Abstract
A high throughput protein biomarker discovery tool has been developed based on multiplexed proximity ligation assays in a homogeneous format in the sense of no washing steps. The platform consists of four 24-plex panels profiling 74 putative biomarkers with sub-pm sensitivity each consuming only 1 μl of human plasma sample. The system uses either matched monoclonal antibody pairs or the more readily available single batches of affinity purified polyclonal antibodies to generate the target specific reagents by covalently linking with unique nucleic acid sequences. These paired sequences are united by DNA ligation upon simultaneous target binding forming a PCR amplicon. Multiplex proximity ligation assays thereby converts multiple target analytes into real-time PCR amplicons that are individually quantified using microfluidic high capacity qPCR in nano liter volumes. The assay shows excellent specificity, even in multiplex, by its dual recognition feature, its proximity requirement, and most importantly by using unique sequence specific reporter fragments on both antibody-based probes. To illustrate the potential of this protein detection technology, a pilot biomarker research project was performed using biobanked plasma samples for the detection of colorectal cancer using a multivariate signature.
The quest for early detection of cancer leads us to retroactively mine biobanked plasma samples in hopes of finding better protein biomarkers and biomarker combinations (1–4). Such development of serological biomarkers find vast uses not only for disease detection but also patient monitoring and in general research applications. Thus, identification and employment of multiprotein signatures is a promising and natural step in modern molecular medicine. Novel technologies to enable such studies require several features such as multiplexing, low sample consumption, high sensitivity and of course good immunoassay performance in general. In particular, one needs to extract as much high quality putative biomarker data as possible from a single sample collection with good clinical background information without consuming the entire sample. Multiplexed immunoassays performed on planar arrays or beads are limited in multiplexing capacity by the inherent antibody cross-reactivity which leads to extensive effort devoted to assay development and optimization (5). As each detector antibody in these standard assays carry the same reporter fluorophore, any nonspecific binding of a detector antibody will give rise to false positive signals. In contrast, the proximity ligation assay (PLA)1 utilizes pairs of target specific antibodies linked with DNA strands forming so called proximity probe pairs, which upon simultaneous and pair wisebinding to their respective analyte in a homogeneous solution enables an enzymatic ligation reaction. Thereby, a new PCR amplicon is formed composed of both reporter sequences of the proximity probes (6, 7). This reporter molecule reflects the identity of the protein through sequence encoding in multiplex and its amount corresponds to the protein analyte concentration. Previous smaller scale reports have shown that the homogeneous proximity ligation assay is a promising tool for such studies (8–10). Here we report the further development in both throughput, multiplexing, and especially assess the immunoassay performance in greater detail. When building content in multiplexed immunoassays the availability of binding reagents for target proteins are often a limiting step. Because PLA can use both matched monoclonal antibodies or just a single batch of an affinity purified polyclonal antibody raised against the whole native antigen split in two aliquots, the potential repertoire of multiplex PLAs should be greater than for conventional multiplex assays. We show herein that multiplex PLA panels can be developed without the need for extensive antibody selection, optimization, and reselection. In this report we have used new antibody-oligonucleotide conjugation chemistry, previously not used for PLA, to build four 24-plex assays including spike-in standard controls, and validated their performance in human diseased and control plasma samples. Quantification of the PLA reaction products using high throughput nanoliter microfluidic real-time PCR enabled rapid putative biomarker profiling of 74 cases of colorectal cancer versus 74 matched control samples using all four panels.
EXPERIMENTAL PROCEDURES
Subjects
Subjects undergoing a sigmoidoscopy or colonoscopy either following symptoms consistent with colorectal cancer (CRC) or patients attending surveillance programs because of hereditary CRC (hereditary nonpolyposis colorectal cancer and familial adenomatous polyposis) were included in a cross sectional study. A total of 5165 subjects were included (11, 12) and according to the Helsinki II Declaration, oral and written consent was given from each subject. The study was approved by The Regional Ethical Committee (KF 01–080/03). A case-control study was designed for the present study by randomly choosing 74 biobanked stage I–IV (13) CRC samples and 74 age and gender matched individuals with no pathological findings by endoscopy and/or no self reported diseases or medication. Main clinical characteristics of the samples included in the study are presented in Table I.
Table I. Clinical characteristics for the 74 CRC patients included in the study. AJCC: American Joint Committee on Cancer (13).
| Characteristics | Subjects, n (%) |
|---|---|
| Gender | |
| Female | 36 (49) |
| Male | 38 (51) |
| Age group | |
| 40–49 | 3 (4) |
| 50–59 | 10 (14) |
| 60–69 | 17 (23) |
| 70–79 | 24 (32) |
| 80–89 | 19 (26) |
| 90–99 | 1 (1) |
| Cancer stage | |
| AJCC classification | |
| I | 8 (11) |
| II | 30 (40) |
| III | 16 (22) |
| IV | 15 (20) |
| Not specified | 5 (7) |
Samples
EDTA blood samples were collected from all subjects at time of endoscopy following standard operating procedure (11). The samples were centrifuged at 2500 × g for 10 min at 4 °C. Following centrifugation the plasma was aspirated, aliquoted and stored at −80 °C.
Antibodies and Oligonucleotide Sequences
In a literature search target assays were selected using criteria such as potential biomarkers for CRC, general cancer markers, availability of appropriate antibodies, and reported plasma levels.
A total of 80 antibody pairs against both high and low abundant proteins in plasma were selected for configuration of the multiplex panels 1–4 (see Table II). The few instances of antibody-oliognucleotide conjugation failure resulted in removal of these assays. In addition, antibodies against green fluorescent protein (GFP), phycoerythrin (PE), and allophycocyanin (APC) were included as internal control standards along with an oligonucleotide amplicon. The majority were affinity-purified polyclonal antibodies raised against the whole native recombinant proteins except for the monoclonal antibodies CEA, CA19–9, CA 15.3, CA242, and CA125. More detailed information about antibody and antigens sources can be found in supplemental Table S1.
Table II. Configuration of the four panels used for multiplex PLA biomarker screening.
| Panel | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Dilution | 1:1 | 1:1 | 1:10 | 1:100 |
| Sequence | ||||
| 1 | Carbonic Anhydrase IX | Survivin | Spondin-2/Midin | Clusterin |
| 2 | MCP-1 (CCL2) | Desmin | EPCAM | No assay |
| 3 | Mesothelin | HGF R/cMET | NSE (enolase 2) | sP-selectin |
| 4 | IL-6 | IL-17 | Transaldolase 1 | MMP-2 |
| 5 | No assay | IL-23 | No assay | MMP-9 |
| 6 | oit1/FAM3D | FoxP3 | Galectin-3 | Tenascin-C |
| 7 | CEACAM 5 pAb | CEACAM 5 pAb | TIMP1 | TIMP1 |
| 8 | VEGF | CEACAM1 | EGFR | CRP |
| 9 | No assay | TGFb1 | TFF3 | PAI-1/Serpin |
| 10 | No assay | TGFb3 | u-PA | sVCAM/CD106 |
| 11 | CEA mAb | KLK3 (PSA) | CEA mAb | IGF-I |
| 12 | CA125 | Cystatin SN | Transferrin Rec. (CD71) | Cathepsin B |
| 13 | Her4 | Prolactin | CA 15.3 | Angiogenin |
| 14 | HGF/SF | DcR3 | CA19–9 | IGF2 |
| 15 | FABP2 | IL8 | Tropomyosin (CH1) | Tetranectin |
| 16 | Oligo PCRa | Oligo PCRa | Oligo PCRa | Oligo PCRa |
| 17 | APCa | APCa | APCa | APCa |
| 32 | GFPa | GFPa | GFPa | GFPa |
| 35 | IL8 | CTGF | OPN | Cystatin C |
| 38 | PEa | PEa | PEa | PEa |
| 45 | Her2 | GDNF | MIF | IGFBP-1 |
| 51 | Her3 | Fractalkine | No assay | GRP78/HSPA5/BiP |
| 53 | TNFa | MIP-1 | SLPI | Kallikrein 11 |
| 57 | IL-1a | CA 242 | S100A8 | YKL-40 |
a Internal control used for normalization and reaction quality control.
The 40-mer 3′ and 5′ oligonucleotide sequences used for the antibody conjugation consisted of a central 20 bp connector specific sequence, denoted universal, and a flanking 20 bp sequence for primer targeting in PCR amplification and quantitative PCR (see Fig. 1, supplemental Table S2). To generate the variable primers and probe sequences, a selection from a library containing more than 2000 unique DNA sequences was evaluated by performing in silico testing to limit cross-hybridization and formation of hairpin loops using Zuker mFold (data not shown). A set of 62 sequences passed the in silico screen as suitable for conjugation from which 24 were used in this study. Connector oligonucleotide, the 24 oligonucelotide pair sequences, forward primers (PreAmp-F, qPCR-F) and reverse primers (PreAmp-R, qPCR-R) were obtained from Biomers(GmbH, Germany) (See supplemental Table S2 for more detailed information).
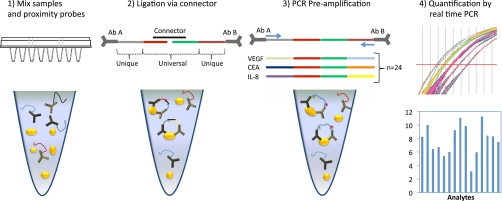
Fig. 1.
Schematic description of multiplex homogenous proximity ligation assay. (1) The samples are incubated with 23 pairs of proximity probes in 1 μl sample. (2) Upon proximal binding of the pairs of PLA probes to a common antigen the oligonucleotide sequences are ligated using a connector oligonucleotide. (3) The ligated products are amplified in multiplex using all pre-amplification primers simultaneously. (4) This is followed by quantification of each biomarker by quantitative real time PCR using only the unique primer pair for a given analyte.
Data Management and LIMS
In order to manage and conveniently share project data, a web-based laboratory information management system (LIMS), developed in Java language, and based on a relational database management system (MySQL), was developed (Integromics S.L., Madrid, Spain). The LIMS data model was designed within the EU-funded FP7 project PROACTIVE. The resulting LIMS consists of interconnected entities, and allows efficiently collecting, storing, and recalling the generated high throughput data, together with parameters characterizing the experiment, as well as relevant clinical information about the patient sample (supplemental Fig. S7).
Data visualization tools, implemented using the freely available Google Chart Tools API (Application Programming Interface), were used for primary quality controls (supplemental Fig. S8) and provided insight into patterns in high-dimensional data (supplemental Fig. S9).
Proximity Probe Preparation
Proximity probes were prepared by linking a single batch of affinity purified polyclonal antibody or matched monoclonal antibody pairs to 3′-hydroxyl free and 5′-phosphate free 40-mer oligonucleotide sequences thereby forming unique amplicons representing each target protein. The antibody-oligonucleotide conjugates included in the biomarker panels were generated by Innova Biosciences (Cambridge, UK) using their Lightning-LinkTM technology. Conjugation quality was analyzed by SDS-PAGE and a few failures resulted in four empty assays because of antibody formulations incompatible with conjugation.
Biomarker Panel Evaluation
Prior to evaluation of PLA panels, the proximity probes were pooled together with a probe mix diluent (Olink Bioscience, Uppsala, Sweden) at a final concentration of 2 nm and stored at 4 °C. The 24-plex assays for each panel were evaluated for performance with respect to sensitivity, dynamic range, linearity of dilution and recovery in plasma. Dilution series of antigen ranging from 2 pm to 200 pm in 1 μl samples were prepared in PLA buffer (Olink Bioscience, available upon request) and assayed in multiplex to assess sensitivity and dynamic range. The incubations of sample and Proximity-probes totaled 4 μl and was performed in a 96 well plate tightly sealed with a plastic film at 4 °C over night. For each dilution series a control sample without antigen was included to set the background level. Recovery values were determined by spiking 20 pm antigen in 90% plasma samples. Furthermore, antibody cross-reactive events were examined in buffer by comparing 24-plex antigen mixes to 4 or 5 antigen submixes and specificity was also assessed in a complex sample matrix containing 100% chicken plasma. A linearity of dilution experiment was performed to evaluate possible inhibitory factors present in plasma as well as to ensure that the analytes measured were within their linear range. Two fivefold dilutions were prepared in two plasma samples for each panel and the starting concentration of the plasma was 100% in panel 1 and 2, 10% in panel 3 and 1% in panel 4. For those assays where the signals were not reduced sufficiently by simple dilution, free antibody titrations were performed in order to set the assay within the linear range of its standard curve, that is below the inherent hook-effect of this homogeneous immunoassay (6, 7).
Biomarker Profiling—Multiplex Proximity Ligation Assay
Depending on the panel examined, the plasma concentration was 100% (Panel 1 and 2), 10% (Panel 3) or 1% (Panel 4) in the PLA analyses, a total of 148 plasma samples were analyzed.
Multiplex PLA was performed by mixing 1 μl sample with plasma dilution buffer (Olink Bioscience, available upon request) at a 1:1 ratio and an internal control standard spike-in mix of 200 pm GFP, 20 pm PE, 40 pm APC, and 40fM of a DNA sequence (oligo PCR) to asses reaction quality. The mixture was then incubated for 20 min at RT. Next, 2 μl of proximity probe mixture containing 1× Probe Mix Diluent (Olink Bioscience, available upon request), 1% bovine serum albumin and 0.1% Triton X-100 was added to 2 μl of each prediluted sample followed by an incubation at 4 °C over night to allow the probes to bind to the analytes. Next, ligation of the bound probes was performed by incubating 96 μl of reaction mixture containing 1× T4 ligation buffer supplemented with 100 nm connector oligonucleotide and 0.006 units of T4 DNA ligase (Fermentas, Burlington, Ontario) with 4 μl of probed samples for 10 min at 37 °C followed by heat inactivation at 65 °C for 10 min. To digest the connector oligonucleotide, 1 Unit of uracil-DNA excision mix (Epicenter Madison, WI, USA) was added and incubated at 37 °C for 20 min and heat inactivated at 70 °C for 10 min. Finally, pre-amplification was performed in a total volume of 25 μl by mixing 20 μl of ligated product with 5 μl pooled PCR mix (1× PCR buffer (Invitrogen), 15 mm MgCl2 (Invitrogen), 1 mm dNTP (Invitrogen), 0.2 μm of each forward and reverse Pre-AMP primer in 24-plex (See supplemental Table S2), 7.5 units Platinum Taq polymerase (Invitrogen)) for 17 cycles that included an initial incubation for 10 min at 95 °C followed by 2 cycles for 15 s at 95 °C, 10 min at 46 °C, 2 min at 60 °C, and 15 cycles for 15 s at 95 °C, 2 min at 54 °C for and 2 min at 60 °C. The products were finally diluted fivefold in 1× Tris-EDTA buffer prior to detection by real-time PCR.
Real Time qPCR
The qPCR reactions were performed either on an ABI 9700 HT Fast (Applied Biosystems) instrument or the BioMarkTM micro fluidic system from Fluidigm. Regardless of instrument used, each protein assay used separate qPCR reactions with individual primer pairs (Frw and Rew qPCR primer, see supplemental Table S2). For the 9700 HT Fast system the diluted DNA products were first incubated for 30 min at 37 °C in a mixture containing1.4 × Fast Universal Master Mix (Applied Biosystems), dH2O and 0.05 Units of uracil-DNA excision mix (Epicenter) for partial digestion of primers used in the pre-amplification step to reduce interference during the qPCR. The q-PCR reaction was performed on a 384-well plate by transferring 7 μl of the pre-amplified sample mix and 3 μl of 3 μm primer (Biomers), dH2O and 0.8 μm TaqMan probe (Applied Biosystems) to a total sample volume of 10 μl per well. The thermal cycler program was initiated with 5 min at 95 °C followed by for 45 cycles for 15 s at 95 °C and for 1 min at 60 °C.
The Fluidigm system required priming of the 48 × 48 Dynamic Chip array prior to use by injecting 300 μl control line fluid into each accumulator on the chip and run on an integrated fluidic circuit controller for 20 min performed according to manufacturers protocol. Next, an assay mix containing 1× assay loading reagent (Fluidigm, San Francisco, CA) and 2.5 μm of TaqMan probe (Applied Biosystems, Foster City, CA) was prepared and mixed with 9 μm of each primer pair. A sample master mix consisting of 1.4× sample loading reagent, 1.6× of Fast Universal Master Mix (Applied Biosystems) and 0.001 Units of uracil-DNA excision mix was prepared and mixed together with 1.8 μl diluted DNA products to a total volume of 6.5 μl. Thereafter, 5 μl of the assay mix and sample mix were added to their respective wells on the chip and run using the integrated fluidic circuit controller software for 60 min in order to load the samples and the assays into the chip. Each assay was run in duplicates and the micro fluidic compartment was loaded with 9 nL sample mix with 1 nL assay mix. Finally, the chip was run on the Biomark instrument running for 5 min at 95 °C followed by 40 cycles for 15 s at 95 °C and for 1 min at 60 °C.
Data Acquisition and Analysis
The qPCR data was analyzed with the RQ Manager 1.2 software (Applied Biosystems) or BioMark system software (Fluidigm). Thereafter, the recorded Ct values were converted to a linear scalefrom a log2-scale providing an estimation of the number of amplicons, X = 2^(30-Ct), subtracting the Ct value of the analyzed point from the threshold number for fluorescence detection theoretically assuming a single amplicon molecule. The data of each individual sample was normalized against its spike-in control GFP value before further statistical analysis. Recovery values were calculated by the formula [% Recovery = Amplicons (Spiked Ag)−Amplicons (Plasma)/Amplicons (Ag)−Amplicons (buffer)]. Sensitivity was estimated for all assays by the formula [Sensitivity = Concentration of spike-in closest to background (Molar)/2d̂Ct where dCt] is the signal over background level.
Enzyme-linked Immunosorbent Assay (ELISA)
For quality assessment and validation of the PLA technology to an orthogonal standard method the protein level for TIMP-1, CEA, CA242, IL-8, SLPI, and VEGF were determined by ELISA. TIMP-1 was measured by a rigorously validated in-house assay (14). The concentration of the other analytes was measured by commercially available ELISA kits; CEA (IBL, Hamburg, Germany), CA242 (Fujirebio Diagnostics, Gothenburg, Sweden), IL8, SLPI, and VEGF (R&D Systems, Minneapolis, MN) according to the manufacturers recommendations. Prior to ELISA measurements, the plasma samples were diluted in order to yield values within the linear range of the standard curve for each assay. The protein levels were determined using a PowerWave x1 microplate reader (Bio-Tek Instruments, Germany) measuring TIMP-1 and CA242 at 405 nm and CEA, IL-8, VEGF, and SLPI at 450 nm. All samples were determined in duplicate and the mean values were used for statistical analysis. Samples with intra-assay CV>10% were re-analyzed. A four-parameter-fitted standard curve was generated using KC4TM Software (Bio-Tek Instruments, Germany), by which the concentrations of the six biomarkers were calculated.
Statistical Tests
For univariate analysis serological markers levels were given in log-scale and used for further calculations. The distribution was analyzed for each biomarker by Quantile-Quantile plots (Q-Q) to verify normal distribution which is a prerequisite for the statistical analyses by t test. For the descriptive statistics median, minimum, maximum, actual mean, 95% confidence interval, and standard deviation were calculated. The statistical analyses were based on a case-control design and differences were compared using a paired t test. p values below 0.05 were considered significant. The correlation of PLA data to ELISA was calculated using standard procedure Spearman's rank correlation coefficient. All univariate data analyses were performed using the SAS (v 9.1, SAS Institute, USA) and GraphPad Prism (v 4.03 GraphPad Software, USA).
Multivariate analysis for the potential of detecting CRC was performed using random forest classifier design as implemented in the random Forest (15) package in R (16) and in-house scripts. As a crude filter to eliminate uninformative assays, prior to the analysis, assays with a variance of the raw Ct value not larger than either the variance of PE or GFP in the corresponding panel were removed. Binary classifiers of healthy versus CRC samples were then built using the remaining 54 assays using repeated hold-out of matched samples. In particular, 15 hold-out matched cases were selected at random and a random forest classifier was induced using the remaining 59 cases. Variable importance as measured by the mean decrease gini index (17) was computed and the 15 hold-out cases were used as an external test set, providing an unbiased error rate estimate. The random selection was repeated 15 times, each repetition producing an error rate estimate and estimates of variable importance. The expected error rate was then estimated as the mean of the error rate in the 15 repetitions. In addition, following the procedure introduced by Nadeau and Bengio (18), a conservative estimate of the variance of the estimated expected error rate was computed by first splitting the 74 cases into two partitions and then estimating the expected error rate as above in each partition. This was repeated 10 times, and the mean variance between the two partitions was used as a conservative estimate of the variance of the estimated expected error rate. Finally a conservative confidence interval for the expected error rate was determined under the assumption of normality using this variance estimate (18). The comparison between multivariate and univariate classification was produced by building and estimating the expected error rate, as described above, for random forest classifiers built using only a single assay.
RESULTS
Multiplex PLA Performance
The principle of homogenous multiplex proximity ligation assay has previously been described in detail (8) and is outlined in Fig. 1. To further the development of PLA as a high throughput biomarker screening technology, enhancements to the experimental procedure were made including oligonucleotide-antibody conjugation procedures, probe sequence design, primer configurations and pre-amplification strategy in order to accommodate microfluidic nano liter qPCR. The sequence design for the PLA probes included a universal center region for all assays allowing ligation of all 3′-probe sequences to the 5′-probe sequences templated by one and the same splint, followed by pre-amplification by PCR of all sequences in 24-plex using a mixture of 24 pre-amp primer pairs. The universal center region, which also contained a binding site for a common TaqMan probe, was flanked by unique PCR primer sites for each probe. The pre-amplification was essential to enable the use of a final low volume nano liter qPCR where the quantification of each specific analyte was performed using only one qPCR primer pair specific for each analyte, for example IL-8 Rew and IL-8 Frw. In this study we have extended the multiplex capabilities to generate data on 74 human protein biomarkers. Four multiplex panels were developed, each detecting 24 analytes, including three nonhuman internal control proteins and one DNA sequence standard (Table II). The spike-in references were used for normalizing the data and to assess reaction quality. These panels were evaluated for assay performance and validated in plasma samples of 74 patients with colorectal cancer and in a sample set of 74 age and gender matched healthy individuals.
The performance of each assay was studied by preparing dilution series ranging from 2 pm to 200 pm of spike-in antigens in 24-plex (Fig. 2). The results demonstrate that for 47 out of 54 (87%) assays, for which the recombinant antigen was available could detect protein levels below 2 pm (Fig. 2, supplemental Fig. S1). Furthermore, to ensure that the analytes in plasma samples were measured within their linear range, two fivefold dilutions of plasma samples were performed (Fig. 2). For most assays a good linearity was observed although some high abundant protein assays required transferring them to a panel with greater dilution. As expected, assay characteristics such as sensitivity varied among assays. However, sensitivity estimations from signal to noise ratios at 2 pm antigen concentration demonstrated sensitivities down to 26 fM (Fig. 3, Table III) implying the potential of multiplex PLA even when using simple single batches of affinity purified polyclonal antibodies to study low abundance proteins which are difficult to measure using traditional immunoassay technology. Four of the matched monoclonal antibody sets used(CA15.3, CA19–9, CA125, and CA242) are also available as sandwich Enzyme Immuno Assays (EIA), and in a sensitivity comparison between multiplexed PLA and EIA, PLA displayed a greater than 10-fold improvement for three of them while consuming still only 1 μl sample (supplemental Fig. S2).
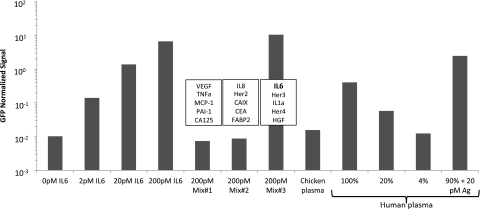
Fig. 2.
Evaluation of assay performance for 4 × 24-plex homogenous PLA. Exemplified by data for the detection of interleukin 6(IL-6) where the recombinant antigens are present at concentrations ranging from 2–200 pm in buffer. To illustrate specificity, assay evaluation was also performed in smaller antigen subset mixes selected from all antigens in a panel for which IL-6 was only present in mix #3. Assay specificity was also assessed in 100% chicken plasma. Linearity of the assay was further tested by performing two fivefold dilutions in PBS with 0.1% BSA from 100% to 4% plasma and recovery experiments calculated by spiking 20 pm pure antigen in plasma, data normalized against GFP.
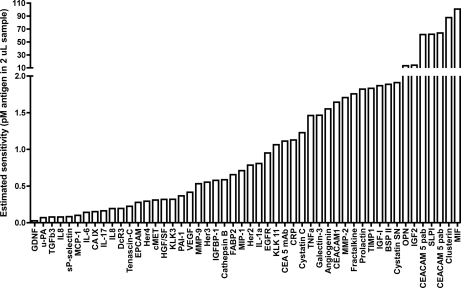
Fig. 3.
Estimated assay sensitivity by back-calculating from the signal over background at the point of the standard curve nearest background (0 pm) value for the 50 analytes included in panel 1–4 for which antigen was available. Values shown ranged from 26 fM up to 110 pm.
Table III. PLA performance.
| Panel | 1 | 2 | 3 | 4 | Total |
|---|---|---|---|---|---|
| Assay detects protein in healthy plasma (total) | 14 (17) | 14 (20) | 16 (18) | 16 (19) | 60 (74) |
| Number of assays with available antigen that lie within estimated sensitivity range belowa | |||||
| 10 fM - 100 fM | 1 | 2 | 1 | 1 | 5 |
| 100 fM - 1 pm | 10 | 6 | 2 | 5 | 23 |
| 1 pm - 10 pm | 3 | 4 | 2 | 6 | 15 |
| 10 pm - 100 pm | 1 | 1 | 3 | 2 | 7 |
| CV inter-sample variation of PE (n = 148 samples) | |||||
| Raw | 52% | 67% | 44% | 54% | |
| Normalized (GFP) | 18% | 18% | 10% | 11% | |
| Recovery of spiked-in PE detection (median) | |||||
| Raw | 20% | 24% | 70% | 114% | |
| Normalized (GFP) | 149% | 170% | 84% | 119% | |
a See Fig. 3 for details.
To investigate the specificity of the multiplexed assay, the antigens were measured in 24-plex assays, comparing signals and background for having either all available antigen present or having 4–5 antigens present in small subset mixes (Fig. 2, supplemental Fig. S1, supplemental Table S5). This type of analysis would reveal potential cross-reactivity of PLA-probes as wells as cross-reactivity of the DNA sequence reporter systems. In 71 out of 74 (96%) of the assays no cross-reactivity was observed implying high specificities of the assays. The unspecific signal observed for three assays was most likely because of contamination of the few antigens derived from crude nonrecombinant sources and only partially purified. These were cancer antigen standards from ELISA kits. Furthermore, in the presence of a mixture containing 100% chicken plasma no significant nonspecific events occurred when assayed with any of the 24-plex panels (supplemental Fig. S1). Specificity was further demonstrated by the capability of the method to discriminate between protein levels of prostate specific antigen (PSA) between male and female samples analyzed in the cohort of 148 plasma samples (supplemental Fig. S3).
A total of 60 assays out of 74 (81%) were able to detect its target protein in plasma when assayed in multiplex demonstrating the capability of the technology to target a wide range of both low and high abundant plasma proteins (Table III). When evaluating results from recovery experiments discrepancy between sample diluents and sample matrix was observed suggesting that some inhibitory factors affecting assay performance may be present, which was not observed upon plasma dilution. However, recovery evaluation could not be assessed for all the assays as some plasma levels exceeded our standard spike-in antigen concentration. Three nonhuman proteins spike-in references GFP, PE, and APC and a ligation independent oligonucleotide were used as reaction quality controls and data normalization. Assay precision was dramatically improved as assesed by inter-sample variability of PE signals (consistently spiked-in at the same concentration in all samples), from an overall average of 54.4% before normalization, to 14.3% following spike-in normalization by division with the GFP PLA-signal of the same reaction. This novel nonhuman protein spike-in feature, also illustrates the power of multiplexing to normalize data sets and thereby improve data quality. Greater spike variability across samples was seen with the nondiluted panels compared with the diluted ones as summarized in Table III. The precision of each individual assay was investigated by running quadruplicate PLAs for both background and two plasma samples, revealing an average coefficient of variation across all assays of 11%. See supplemental Table S3 for the individual data.
Profiling of Colorectal Cancer Plasma Proteins by Multiplex Proximity Ligation Assay
We analyzed human plasma samples from 74 patients with stage I–IV CRC and 74 healthy gender and age matched controls in order to validate the 24-plex PLA panels in biological samples. Each PLA reaction was analyzed using high throughput real time instrumentation only consuming nanoliters of reaction volume and the raw data was converted to linearized values and normalized to the internal standard protein GFP. We initially evaluated whether any of these tumor markers were significantly elevated or reduced in cancer patients compared with healthy controls. The comparison was made by performing paired t test (p < 0.05). Among the tumor markers with the greatest significance of differences were CEA, TIMP1, CA242 and CA-19.9, which are all well characterized biomarkers known to be highly expressed in colorectal cancer (supplemental Table S4 and supplemental Fig. S4) confirming previous published results (11, 12). Interestingly, the biomarker IL-8 was observed to be significantly elevated in CRC compared with healthy controls. Thus, our results also demonstrated the need for highly sensitive protein detection methods because IL-8 could hardly be detected by our ELISA (only 10 out of 148 samples) but was consistently detected by multiplex PLA (supplemental Fig. S5). This highlights the feasibility of homogenous multiplex PLA to discover potentially new proteins to be used as biomarkers in CRC.
PLA biomarker data was validated by correlation to standard ELISA on six proteins across the sample cohort and an overview of Spearman's rank correlation coefficients are summarized in Table IV. Correlation values for biomarker CEA mAb was 0.83 for panel 1 demonstrating good precision of the multiplex PLA technology. The poor intertechnology correlation observed for CEA pAb and SLPI is likely because of those PLAs having very weak signals in plasma and are thus not capable of detecting the antigen. Also, one should also take into account the use of different antibodies for all assays of the two technologies compared, except for CA242 which requires further investigation. Because of the low sensitivity of IL-8 ELISA the correlation to PLA was only based on the 10 samples that could be measured by ELISA (Table IV, supplemental Fig. S5).
Table IV. ELISA-PLA correlation.
| Marker (Panel no.) | Number of samples | Spearman r | 95% CI |
|---|---|---|---|
| CEA mAb (1) | 121 | 0.8187 | 0.7473 to 0.8713 |
| CEA pAb (1) * | 121 | 0.3397 | 0.1664 to 0.4927 |
| CEA pAb (2) * | 121 | 0.1743 | −0.009750 to 0.3469 |
| CEA mAb (3) | 121 | 0.7472 | 0.6532 to 0.8186 |
| TIMP-1 (3) | 146 | 0.6374 | 0.5262 to 0.7271 |
| TIMP-1 (4) | 146 | 0.527 | 0.3946 to 0.6380 |
| VEGF (1) | 148 | 0.4858 | 0.3478 to 0.6032 |
| CA242 (2) | 142 | 0.1751 | 0.005735 to 0.3347 |
| SLPI (3) * | 148 | −0.6195 | −0.7123 to −0.5054 |
| IL-8 (1) | 10 | 0.7939 | – |
| IL-8 (2) | 10 | 0.8061 | – |
Multivariate Modeling of Quantitative PCR Profiles
Random Forest classifiers were constructed by resampling using 59 cases for training and 15 for test (see Materials and Methods). The estimated expected error rate for random Forest classification of the present data set using 59 cases for design was 29% with the conservative 95% confidence interval (22%, 37%). Because the interval does not cover 50%, even at this early stage it is possible to rule out that the classification models are guessing (no signal picked up in the data). Furthermore, importance of individual assays in obtaining an informative classification was quantified by computing the mean gini decrease score in each of 15 random hold-outs of 15 cases. The Gini Index is a measure of impurity in a node in the decision tree, for two-class problems the score ranges from 0 (for a pure node having examples from only one class) to 0.5 (when half of the examples come from respective class). The decrease Gini Index is computed as the decrease in Gini Index when a variable is used to split the examples (weighted by the fraction of examples sent to each child node). These scores are computed for each variable in every tree the variable is used and averaged to produce the final measure of variable importance.
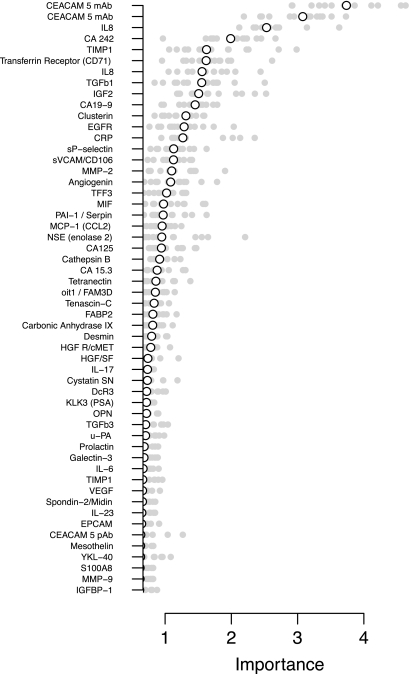
Fig. 4 shows the importance scores obtained, sorted by the median importance across different hold-outs. Among the 54 assays, CEA and IL-8 are clearly the most important according to this procedure, followed by CA242 and TIMP1. However, only CEA and IL-8 are clearly important in all random repetitions. It should be noted that, as expected, the two monoclonal CEA assays from different PLA-panels that are found to be important are also highly correlated, and tend to be mutually exclusive in the ensemble of decision trees that comprise the random forest. The finding that these well known CRC markers (12) as the most relevant, illustrates that the multiplex PLA procedure for biomarker research works and has the potential to also find more novel markers in the future as more assays are developed.
Fig. 4.
Variable importance for the 54 assays used in random forest classification in 74 CRC patients and 74 matched healthy controls. Importances in 15 individual random hold-outs are shown as dots and the median value as a circle.
In addition, to demonstrate the potential of multivariate detection of CRC in plasma samples, univariate classifiers (also in the form of random forests) were computed by repeated hold-out as above (supplementary Fig. S6). The best performance both in terms of false negative rate and false positive rate was obtained using all assays for classification. In the univariate analysis, CEA and TIMP1 are among the most important for univariate discrimination, whereas in the multivariate model TIMP1 is less important for discrimination. Conversely, an assay such as HGF/SF is found to be significantly different in the univariate analysis, but is shown to be irrelevant to classification using this method. This demonstrates that simply aggregating univariate markers does not discover the best multivariate signature.
DISCUSSION
The PLA principle has been implemented in several challenging protein detection applications such as in situ work providing localized detection of protein-protein interactions (19–21), pathogen detection (22), limited tissue lysate analysis (23), and protein-DNA interactions (24) among others (25). Reported herein, we have developed the multiplex proximity ligation assay into a high throughput technology capable of profiling 74 putative biomarkers consuming only 2 μl sample in total for all four panels with excellent sensitivity and verified high specificity. The assays are configured into four panels each containing 17–20 analytes along with four controls all evaluated for immunoassay performance. We also validated the technology in a biomarker research setting by assaying biobanked plasma from 74 colorectal cancer patients and 74 matched healthy controls. In comparison to previous multiplex PLA reports, we have increased the multiplexing level from 8 to 24, and made deeper assessments of assay performance especially of specificity using real-time PCR instrumentation of greater throughput. A new and improved method for oligonucleotide-antibody coupling was used based on the lightning-link technology, which can complete the conjugation in a one-tube reaction. No further downstream purification was necessary to separate free oligonucleotide from conjugate, thereby eliminating a large hurdle in assay development as previous reports used size-exclusion chromatography for each coupling reaction.
The new sequence design was optimized for pre-amplification and q-PCR. A mix of 24 primer-pairs containing uracil residues was used in pre-amplification and their partial uracil-N-glycosylase degradation removed potential carry-over to the q-PCR stage, while maintaining efficient pre-amplification (data not shown).
Data normalization by use of nonhuman internal reference control proteins is a new feature of PLA reported herein showing excellent ability to improve assay precision and reduce effects of enzymatic inhibition of the DNA ligase by the presence of human plasma. We used several internal controls to evaluate the use of a reference and finally used the GFP spike-in to normalize our putative biomarker data. We included the monoclonal CEA assay in two panels and found the results from the plasma sample studies mutually exclusive in the multivariate analysis indicating good assay precision and also high specificity as the assay reports the same data irrespective of which other proteins were simultaneously detected in this multiplexed procedure. Precision was further investigated for each individual assay in multiplex by performing four technical replicates of two different plasma samples, revealing a good average precision of the entire assay procedure of 11% coefficient of variation.
Current approaches for multiplex protein detection for biomarker research are hampered by slow assay development pace and difficulties in multiplexing because of increased antibody cross-reactivity with increased levels of multiplexing (5, 26–28). By successfully applying readily available affinity purified polyclonal antibodies as the target specific binding reagent and eliminating antibody cross-reactivity through the unique dual nucleic acid reporter system of PLA we show the potential of this technology for removing some of the major hurdles in high content immunoassay development. During the development of this work, we scaled the assay from 9 to 24-plex without loosing specificity and frequently move analytes in between the four panels to fit our daily analysis needs, illustrating a mix-and-match principle of multiplex assay configuration. Assay specificity was investigated by several procedures, one using recombinant antigen submixes of a few of the analytes in a panel and gaining only signals from expected amplicons, second by assessing detectability in human plasma and linearity of dilution, and third, no nonspecific signals observed in a very complex matrix such as chicken plasma. Such specificity investigations should be recommended for all multiplex immunoassay development. As an important note, no assay optimization at the immunoassay level, or antibody reselection, was performed for the sake of specificity improvement illustrating the scalability of multiplexed PLA. Here, the sensitivity of PLA has also been shown to possess the ability to reveal low abundance biomarkers of significant importance that are difficult to detect with conventional assays as illustrated by the IL-8 example showing both biomarker potential with PLA and difficulty to detect with ELISA. In order to quantify this large number of real-time PCR amplicons, we took advantage of recent developments in microfluidic nano-liter real time PCR by using the Biomark instrument with a capacity of generating up to 9216 data points in one run (96 samples × 96 assays). Although this nucleic acid quantification platform works well in combination with multiplexed PLA, we also envision other technologies useful, such as DNA-tag counting by next generation DNA sequencing and DNA microarray hybridization.
Multiplexed PLA was applied in this study on a selected number of plasma samples obtained from our large endoscopy study (12). We selected 74 samples representing CRC patients with various stages of the CRC disease and then designed a control group consisting of age- and gender-matched healthy individuals with no pathological findings by endoscopy, and no self-reported diseases or medication. Although several the analytes showed potential in their discrimination between CRC patients and the controls, we do not consider these experiments as sufficient to justify any conclusions regarding sensitivity and specificity as CRC detection markers, knowing that far more samples should be included before such conclusions can be drawn. However, with the successful validation of the multiplexed PLA technology, we are now initiating a large study including all the samples from our endoscopy study (12) and selected analytes. We envision that the final result will be an algorithm combining several analytes as for example, CEA and TIMP-1.
In conclusion, the ability of PLA to use single batches of polyclonal antibodies with sufficient specificity for detecting target proteins in plasma even in multiplex outlines a path forward to rapidly generating hundreds of sensitive immunoassays for large scale biomarker pursuits in biobanked human plasma samples.
Footnotes
* This work was funded by the EU FP-7under the PROACTIVE project and the Danish Endoscopy Study Group provided the plasma sample material.
 This article contains supplemental Figs. S1 to S9 and supplemental Tables S1 to S5.
This article contains supplemental Figs. S1 to S9 and supplemental Tables S1 to S5.
1 The abbreviations used are:
- PLA
- proximity ligation assay
- CRC
- colorectal cancer
- ELISA
- enzyme linked immunosorbent assay
- EIA
- enzyme immuno assay
- qPCR
- quantitative real-time polymerase chain reaction
- PE
- phycoerethyrin
- APC
- allophycocyanin
- GFP
- green fluorescent protein
- CEA
- carcinoembryonic antigen.
REFERENCES
- 1. Schrohl A. S., Würtz S., Kohn E., Banks R. E., Nielsen H. J., Sweep F. C., Brünner N. (2008) Banking of biological fluids for studies of disease-associated protein biomarkers. Mol. Cell Proteomics 7, 2061–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson L. (2005) Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J. Physiol. 563(Pt 1), 23–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Templin M. F., Stoll D., Schrenk M., Traub P. C., Vöhringer C. F., Joos T. O. (2002) Protein microarray technology. Trends Biotechnol. 20, 160–166 [DOI] [PubMed] [Google Scholar]
- 4. Polanski M., Anderson N. L. (2007) A list of candidate cancer biomarkers for targeted proteomics. Biomark. Insights 1, 1–48 [PMC free article] [PubMed] [Google Scholar]
- 5. Ellington A. A., Kullo I. J., Bailey K. R., Klee G. G. (2009) Antibody-based protein multiplex platforms: technical and operational challenges. Clin. Chem. 56(2), 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S. M., Ostman A., Landegren U. (2002) Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20, 473–477 [DOI] [PubMed] [Google Scholar]
- 7. Gullberg M., Gústafsdóttir S. M., Schallmeiner E., Jarvius J., Bjarnegård M., Betsholtz C., Landegren U., Fredriksson S. (2004) Cytokine detection by antibody-based proximity ligation. Proc. Natl. Acad. Sci. U.S.A. 101, 8420–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fredriksson S., Dixon W., Ji H., Koong A. C., Mindrinos M., Davis R. W. (2007) Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat Methods. 4, 327–329 [DOI] [PubMed] [Google Scholar]
- 9. Fredriksson S., Horecka J., Brustugun O. T., Schlingemann J., Koong A. C., Tibshirani R., Davis R. W. (2008) Multiplexed proximity ligation assays to profile putative plasma biomarkers relevant to pancreatic and ovarian cancer. Clin. Chem. 54(3), 582–589 [DOI] [PubMed] [Google Scholar]
- 10. Chang S. T., Zahn J. M., Horecka J., Kunz P. L., Ford. J. M., Fisher G. A., Le Q. T., Chang D. T., Ji H., Koong A. C. (2009) Identification of a biomarker panel using a multiplex proximity ligation assay improves accuracy of pancreatic cancer diagnosis. J. Transl. Med. 7, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nielsen H. J., Brünner N., Frederiksen C., Lomholt A. F., King D., Jørgensen L. N., Olsen J., Rahr H. B., Thygesen K., Hoyer U., Laurberg S., Christensen I. J. (2008) Plasma tissue inhibitor of metalloproteinases-1 (TIMP-1): A novel biological marker in the detection of primary colorectal cancer. Protocol outlines of the Danish-Australian endoscopy study group on colorectal cancer detection. Scand J Gastroenterol. 43, 242–248 [DOI] [PubMed] [Google Scholar]
- 12. Nielsen H. J., Brünner N., Jørgensen L. N., Olsen J., Rahr H. B., Thygesen K., Hoyer U., Laurberg S., Stieber P., Blankenstein M. A., Davis G., Dowell B. L., Christensen I. J. (2010) Plasma TIMP-1 and CEA in detection of primary colorectal cancer: Aprospective, population based study of 4,509 high risk individuals. Scand. J. Gastroenterol. 46(1), 60–69 [DOI] [PubMed] [Google Scholar]
- 13. O'Connell J. B., Maggard M. A., Ko C. Y. (2004) Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J. Natl. Cancer Inst. 96(19), 1420–1425 [DOI] [PubMed] [Google Scholar]
- 14. Holten-Andersen M. N., Murphy G., Nielsen H. J., Pedersen A. N., Christensen I. J., Høyer-Hansen G., Brünner N., Stephens R. W. (1999) Quantitation of TIMP-1 in plasma of healthy blood donors and patients with advanced cancer. Br. J. Cancer 80, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liaw A., Wiener M. (2002) Classification and regression by random forest. R. News 2(3), 18–22 [Google Scholar]
- 16. R Development Core Team R: A language and environment forstatistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2009. ISBN 3–900051-07–0, URL http://www.R-project.org [Google Scholar]
- 17. Calle M. L., Urrea V. (2010) Letter to the Editor: Stability of random forest importance measure. Briefings Bioinformatics 7–10, 12(1), 86–90 [DOI] [PubMed] [Google Scholar]
- 18. Nadeau C., Bengio Y. (2003) Inference for the generalization error. Machine Learning 52, 239–281 [Google Scholar]
- 19. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3(12), 995–1000 [DOI] [PubMed] [Google Scholar]
- 20. Jarvius M., Paulsson J., Weibrecht I., Leuchowius K. J., Andersson A. C., Wählby C., Gullberg M., Botling J., Sjöblom T., Markova B., Ostman A., Landegren U., Söderberg O. (2007) In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol. Cell Proteomics 6(9), 1500–1509 [DOI] [PubMed] [Google Scholar]
- 21. Leuchowius K. J., Jarvius M., Wickström M., Rickardson L., Landegren U., Larsson R., Söderberg O., Fryknäs M., Jarvius J. (2010) High content screening for inhibitors of protein interactions and post-translational modifications in primary cells by proximity ligation. Mol. Cell Proteomics 9(1), 178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gustafsdottir S. M., Nordengrahn A., Fredriksson S., Wallgren P., Rivera E., Schallmeiner E., Merza M., Landegren U. (2006) Detection of individual microbial pathogens by proximity ligation. Clin. Chem. 52, 1152–1160 [DOI] [PubMed] [Google Scholar]
- 23. Bjarnegard M., Enge M., Norlin J., Gustafsdottir S., Fredriksson S., Abrahamsson A., Takemoto M., Gustafsson E., Fässler R., Betsholz C. (2004) Endothelium-specific ablation of PDGFB leads to pericyte loss and glomerular, cardiac and placental abnormalities, Development 131, 1847. [DOI] [PubMed] [Google Scholar]
- 24. Gustafsdottir S. M., Schlingemann J., Rada-Iglesias A., Schallmeiner E., Kamali-Moghaddam M., Wadelius C., Landegren U. (2007) In vitro analysis of DNA-protein interactions by proximity ligation. Proc. Natl. Acad. Sci. U.S.A. 104, 3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swartzman E., Shannon M., Lieu P., Chen S. M., Mooney C., Wei E., Kuykendall J., Tan R., Settineri T., Egry L., Ruff D. (2010) Expanding applications of protein analysis using proximity ligation and qPCR. Methods 50(4), S23–26 [DOI] [PubMed] [Google Scholar]
- 26. de Jager W., Rijkers G. T. (2006) Solid-phase and bead-based cytokine immunoassay: a comparison. Methods 38(4), 294–303 [DOI] [PubMed] [Google Scholar]
- 27. Elshal M. F., McCoy J. P. (2006) Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 38(4), 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zichi D., Eaton B., Singer B., Gold L. (2008) Proteomics and diagnostics: Let's get specific, again. Current Opinion Chem. Biol. 12(1), 78–85 [DOI] [PubMed] [Google Scholar]