DOI] [PubMed] [Google Scholar]
- 17. Gewinner C., Wang Z. C., Richardson A., Teruya-Feldstein J., Etemadmoghadam D., Bowtell D., Barretina J., Lin W. M., Rameh L., Salmena L., Pandolfi P. P., Cantley L. C. (2009) Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 16, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Banfić H., Tang X., Batty I. H., Downes C. P., Chen C., Rittenhouse S. E. (1998) A novel integrin-activated pathway forms PKB/Akt-stimulatory phosphatidylinositol 3,4-bisphosphate via phosphatidylinositol 3-phosphate in platelets. J. Biol. Chem. 273, 13–16 [DOI] [PubMed] [Google Scholar]
- 19. Franke T. F., Kaplan D. R., Cantley L. C., Toker A. (1997) Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668 [DOI] [PubMed] [Google Scholar]
- 20. Gray A., Van Der Kaay J., Downes C. P. (1999) The pleckstrin homology domains of protein kinase B and GRP1 (general receptor for phosphoinositides-1) are sensitive and selective probes for the cellular detection of phosphatidylinositol 3,4-bisphosphate and/or phosphatidylinositol 3,4,5-trisphosphate in vivo. Biochem. J. 344, 929–936 [PMC free article] [PubMed] [Google Scholar]
- 21. Ma K., Cheung S. M., Marshall A. J., Duronio V. (2008) PI(3,4,5)P3 and PI(3,4)P2 levels correlate with PKB/akt phosphorylation at Thr308 and Ser473, respectively; PI(3,4)P2 levels determine PKB activity. Cell. Signal. 20, 684–694 [DOI] [PubMed] [Google Scholar]
- 22. Thomas C. C., Deak M., Alessi D. R., van Aalten D. M. (2002) High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 12, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 23. Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M., Yaffe M. B., Boussiotis V. A., Gertler F. B. (2004) Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7, 571–583 [DOI] [PubMed] [Google Scholar]
- 25. Dowler S., Currie R. A., Downes C. P., Alessi D. R. (1999) DAPP1: a dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem. J. 342, 7–12 [PMC free article] [PubMed] [Google Scholar]
- 26. Tiwari S., Choi H. P., Matsuzawa T., Pypaert M., MacMicking J. D. (2009) Targeting of the GTPase Irgm1 to the phagosomal membrane via PtdIns(3,4)P(2) and PtdIns(3,4,5)P(3) promotes immunity to mycobacteria. Nat. Immunol. 10, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith K., Humphreys D., Hume P. J., Koronakis V. (2010) Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin-pedestal formation. Cell Host Microbe 7, 13–24 [DOI] [PubMed] [Google Scholar]
- 28. Downes C. P., Leslie N. R., Batty I. H., van der Kaay J. (2007) Metabolic switching of PI3K-dependent lipid signals. Biochem. Soc. Trans. 35, 188–192 [DOI] [PubMed] [Google Scholar]
- 29. Catimel B., Schieber C., Condron M., Patsiouras H., Connolly L., Catimel J., Nice E. C., Burgess A. W., Holmes A. B. (2008) The PI(3,5)P2 and PI(4,5)P2 interactomes. J. Proteome Res. 7, 5295–5313 [DOI] [PubMed] [Google Scholar]
- 30. Catimel B., Yin M. X., Schieber C., Condron M., Patsiouras H., Catimel J., Robinson D. E., Wong L. S., Nice E. C., Holmes A. B., Burgess A. W. (2009) PI(3,4,5)P3 Interactome. J. Proteome Res. 8, 3712–3726 [DOI] [PubMed] [Google Scholar]
- 31. Krugmann S., Anderson K. E., Ridley S. H., Risso N., McGregor A., Coadwell J., Davidson K., Eguinoa A., Ellson C. D., Lipp P., Manifava M., Ktistakis N., Painter G., Thuring J. W., Cooper M. A., Lim Z. Y., Holmes A. B., Dove S. K., Michell R. H., Grewal A., Nazarian A., Erdjument-Bromage H., Tempst P., Stephens L. R., Hawkins P. T. (2002) Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol. Cell 9, 95–108 [DOI] [PubMed] [Google Scholar]
- 32. Pasquali C., Bertschy-Meier D., Chabert C., Curchod M. L., Arod C., Booth R., Mechtler K., Vilbois F., Xenarios I., Ferguson C. G., Prestwich G. D., Camps M., Rommel C. (2007) A chemical proteomics approach to phosphatidylinositol 3-kinase signaling in macrophages. Mol. Cell. Proteomics 6, 1829–1841 [DOI] [PubMed] [Google Scholar]
- 33. Shinohara M., Terada Y., Iwamatsu A., Shinohara A., Mochizuki N., Higuchi M., Gotoh Y., Ihara S., Nagata S., Itoh H., Fukui Y., Jessberger R. (2002) SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 416, 759–763 [DOI] [PubMed] [Google Scholar]
- 34. Kimber W. A., Trinkle-Mulcahy L., Cheung P. C., Deak M., Marsden L. J., Kieloch A., Watt S., Javier R. T., Gray A., Downes C. P., Lucocq J. M., Alessi D. R. (2002) Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem. J. 361, 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Batty I. H., Fleming I. N., Downes C. P. (2004) Muscarinic-receptor-mediated inhibition of insulin-like growth factor-1 receptor-stimulated phosphoinositide 3-kinase signalling in 1321N1 astrocytoma cells. Biochem. J. 379, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Batty I. H., Downes C. P. (1994) The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary, selective, consequences of inositol depletion in 1321N1 cells. Biochem. J. 297, 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 40. Hilpelä P., Oberbanscheidt P., Hahne P., Hund M., Kalhammer G., Small J. V., Bähler M. (2003) SWAP-70 identifies a transitional subset of actin filaments in motile cells. Mol. Biol. Cell 14, 3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Y., Toth M., Hamman M. S., Monahan S. J., Lodge P. A., Boynton A. L., Salgaller M. L. (2002) Serological cloning of PARIS-1: a new TBC domain-containing, immunogenic tumor antigen from a prostate cancer cell line. Biochem. Biophys. Res. Commun. 290, 830–838 [DOI] [PubMed] [Google Scholar]
- 42. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 43. Ong S. E., Foster L. J., Mann M. (2003) Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 [DOI] [PubMed] [Google Scholar]
- 44. Brown M. D., Sacks D. B. (2006) IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol. 16, 242–249 [DOI] [PubMed] [Google Scholar]
- 45. Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005) IQGAP1: a key regulator of adhesion and migration. J. Cell Sci. 118, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 46. Lemmon M. A., Ferguson K. M. (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 47. Narayan K., Lemmon M. A. (2006) Determining selectivity of phosphoinositide-binding domains. Methods 39, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 49. DiNitto J. P., Lambright D. G. (2006) Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta 1761, 850–867 [DOI] [PubMed] [Google Scholar]
- 50. Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 51. Isakoff S. J., Cardozo T., Andreev J., Li Z., Ferguson K. M., Abagyan R., Lemmon M. A., Aronheim A., Skolnik E. Y. (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17, 5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wakamatsu I., Ihara S., Fukui Y. (2006) Mutational analysis on the function of the SWAP-70 PH domain. Mol. Cell. Biochem. 293, 137–145 [DOI] [PubMed] [Google Scholar]
- 53. Campa F., Yoon H. Y., Ha V. L., Szentpetery Z., Balla T., Randazzo P. A. (2009) A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J. Biol. Chem. 284, 28069–28083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. (2002) ARAP1: a point of convergence for Arf and Rho signaling. Mol. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 55. Brandt D. T., Grosse R. (2007) Get to grips: steering local actin dynamics with IQGAPs. EMBO Rep. 8, 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. White C. D., Brown M. D., Sacks D. B. (2009) IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 583, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tirnauer J. S. (2004) A new cytoskeletal connection for APC: linked to actin through IQGAP. Dev. Cell 7, 778–780 [DOI] [PubMed] [Google Scholar]
- 58. Cain R. J., Ridley A. J. (2009) Phosphoinositide 3-kinases in cell migration. Biol. Cell 101, 13–29 [DOI] [PubMed] [Google Scholar]
- 59. Stephens L., Milne L., Hawkins P. (2008) Moving towards a better understanding of chemotaxis. Curr. Biol. 18, R485–494 [DOI] [PubMed] [Google Scholar]