Abstract
Lysate microarrays (reverse-phase protein arrays) hold great promise as a tool for systems-level investigations of signaling and multiplexed analyses of disease biomarkers. To date, however, widespread use of this technology has been limited by questions concerning data quality and the specificity of detection reagents. To address these concerns, we developed a strategy to identify high-quality reagents for use with lysate microarrays. In total, we tested 383 antibodies for their ability to quantify changes in protein abundance or modification in 20 biological contexts across 17 cell lines. Antibodies yielding significant differences in signal were further evaluated by immunoblotting and 82 passed our rigorous criteria. The large-scale data set from our screen revealed that cell fate decisions are encoded not just by the identities of proteins that are activated, but by differences in their signaling dynamics as well. Overall, our list of validated antibodies and associated protocols establish lysate microarrays as a robust tool for systems biology.
One of the primary goals of systems biology is to uncover and model the complex relationships between proteins in living cells and organisms. Data-driven approaches to addressing this problem require ways to obtain quantitative information on protein abundance and post-translational modifications (PTMs)1 in a systematic and high-throughput fashion. Several different immunoaffinity-based methods have been used in systems-level studies to determine the amounts, subcellular locations, and PTM levels of proteins in complex biological samples. Antibody-based technologies that are compatible with multiplexing include flow cytometry (1), microsphere-based assays (2, 3), immunocytochemistry coupled with automated microscopy (4), miniaturized Western blotting (5), and antibody microarray-based methods such as direct-detection microarrays (6, 7), sandwich-style microarrays (8–11), and “reverse-phase” or lysate microarrays (12, 13). Compared with their low-throughput counterparts, high-throughput technologies are often constrained by smaller sample sizes, lack of separation steps, and an inability to tailor the assay to each antibody. To ensure uniformly high data quality across a large number of analytes and biological samples, careful characterization of each antibody is critical. Studies in which the quantitative data are used to train computational models impose an even higher standard.
Among high-throughput approaches, lysate microarray technology is particularly well suited for systems-level investigations. Thousands of biological specimens can be arrayed onto hundreds of membrane-coated slides, each of which can be queried with a different detection antibody. This format allows dense sampling of information at a protein level and in a high-throughput fashion. Although several groups have used this technology to study biological systems (12, 14) and although standardized protocols have been published (15), lysate microarrays have not yet gained wide-spread adoption, largely owing to questions regarding data quality and the limited availability of highly validated detection antibodies. Previous studies have recognized the need for rigorous antibody characterization and have used quantitative immunoblotting (Western blotting) to validate large collections of antibodies (13, 16). These studies showed that the reactivity of antibodies on lysate microarrays differs from that on traditional immunoblots, even when the same antibodies and lysates are used under otherwise identical conditions. In our own work (13), which focused on a single cell line, we started with a set of 61 commercial antibodies and found that only 12 of them yielded data on lysate microarrays that matched those collected by quantitative Western blotting. Whereas our approach was successful at discovering functional detection antibodies, it was time-intensive and not easily scaled. It also resulted in a discouragingly small number of antibodies that were validated for use with a single cell line. This highlighted a need to develop a much more efficient strategy to identify suitable antibodies that could be used across a broad range of cell types.
Here, we present a novel and efficient way to systematically identify and validate detection antibodies for use with lysate microarrays (see Fig. 1A). A set of relevant candidate antibodies is first chosen within a broad biological area of interest. These antibodies are then screened against a wide variety of “biological contexts” using lysate microarrays. Each context represents a specific combination of cellular background and treatment conditions. Based on the statistical significance of the resulting measurements, promising antibody-context pairs are further evaluated by quantitative Western blotting. If the two data sets agree, the antibody is considered validated for use with that cellular background. Using this strategy, we screened 383 commercial antibodies and successfully validated 82 of them in one or more biological context. This list of antibodies and the associated protocols represents a valuable resource to the scientific community and should facilitate more widespread use of this technology. Although this study focused on characterizing antibodies for lysate microarrays, our overall strategy is general and can be applied to other high-throughput immunoaffinity assays as well.
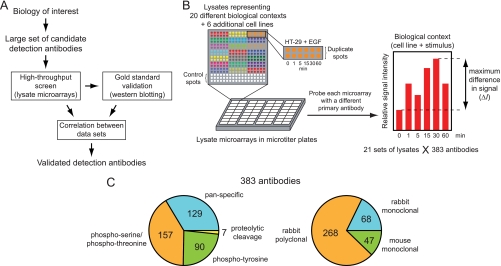
Fig. 1.
Design of a novel screen for functional detection antibodies for lysate microarrays. A, General strategy for identifying detection antibodies, combining a primary high-throughput screen of candidate detection reagents with a secondary gold-standard assay. Validation of antibodies depends on concordance between the measurements from both techniques. B, Schematic of microarray screen. Lysates from many different biological contexts are arrayed onto nitrocellulose pads, assembled into microtiter plates, and probed with each of a large collection of primary antibodies. Microarray “hits” are assigned based on the statistical significance of the maximum observed signal spread, ΔI, across each lysate set. C, Breakdown of antibodies included in the screen by target epitope class and host species.
EXPERIMENTAL PROCEDURES
Cell Culture
HMEC cells were cultured in HuMEC basal serum-free medium (Invitrogen) supplemented with HuMEC supplement kit (Invitrogen, Carlsbad, CA), 100 I.U./ml penicillin and 100 μg/ml streptomycin (Mediatech, Herndon, VA), and 1 ng/ml cholera toxin (Sigma-Aldrich, St. Louis, MO). Jurkat cells were cultured in RPMI 1640 (Mediatech) supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT), 2 mm glutamine (Mediatech), 100 I.U./ml penicillin, and 100 μg/ml streptomycin. HT-29 cells were cultured in McCoy's 5A Medium (ATCC, Manassas, VA) supplemented with 10% FBS, 100 I.U./ml penicillin, and 100 μg/ml streptomycin. All other cell lines were cultured in Dulbecco's modification of Eagle's medium (Mediatech) supplemented with 10% FBS, 2 mm glutamine, 100 I.U./ml penicillin, and 100 μg/ml streptomycin. Medium for FlpIn-293 cell lines additionally contained 150 μg/ml hygromycin B. To generate lysates, cells were serum-starved for 24 h, stimulated with cytokine or small molecule for the prescribed period of time, washed with ice-cold phosphate-buffered saline (PBS), and lysed in 2% SDS buffer. Cell lysates were cleared by filtration through 0.2 μm filter plates (Pall Corporation, East Hills, NY) and stored at −80 °C. Lysate concentrations were determined using the Micro BCA assay kit (Pierce Biotechnology, Rockford, IL) and lysates from each time course treatment were diluted to the same concentration in 2% SDS buffer. To ensure complete protein denaturation, all lysates were then boiled for 5 min at 95 °C prior to microarraying or Western blotting. Detailed information about all cell lysates generated in this study can be found in supplemental Table S1.
Microarray Fabrication
Custom lysate microarrays were printed by Aushon Biosystems (Billerica, MA) on 16-pad nitrocellulose-coated glass slides (Grace Bio-Labs, Bend, OR). Lysates were arrayed at 250 μm spacing using solid 110 μm pins, which resulted in an average feature diameter of 180 μm when visualizing spot protein content (data not shown). Lysates from time course treatments and unstimulated cell lines were arrayed in technical duplicates. In addition, each array contained six, eight-point, twofold serial dilutions of control lysates (supplemental Experimental Procedures), as well as six spots containing lysis buffer only, for a total of 306 microarray spots per nitrocellulose pad. Following microarray printing, slides were stored dry, in the dark, and at room temperature until further processing.
Microarray Probing
To remove the buffer and detergent contained in each microarray spot, slides were washed three times for 5 min each with 1× PBS/0.1% Tween-20 (PBST), incubated in Tris/HCl (pH 9) for 24 h, washed again with PBST, and centrifuged dry. Silicon gaskets and bottomless 16-well plates (Grace Bio-Labs) were then attached and slides were blocked with 5% BSA/PBST for 1 h at 4 °C. Microarrays were incubated in a mixture of 1:1000 anti-β-actin antibody and 1:500 pan- or phosphospecific antibody in 5% BSA/PBST at 4 °C for 24 h. Following washing, slides were incubated in a mixture of 1:1000 anti-rabbit-680 and 1:1000 anti-mouse-800 antibodies (17) in 5% BSA/PBST for 24 h at 4 °C. Silicon gaskets and bottomless plates were removed, slides were washed again, and centrifuged dry. Microarrays were scanned in the 680 nm and 800 nm channels using an Odyssey imager (LI-COR, Lincoln, NE) at 21-μm resolution. Each slide was scanned at a range of scanner sensitivities to account for the large differences in signal intensity between the 16 antibodies tested on each slide.
Analysis of Microarray Data
All data processing and analysis steps were carried out using custom-built code for Matlab® 7.4 (The Mathworks, Natick, MA). We first corrected microarray data for nonlinearity using antibody-specific calibration curves, as described previously (13). Signal intensities within each microarray were then mean-normalized to enable statistical comparisons across different arrays. Signal intensities from target proteins were subsequently normalized using the β-actin signal intensities from the same microarray spots to account for any differences in lysate concentration or spotting. Lastly, data from duplicate spots were averaged. Data from each antibody and biological context were organized into vectors in two separate ways. For the 20 time course treatments, each data vector consisted of six data points, corresponding to the six different time points of treatment. For comparisons across cell lines, each vector consisted of 17 data points, corresponding to the 17 unstimulated cell lines. We calculated two measures of signal up-regulation for each vector: (1) signal difference (ΔI) was defined as the difference between the highest and lowest signal intensity; and (2) fold-up-regulation was calculated as the ratio of the highest and lowest signal intensities in each data vector.
To derive a statistical threshold for significant signal difference, ΔIthreshold, histograms of differences in signal intensity were prepared for either biological or analytical replicates in our data set. For biological noise, we made use of the fact that, in several instances, a given cell line was subjected to more than one stimulation condition, but separate “0-min” samples were prepared in each case (supplemental Table S1). For example, we collected time courses of HeLa cells treated with anisomycin, EGF, insulin, and TNFα, but the 0-min time points remained untreated in all four sets of lysates. Taking into account all pairwise combinations within each cell line, our data set contained a total of 16 sets of biological duplicates. For analytical replicate noise, we used data from all duplicate microarray spots. We then plotted the distribution of differences in signal intensity between duplicates, and fitted these data to an exponential distribution. ΔIthreshold was calculated by solving the cumulative distribution function of this exponential for the value 1- αsingle, where αsingle is the significance level for individual comparisons and relates to the significance level for multiple comparison, αmultiple, according to the following relationship: αmultiple = 1 − (1 − αsingle) (Dunn-Ŝidák correction; n = 6 for “time courses” data set; n = 17 for “cell lines” data set). Hits within the time courses data set were defined as those vectors exceeding ΔIthreshold at αmultiple = 0.01 and showing greater than a 1.5-fold change in signal. Because ΔIthreshold does not capture the systematic variation in signal across cell lines, hits within the cell lines data set were defined as the 50 top-scoring (highest ΔI) non-PTM-specific antibodies.
Self-organizing Map Analysis (SOM)
The SOM analysis was performed in Matlab® using the SOM Toolbox (18). The parameters for the SOM analysis were as follows: topology of the map was chosen to be sheet, distance metric was cosine correlation, and the number of map units was chosen to be 66. We used the batch learning algorithm, and the neighborhood function was chosen to be Gaussian with the parameters given by Vesanto et al. (18). We used the U-matrix method to identify a group of map units that represent a cluster (19). For each cluster, we computed statistical significance using a permutation test method (20). First, we computed correlation distances for all combinations of time courses in a cluster. If the two profiles correlated perfectly, their distance was assigned to be zero, whereas perfect negative correlation resulted in the distance value of two. We then computed the mean of these pairwise comparisons. This procedure was followed by choosing an equal number of time courses randomly from the entire data set and computing pairwise correlation distances of all combinations. We repeated this process 5000 times and calculated a p value by counting the number of times a randomly chosen cluster produced a mean distance less than or equal to the mean distance of the original cluster, and dividing this number by 5000. Large p values suggest the original cluster may have arisen simply by chance.
RESULTS
Design of a High-throughput Screen for Functional Detection Antibodies
An antibody can be used with lysate microarrays if it meets two criteria: (1) it produces a significant difference in signal across the samples of interest, and (2) these differences correspond to changes in the levels of the target antigen. We reasoned that an antibody is likely to satisfy these criteria in some biological contexts, but not in others, as its antigen may be abundant in some cells or tissues, but not in others, or its levels may remain unchanged across the available samples in a given experiment. We therefore assumed that antibody validation efforts would ultimately be context-specific, but that some antibodies might perform well in many different settings.
To enable rapid and context-dependent assessment of antibody performance, we designed the following high-throughput screen using lysate microarrays (Fig. 1B). Lysates from many different “biological contexts” are microarrayed onto glass-supported nitrocellulose pads and the resulting arrays are assembled into a microtiter plate format (one array per well). Each biological context constitutes a set of related lysates in which a single cell line has been treated for different lengths of time with a molecular stimulus (growth factor or pharmacological agent). Each well is probed with a candidate detection antibody, chosen only on the basis of vendor-supplied information. Following incubating the arrays with an appropriate dye-labeled secondary antibody, the arrays are scanned for fluorescence and the intensities of the microarray spots are quantified. For each combination of detection antibody and biological context, the maximum difference in signal between individual lysates is calculated and this metric is used to separate “hit” from “nonhit” antibodies.
To test antibodies for their ability to detect dynamic changes in antigen levels, we started by generating lysates from 20 different biological contexts of interest, each consisting of unstimulated cells (serum-starved) and cells stimulated for five different lengths of time with either a growth factor or a small molecule. We focused on cell lines and treatment conditions that are commonly used in system-level studies of signal transduction, that are easily reproduced, and that span the broad cellular processes of growth, migration, stress response, and apoptosis. In addition to five commercially available cell lines, our set included six isogenic lines derived from HEK 293 cells that each express a different receptor tyrosine kinase (RTK) (21). These cell lines were included to assess the ability of antibodies to capture differential activation of the same signaling proteins within the same genetic background. Altogether, these 20 biological contexts included 11 distinct cell lines and provided us with the “time courses” data set (see below). To identify antibodies that detect variations in protein abundance across different cell lines, we included lysates from six additional, untreated lines. Together with the untreated samples from the first 11 cell lines, this set of lysates provided us with the “cell lines” data set (see below). All 21 sets of lysates (representing 126 independent samples) were printed as technical duplicates on glass-supported nitrocellulose pads and assembled into a microtiter plate format. Additional control spots (lysis buffer and dilution series of selected lysates) were included in each microarray to ensure data quality and to enable data processing (see Experimental Procedures). Detailed information about all lysate sets used in this study can be found in supplemental Table S1 available online.
To maximize the likelihood of identifying functional antibodies, we focused on antibodies that recognize proteins involved in the cellular processes induced by our treatment conditions: cell growth, proliferation, stress response, and apoptosis. We also used our prior knowledge of network connectivity to refine our choice of antibodies to screen. For example, as several sets of lysates were derived from cells treated with RTK ligands, we included antibodies that report on the activation of proteins in the canonical Ras/MAPK, PI3K/Akt, PLCγ, and STAT signaling pathways (22). In total, 383 commercially available antibodies were obtained for this study (Fig. 1C), 254 of which are PTM-specific and 129 of which recognize both modified and unmodified proteins (“pan-specific”). Among the PTM-specific antibodies, 90 recognize sites of tyrosine phosphorylation (single or multiple), 157 recognize single or multiple phosphorylation sites that include at least one serine or threonine residue, and 7 detect proteolytic cleavage events. To test if antibody performance depends on the host of origin, we included mono- and polyclonal antibodies derived from rabbits, as well as monoclonal antibodies derived from mice. A complete list of all the antibodies used in this study can be found in supplemental Table S2.
Lysate Microarray Screen Provides a Quantitative Measure of Antibody Performance
To assess antibody performance, we probed our lysate microarrays in single wells of microtiter plates with each of the 383 primary antibodies using a single, standardized set of conditions that had previously been optimized (Experimental Procedures). Although it is possible that some antibodies requiring specialized conditions would be missed using this approach, we expect this to be rare as we have not yet encountered any antibodies that yield high quality data under specialized conditions but fail under the general conditions of our optimized protocol.
To correct for variation in lysate concentration or microarray spotting, we pooled each antibody with an anti-β-actin antibody derived from a different host species. This provided a way to measure the amount of lysate deposited in each spot. For signal detection, we incubated the microarrays with a mixture of two infrared dye-labeled secondary antibodies (17) and scanned the slides in both fluorescent channels (Fig. 2A). This detection strategy substantially reduces assay nonlinearity that is often introduced by methods that rely on enzyme-driven signal amplification. Our screening approach is both economical and scalable: up to 100 antibodies can be tested in parallel, using only 0.2 μl of a 1 mg/ml stock solution to probe each microarray (100 μl volume).
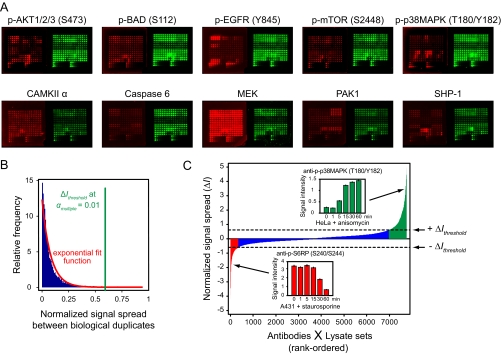
Fig. 2.
Identification of microarray hits based on observed assay variability. A, Sample of ten representative lysate microarrays generated in our screen. Each array was probed with a different primary antibody (red arrays), pooled with a β-actin-detecting antibody of a different species (green arrays) to normalize for protein concentration. Top row: five antibodies detecting post-translational modifications. Bottom row: five antibodies detecting total levels of target proteins (pan-specific antibodies). B, Distribution of biological noise in the screen. The vertical green line marks the threshold for statistical significance of signal spread at a significance level of α = 0.01, corrected for multiple testing. C, Plot of rank-ordered, normalized signal spread, ΔI, for all antibody-context pairs. Positive values indicate primarily up-regulation of antigen levels and negative values indicate primarily down-regulation. Red: significant down-regulation of signal. Blue: statistically insignificant change in signal. Green: significant up-regulation of signal. Insets show two examples of signal up- and down-regulation across time course treatments.
Over 200,000 spot intensities were extracted from the microarray images and all subsequent data processing and analysis steps were carried out in an automated fashion using custom-built code (Experimental Procedures). We corrected the data for nonlinearity using antibody-specific calibration curves derived from serial dilutions of control lysates, and normalized all signals relative to their respective β-actin signal intensities (internal standard). To enable statistical comparisons across different arrays, we divided all signal intensities by the mean intensity of each microarray. Finally, we averaged biological duplicates. To capture the performance of each of the 383 antibodies in each of the 21 biological contexts (lysate sets), we organized the microarray data into vectors that contain signal intensities from either the six time points (time courses data set) or 17 cell lines (cell lines data set) for each antibody and lysate set (supplemental Table S3). Our data thus encompass 383 × 21 = 8043 vectors of either 6 or 17 elements. Each vector represents a different antibody-context pair and hence must be evaluated separately for antibody performance.
We previously showed that the signal ratio between two samples as measured by lysate microarrays is often smaller than the actual ratio of antigen levels between the two samples (13). This is because the lysate microarray signal comprises an antigen-specific component and a component arising from antibody cross-reactivity. If the component arising from cross-reactivity dominates the overall signal, the antigen-specific component is lost in the noise of the assay. Thus, as a first step in validating an antibody-context pair, we first determined the difference, ΔI, between the highest and lowest signal intensities within each data vector, and used ΔI as a metric to separate hit antibody-context pairs from nonhit pairs. Because the microarray data are subject to variation arising from both analytical and biological noise, we expected to observe nonzero values of ΔI for essentially all data vectors. In addition, we reasoned that microarray signals from different cell lines might be subject to systematic variation, as the degree of antibody cross-reactivity likely differs across cell lines. Indeed, we found that over 80% of vectors (6494/8043) exhibited a ΔI that was >10% of the average microarray intensity, and over 97% of vectors (7831/8043) exhibited a ΔI of >1%. In the following analyses, time courses data and cell lines data are treated separately. We will start by focusing on the time courses data.
Identification of Antibodies That Report on Time-dependent Changes in Antigen Levels
To separate vectors with significant changes in signal from those for which the nonzero value of ΔI can be accounted for by analytical or biological noise, we analyzed the overall distribution of noise inherent in our measurements to derive a threshold for statistical significance. Because different samples within each vector are biologically distinct (having been prepared in separate tissue culture vessels), any random variation between them must reflect both biological and analytical noise. We therefore prepared a histogram that shows the distribution in signal spread between biological duplicates contained within our time courses data set (see Experimental Procedures). As each microarray was mean-normalized, we were able to compare signal intensities across different antibodies. The resulting histogram of total assay noise was then fit to an exponential distribution (Fig. 2B).
To focus our antibody validation efforts on the strongest hits, and to minimize the number of follow-up experiments performed on false-positive hits, we chose a stringent significance level of α = 0.01. For the six-element vectors of the time courses data set, this corresponds to a statistical cutoff of ΔIthreshold = 0.593. In other words, a vector with a signal difference >59.3% of the average microarray intensity was considered a hit with p < 0.01. Using this statistical criterion, we identified 1084 microarray hits (antibody-context pairs). Upon closer inspection, we noticed that, in a small number of cases, all of the elements in a vector had very high signal intensities, even though they displayed only a small relative change in signal. These vectors exceeded our statistical threshold but are not likely to reflect biologically meaningful changes in antigen levels. We therefore removed from our set of microarray hits 59 vectors that showed <1.5-fold change in signal intensity. Altogether, 1025 out of 7660 vectors (13%) passed our stringent selection process.
Importantly, if we had neglected biological noise and identified microarray hits based solely on analytical assay noise, our statistical cutoff would have been set at ΔIthreshold = 0.195 and the majority of vectors in our data set would have scored as hits (4103 out of 7660 or 54%). Thus, both analytical and biological variability must be accounted for in order to obtain a meaningful statistical threshold to evaluate antibody performance.
To visualize the results of our microarray screen, we rank-ordered all of the vectors by their ΔI values (Fig. 2C). We then classified vectors into those showing primarily up-regulation of antigen levels and those showing primarily down-regulation. If the maximum log2 fold-increase in signal relative to the “0 min” time point exceeded the maximum log2 fold-decrease in signal, ΔI was assigned a positive sign (up-regulation); otherwise, it was assigned a negative sign (down-regulation). Overall, we observed more vectors with positive ΔI than negative ΔI. Upon closer inspection, we noticed that the overwhelming majority of vectors at both extremes of the distribution represent PTM events (primarily phosphorylation). The skewed shape of our distribution reflects the fact that most of the treatments used in our study induce protein phosphorylation, and only a few (such as staurosporine) lower phosphorylation levels.
Our analysis of the time courses data set revealed numerous microarray hits in all cell lines and stimulation conditions that we tested, with between 23 and 93 hits per lysate set (biological context). For a given antibody, between 0 and 18 lysate sets scored positive on the microarrays. Taking into account only the 219 antibodies that scored positive in at least one biological context, a median of four lysate sets scored positive per antibody (supplemental Fig. S1A). Importantly, no single antibody scored positive across all 20 biological contexts. To determine if this is because of differential antibody cross-reactivity or differential pathway activation, we analyzed microarray hits within the set of six isogenic RTK-expressing cell lines and found that only a small number of antibodies scored positive in all six sets of lysates (supplemental Fig. S1B). Because the six cell lines are expected to elicit nearly identical cross-reactive signal for a given antibody, these results indicate that the majority of signaling events are specific to a subset of biological contexts. This highlights the need to validate antibodies within the biological context of interest and not simply rely on previous validation efforts carried out under different experimental conditions.
Identification of Antibodies That Report on Differences in Protein Levels Across Diverse Cell Lines
In addition to discovering antibodies that report on dynamic signaling events, we also wanted to identify antibodies that detect differences in protein levels across the 17 cell lines used in our experiments. Because cell lines differ not only in their levels of a given target protein, but also in the levels of most other proteins, they are expected to elicit different degrees of cross-reactive signal on lysate microarrays in addition to different target protein signals. As ΔIthreshold is calculated solely from biological replicate data, systematic variations of cross-reactive binding are not taken into account. Consistent with this reasoning, when we attempted to identify hit antibodies within the cell lines data set based on biological noise (ΔIthreshold = 0.772 at α = 0.01 for 17-element cell lines vectors; see Experimental Procedures), almost all antibodies (321/383, 84%) scored as hits. Notably, this included most of the phosphospecific antibodies, which were not expected to produce strong signals in unstimulated cells. Thus, it is likely that the observed variation in signal largely reflects differences in cross-reactive binding, rather than specific binding. Despite our inability to derive a statistical metric for antibody performance in the cell lines data set, we nevertheless wished to select a subset of antibodies for validation experiments. We therefore rank-ordered antibodies according to their ΔI values in the cell lines data set, removed all PTM-specific antibodies, and selected the 50 highest-scoring antibodies. All 50 antibodies showed large spreads in signal intensity (ΔI ≥ 1.50) as well as strong fold-differences between the highest and lowest signal (≥3.59-fold).
Secondary Validation by Immunoblotting Reveals Determinants of Antibody Performance
Although the antibodies we identified as hits in our microarray screen all elicited significant changes in signal across lysates, they may still exhibit prohibitively high levels of cross-reactivity. Depending on the relative abundances of target and off-target antigens, the sum of these signals may or may not provide biologically interpretable information. We therefore set out to evaluate the primary hits from our microarray screen using a secondary quantitative assay. Quantitative immunoblotting is generally regarded as the gold standard for assessing the specificity of antibody recognition in the context of cellular lysates. Western blots, however, are more time-consuming, require larger sample volumes for each measurement, and incur a high cost in consumables. For these reasons, we deemed it impractical to evaluate all 1025 + 50 = 1075 microarray hits by immunoblotting.
To extensively validate antibodies while minimizing the overall number of Western blots, we focused our efforts on an informed subset of microarray hits. First, to obtain an unbiased overview of antibody performance, we chose four diverse biological contexts from the time courses data set: A431 + EGF (90 hits), HT-29 + insulin (23 hits), HeLa + anisomycin (33 hits), and HeLa + TNFα (26 hits). We then re-tested each of these 172 microarray-positive antibody-context pairs by quantitative Western blotting. The performance of each antibody was considered satisfactory if: (1) it produced a band of the correct molecular weight; and (2) the sum of all off-target bands did not exceed the intensity of the target band (Fig. 3A, supplemental Experimental Procedures). For immunoblots that passed both of these criteria, the target bands were quantified and compared with the corresponding microarray data (Fig. 3B) by calculating the Pearson correlation coefficient (ρ) of the two vectors (Fig. 3C). Based on the overall distribution of correlation coefficients, we considered an antibody to be fully validated for the biological context in question if and only if the microarray and Western blotting data agreed with ρ ≥ 0.75 (supplemental Fig. S2). This threshold ensures concordance between the two assays while accounting for the inherent variability of the immunoblotting step. Of the 172 Western blots that we produced, 89 (52%) exhibited a dominant band of the correct molecular weight. Of these, 68 (76%) had sufficient correlation between microarray and Western blotting data, for an overall validation rate of 40%.
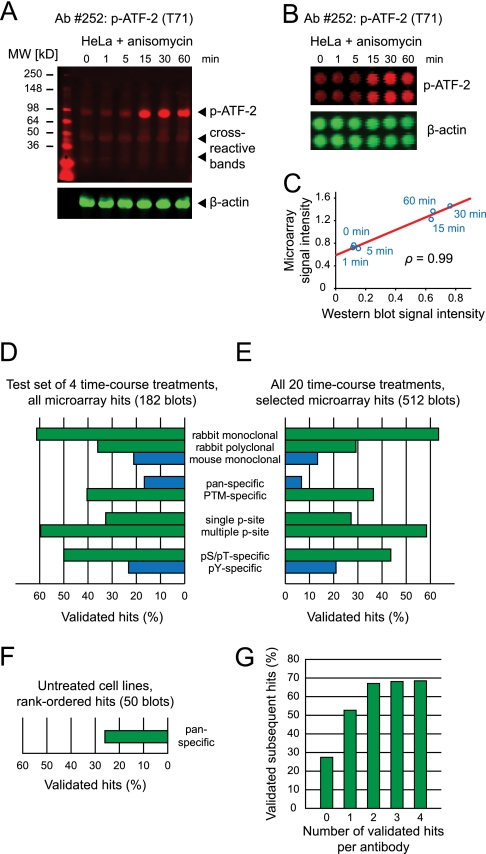
Fig. 3.
Secondary validation by quantitative immunoblotting reveals key determinants of antibody performance. A, Representative Western blot, showing moderate cross-reactivity. Lysates of HeLa cells treated with anisomycin, simultaneously probed with an antiphospho-ATF-2 (T71) antibody (red) and an anti-β-actin antibody (green). β-actin intensity was used to normalize signals for differences in gel loading. B, Lysate microarray images using the same lysates and antibodies as in (A). C, Scatter plot of signal intensities from (A) and (B) reveals degree of correlation between microarray and Western blotting data. D, Determinants of antibody reactivity, derived from an exhaustive Western blotting validation of microarray hits across four time course treatments (A431 + EGF, HT-29 + insulin, HeLa + anisomycin, and HeLa + TNFα). Blue: mouse antibodies, pan-specific antibodies, and pTyr-detecting antibodies generally exhibited lower validation rates. Green: rabbit, PTM-specific, and pSer/pThr-specific antibodies generally showed higher rates of validation. E, Determinants of antibody reactivity, derived from Western blotting validation of a subset of microarray hits across all time course treatments. Results were similar to (D). F, Validation rate of pan-specific antibodies across untreated cell lines. G, Prior validation of an antibody in one or more biological contexts increases the probability that it will perform well in additional biological contexts.
To identify potential predictors of antibody performance, we divided the antibodies into different categories based on their species of origin, method of preparation, or type of antigen (Fig. 3D). For each category, we determined the fraction of Western blots that were deemed acceptable by our criteria. Our results show that rabbit monoclonal antibodies generally perform better than rabbit polyclonal antibodies, which in turn outperform mouse monoclonal antibodies; that PTM-specific antibodies perform better than pan-specific antibodies; and that phosphospecific antibodies directed at sites of serine or threonine phosphorylation perform better than ones directed at sites of tyrosine phosphorylation. Our low success rate with pan-specific antibodies can likely be explained by the fact that the time courses represented in our data set were short in duration (0 to 60 min) and that the stimuli we used have less of an effect on protein synthesis or degradation during this time period than they do on PTM events. Visual inspection of the Western blots revealed that antibodies targeting pTyr epitopes (as opposed to pSer or pThr epitopes) mostly suffered from cross-reactivity with activated RTKs (as assessed by the molecular weight of the cross-reactive band), which are highly expressed in many of the cell lines and which feature numerous pTyr sites. The reason for the poor performance of mouse antibodies in our assay is unclear at this point, but is consistent with a growing trend to develop phosphospecific monoclonal antibodies using rabbits, rather than mice.
Given these results we concluded that the success rate of our immunoblotting validation step could be increased by focusing only on the subset of microarray hits elicited by rabbit antibodies that target post-translational modifications other than tyrosine phosphorylation. This subset comprises 47% of microarray-positive antibodies (103/219) and 53% of microarray hits within the time courses data set (545/1025). Using this information, we performed Western blot validations for each of these 103 antibodies in at least one biological context (231 additional blots). We also assayed 72 antibodies outside of this subset that were of particular biological interest to us, in either one or more lysate sets. For example, we tested several phospho-RTK-detecting antibodies in growth factor-stimulated cell lines. Overall, we observed very similar determinants of antibody reactivity from this larger set of immunoblotting data, with antibodies directed against rabbit, PTM and pSer/pThr epitopes outperforming mouse, pan and pTyr-detecting antibodies (Fig. 3E). In addition, rabbit monoclonal antibodies and multiple-phosphorylation site-specific antibodies performed particularly well, although both categories comprised only a small number of antibodies. It is possible that the precise physicochemical properties of epitopes, such as amino acid content or net charge, may provide additional predictors of antibody performance. In most cases, however, the identities of the antigenic epitopes of the antibodies that we evaluated were proprietary. As a result, we were unable to test this hypothesis. It would likewise be of interest to analyze the microarray performance of antibodies in relation to the specific applications they were recommended for by their manufacturers, such as Western blotting, immunoprecipitation or immunocytochemistry. Most antibodies in our study, however, were not extensively tested for all applications of interest by their respective manufacturers, making it difficult to reach reliable conclusions from such sparse data.
In addition to the time courses data set, we performed Western blot validations for the 50 top scoring antibodies within the cell lines data set. This provided an opportunity to assess antibody cross-reactivity across multiple cell lines. Thirteen antibodies (26%) produced acceptable blots and sufficient correlation with microarray data (Fig. 3F), with most unsuccessful validations attributable to off-target reactivity in a subset of cell lines. Across both the time courses and cell lines data sets, we validated a total of 198 antibody-context pairs, corresponding to 82 distinct antibodies. A summary of the validation status of all antibodies against all lysate sets is provided in supplemental Fig. S3. Our complete microarray and Western blotting data sets are provided in supplemental Table S3 and S4, respectively.
Next, we asked if prior validation of a given antibody in one or more biological contexts increases the probability that it will perform well in other biological contexts as well. Using our lysate microarray and Western blotting data sets, we calculated the percentage of successful validations for a given antibody-context pair as a function of the number of prior validations of the same antibody in other contexts (Fig. 3G). We found a strong increase in the likelihood of antibodies to perform well in additional contexts given prior validations. Overall, one or two prior validations were sufficient to ensure that an antibody would perform well in 60–70% of new biological contexts. This means that our list of 82 validated antibodies provides a valuable resource for future investigations using lysate microarray technology. It nevertheless remains necessary to carefully evaluate each detection antibody in each new setting to guarantee that meaningful information is obtained.
As a further test of antibody specificity and to obtain absolute rather than relative measures of protein abundance, it is possible to use purified protein standards on the arrays (data not shown). This approach has been used successfully in bead-based assays (23). It is usually limited to non-PTM antigens, however, because full-length, correctly modified protein standards are not generally available. In addition, because every antibody exhibits some degree of off-target reactivity, a direct comparison between signal intensities from lysates and purified antigens does not provide a reliable measure of the absolute amount of antigen present in each sample. Purified standards must be added to lysates that have been immunodepleted of each antigen, and this is not practical in the context of high-throughput experiments.
Antibody Reactivity is Independent of Batch, but Varies Substantially Among Different Antibodies Directed at the Same Antigen
Although our screen successfully identified numerous detection reagents, it remained unclear how broadly lysate microarrays could be used to gain quantitative information at the protein level. Having established quantitative criteria for antibody performance, we next wished to address the question of assay generality. Our overall microarray data set, comprising both hit and nonhit antibodies, provides a relatively unbiased base set of sufficient size to extract assay characteristics that should extend beyond the set of antibodies and biological contexts chosen for this study.
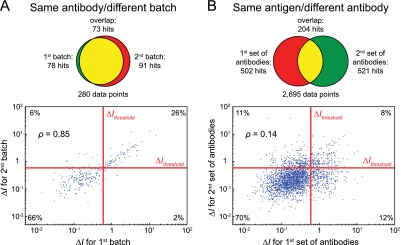
A key aspect of assay generality is whether, once a suitable detection antibody has been found, its reactivity remains robust and reliable. We previously showed that lysate microarrays exhibit extremely low coefficients of variation (<5%) across technical replicates (13). We further asked whether separately manufactured batches of the same antibody perform consistently on lysate microarrays. We first selected 14 phosphospecific antibodies from our screening set (eight polyclonal and six monoclonal). We then obtained from the same manufacturer a second batch, prepared on a different date, of all 14 antibodies and probed an additional 14 microarrays under conditions otherwise identical to the first set of antibodies. We compared ΔI values from the two data sets in the form of a scatter plot (Fig. 4A), and found strong overlap (ρ = 0.85). Notably, even nonhit antibodies in both data sets showed substantial correlation (bottom left quadrant, ρ = 0.47), indicating that antibody reactivity across batches is consistent, even when the signal is close to noise levels. A comparison of microarray hits likewise shows a high degree of concordance between the two data sets: 78 and 91 positive antibody-context pairs were identified, respectively, with an overlap of 73. Upon closer inspection, we found that the 23 combinations of antibodies and lysate sets that tested positive for only one of the two batches had substantially lower signal spreads (ΔI = 1.01 ± 0.10; mean ± S.E.; n = 23) than those antibodies that tested positive in both batches (ΔI = 2.26 ± 0.25 for first batch; ΔI = 2.20 ± 0.26 for second batch; n = 73). These data indicate that our screen identified a consistent set of hit antibodies within the margin of error imposed by data thresholding, and that antibody performance on lysate microarrays is highly robust across experiments and across different lots of the same antibody.
Fig. 4.
Microarray reactivity is antibody-dependent, but not batch- or antigen-dependent. A, Comparison of microarray data using different batches of the same antibodies. Top: Venn diagram showing substantial overlap between microarray hits. Bottom: Scatter plot of signal spreads (ΔI) across all time course microarray data, showing high degree of correlation. Data are represented on a double-logarithmic scale for ease of visualization. B, Comparison of microarray data from different antibodies directed at the same antigens, showing minimal overlap of hits and low degree of correlation. Top and bottom as in (A).
In the context of antibody-based detection methods, a second important aspect of their general applicability is whether or not a suitable antibody can, in principle, be found for each antigen of interest, or if certain intrinsic characteristics of an antigen precludes its detection. To determine whether the identity of the antigen or the antibody determines performance on lysate microarrays, we identified in our screening set 141 pairwise combinations of different antibodies that detect the same antigen. We then compared ΔI values for the two antibodies in the time courses data set in the form of a scatter plot (Fig. 4B). Strikingly, we observed a very weak correlation between the two sets of antibodies (ρ = 0.14), only marginally exceeding the correlation between the same number of randomly chosen pairs of antibodies (ρ = 0.03 ± 0.03, n = 1000). When we focused only on the microarray data that surpassed ΔIthreshold, we again found comparatively little overlap; whereas each set of antibodies contained over 500 individual hits, only 204 hits were common to both sets. This shows that there is very little concordance between different antibodies that target the same antigen.
Taken together, our data demonstrate that reactivity on lysate microarrays is very consistent across different experiments and different batches of antibodies, but is highly antibody-specific. Because the differences in reactivity between antibodies likely reflect different cross-reactive properties, we submit that extensive screening should, in most cases, identify functional detection reagents for almost any antigen of interest, and that success is limited only by the number of available antibodies.
Origin of Nonequivalence Between Microarray and Western Blotting Data
Based on the data collected in this study, we propose a simple model to explain the nonequivalence of microarray and Western blotting data. In our model, the observed signal intensity on lysate microarrays is simply the sum of the signal arising from binding the target antigen and an additional cross-reactive term (supplemental Fig. S4). This off-target signal is either undetectable on Western blots or, if visible, can be ignored because of the size separation step. In either case, this off-target signal remains relatively constant over a series of related lysates (e.g. time-courses of cell stimulation). Depending on the relative magnitudes of both terms, microarray data may appear compressed or even constant in comparison with true target protein levels. Consistent with this model, the Western blotting and microarray data collected in this study generally exhibited strong linear correlations (supplemental Fig. S2A), with offsets from the origin indicating cross-reactivity on the microarrays in the majority of cases (162/198, 82%) (supplemental Fig. S2B).
Our understanding of the origin of the cross-reactive signal remains incomplete. In experiments focusing on individual antibodies, we found that cross-reactive binding was unaffected when nonprotein components were removed from whole-cell lysates prior to microarraying (data not shown). In addition, when microarrays were enzymatically dephosphorylated, phosphospecific antibodies retained their cross-reactive signal intensities but lost all specific signal (data not shown). We therefore propose that antibody-dependent binding to off-target pan-epitopes is the primary cause of signal compression on lysate microarrays. Consistent with this hypothesis, we found that the variability of cross-reactive terms within isogenic but differently stimulated cell lines is significantly smaller than across unrelated cell lines (supplemental Analysis 1), indicating that the pan-protein complement of cell lines is the primary source of differential cross-reactivity. We further speculate that the stochastic arrangement of protein molecules in close proximity on the microarray surface may present chimeric epitopes, comprising two or more separate but spatially adjacent polypeptides, to which an antibody can bind in a cross-reactive manner. In comparison, Western blots may not suffer from this limitation, as size separation presents antibodies with a less complex mixture of proteins at any given location. Conceivably, the high density of identical target epitopes within a Western blot band may even favor specific binding through avidity effects. It will be of great interest in future studies to determine if other assays that do not include a size separation step, such as quantitative immunocytochemistry or flow cytometry, share similar determinants of antibody performance.
Time Courses Fall into Distinct Classes that Match Their Functional Role in Signaling
To demonstrate how the expanded set of detection reagents that we identified in our screen can be used to gain biological insight at a systems level, we used self-organizing maps (SOMs) (24) to analyze our rigorously validated time-course data. This subset of our data consists almost exclusively of PTM events, the vast majority of which were phosphorylation events. The SOM algorithm allows us to cluster the temporal profiles of signaling into groups of similar shapes, thereby identifying signaling patterns conserved across proteins and biological contexts (25).
We began our analysis by converting our data to a z-score scale to enable direct comparisons across measurements. Based on the heuristic principle that the ideal number of map units in a SOM is close to 5√n (where n is the number of vectors to be clustered) (26), we built a map comprising 66 units in 11 rows and 6 columns. Each unit initially contained one naïve reference vector. In the training phase, our SOM algorithm computed distances between each input pattern (data vector) and all reference vectors. The map unit containing the reference vector that correlated most closely with the input pattern was replaced with the arithmetic average of both vectors, whereas topologically close map units were averaged with the input pattern with gradually decreasing weight according to their distance in the map. This process was executed using all 179 complete six-point time courses and repeated 1000 times, leading to self-organization. In the clustering phase, the final map was constructed by assigning each phosphorylation time course to the map unit to whose reference vector it was most similar. We used the correlation distance metric in conjunction with the unified-distance matrix approach to allow robust identification of clusters and component planes. To provide a graphical representation of SOM time course profiles, heat maps of component planes, which represent individual time points, are shown in supplemental Fig. S5A online. Finally, we visualized the result of our SOM analysis in the form of a U-matrix, which describes the mean distances between neighboring map units following the SOM training phase (Fig. 5). Individual distances are shown in supplemental Fig. S5B online. These distances are color-coded: close proximity of two map units is indicated with blue color tones, whereas shades of yellow and red denote dissimilarity. Clusters can be identified in the heat map as blue “valleys” surrounded by yellow or red “mountains.”
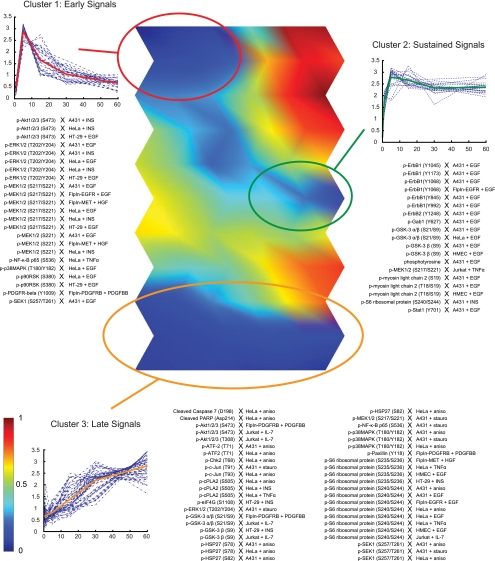
Fig. 5.
Self-organizing map analysis reveals three primary classes of time courses. Shown in the center is a visual representation of the average distances between neighboring map units in a self-organizing map of the time course profiles (U-matrix). Dark blue regions represent clusters of similar dynamic profiles, and regions of early, sustained, and late signaling dynamics (clusters 1, 2, and 3) are highlighted. Early and sustained signals are connected by a transition region of moderately clustered time courses (blue). Shown in panels on the sides are the identities, temporal profiles (dashed lines) and average temporal profiles (solid line) of time courses within each cluster.
Visual inspection of the U-matrix revealed three regions of particularly dense clustering. Based on the shapes of the constituent time courses (supplemental Fig. S5C), we identified these regions as early, sustained, and late signaling events. By grouping neighboring map units that contain time courses of similar shape, we defined boundaries around each region (supplemental Fig. S5D) and confirmed that each cluster was highly significant (p < 2 × 10−4, random permutation test). Upon closer inspection, we observed that the early and sustained clusters were connected by a transition region, which showed a moderate degree of clustering (p = 0.002). When the union of early, sustained, and transition clusters was considered, we likewise observed very significant clustering (p < 2 × 10−4), indicating that there is no fundamental division between early and sustained signals in our data set. Indeed, because these two classes of time courses share the characteristic of rapid onset of signaling, the apparent continuum of signaling dynamics indicates that they are differentially regulated at the level of signal attenuation. In contrast, the late cluster of time courses was strongly separated from both early and sustained signals. This suggests that cells use different mechanisms for the activation of early/sustained and late signaling events, which act over short (∼5 min) or long (>60 min), but not intermediate time scales. Signal attenuation, on the other hand, appears to follow a continuum of variable levels that serve to fine-tune the dynamics of individual signaling proteins.
Biological insight also emerged from direct comparisons of individual signaling proteins across different biological contexts. For example, phosphorylated epidermal growth factor receptor (p-epidermal growth factor receptor) was observed as a sustained signal in seven of nine time courses, whereas none of the six time courses of fibroblast growth factor receptor 1, insulin-like growth factor receptor 1, or PDGFR-β maintained high phosphorylation levels over the course of our 60-min treatments. This may indicate differences in receptor localization dynamics among RTKs, such as endocytic impairment or preferential recycling of epidermal growth factor receptor to the cell surface. We also observed that several signaling proteins were strongly associated with the late cluster. These “constitutive late” signals included phosphorylated ribosomal protein S6 (RPS6) (12 of 15 “late”), heat shock protein 27 (HSP27) (four of six late), cytosolic phospholipase A2 (cPLA2) (three of three late) and the transcription factor ATF-2 (two of two late). The dynamics of these signaling proteins likely reflects the fact that they act mainly as effectors of earlier signals in upstream layers of signal transduction networks. For example, p-RPS6, through its activity on the ribosome, influences protein synthesis (27) and is a key regulator of cell size (28).
Proteins that are not linked to a single cell fate generally showed context-specific signaling dynamics. For example, the phosphorylated MAP kinases MEK1/2, Erk1/2, p90RSK, p38MAPK, and SEK1 were virtually always early signals (24 out of 34 combined time courses), and never late signals (0 out of 34 time courses) under conditions of growth factor stimulation. This is consistent with our current understanding that MAPK phosphorylation is an initial and mostly transient response to ligand stimulation and lies upstream of several different pathways (29). Under conditions of stress, however, we observed a striking reversal of this pattern: 14 of 16 time courses fell into the late cluster and none fell into the early cluster. Importantly, the identity of the cell line played no role in determining signaling dynamics under either set of conditions. These results suggest that phosphorylation of MAP kinases is independent of cell type, but that the dynamics of this pathway encode its dual physiological role as a response to conditions of growth or stress (30).
Whereas phosphorylation of AKT was observed only under the relatively narrow set of conditions that favor cell growth and proliferation, AKT time courses showed a surprisingly broad spectrum of shapes: two of 17 time courses followed early activation, three were intermediate between early and sustained signals, seven showed late activation, and the remaining five fell into noncategorical clusters. Differential activation was observed even in closely related cellular contexts. For example, the time courses of AKT pS473 in response to EGF stimulation of A431, HMEC, and HT-29 cells lay in map units that are topologically distant from each other, with no significant clustering (p = 0.51). A similarly broad spectrum of profiles was observed for the AKT substrate GSK3; its time courses were distributed among late (five time courses), sustained (four time courses), transition region (three time courses) and noncategorical clusters (four time courses). Signaling dynamics within the AKT pathway thus appear to encode cell type-specific responses.
DISCUSSION
Quantitative, data-rich technologies are needed to study how information is transferred and processed in cells, and how defects in signaling networks lead to human disease. Lysate microarray technology provides a powerful tool for systems-level investigations, but has so far been limited by questions regarding data quality and by a scarcity of highly validated detection reagents. Here, we addressed both of these issues by developing a general and efficient way to identify antibodies that are functional on microarrays of cell lysates. Altogether, we screened 383 primary antibodies in each of 21 diverse biological contexts, capturing both changes in protein abundance and signaling dynamics. Based on the observed assay variability, we established stringent statistical criteria for assessing antibody performance, and further characterized microarray hits using a secondary validation step. Our analysis identified 82 unique antibodies, each one of which allows quantitative protein-level measurements in one or more of the biological contexts that we tested (supplemental Fig. S3).
In addition to identifying a large set of high-quality detection reagents that will facilitate future research, an analysis of our screening and validation efforts allowed us to identify specific characteristics of this assay that can be extended beyond the particular set of antibodies, cell lines and treatment conditions that we used in this study. For example, we identified several determinants of antibody quality: the best reagents were monoclonal antibodies derived from rabbits, were PTM-specific, and were directed at sites of serine or threonine phosphorylation. Other antibodies were less likely to perform well on lysate microarrays, although functional reagents were identified in every category that we tested. In addition, antibodies that had been rigorously validated in one or more biological context were much more likely to perform well in other contexts. This means that the 82 commercial antibodies that we validated in this study (supplemental Fig. S3) constitute a highly enriched set of reagents that should prove generally useful in a wide variety of other biological contexts. Finally, we established robust protocols that exhibit minimal variation across replicate experiments and different batches of the same antibodies. Importantly, we found that assay performance was highly dependent on the identity of the antibody, but not on the antigen itself. Overall, we did not find any constraints on the number of antibodies that can be validated or the type of protein that could be detected. Our study can therefore be used as a blueprint for future screening efforts to identify suitable detection reagents for this and other high-throughput immunoassays. We envision that the resource provided by this study could serve as the nucleus of an ever-expanding repository to which researchers contribute antibody validation information.
Above all, the value of any proteomic technology lies in the biology that can be uncovered using it. By performing a self-organizing map analysis of our time course data, we showed how a large set of highly validated detection reagents can be used to generate reliable information, and how this information can then be mined to gain insight into the logic and organization of signaling dynamics on a systems level. Our unbiased analysis identified a core set of time-dependent profiles, corresponding to early, sustained, and late signaling events. We found that, in addition to the identity of signaling proteins that are activated, the precise dynamics of post-translational modification events play an important role in conferring specificity to signaling in different biological contexts. Our results thus expand on previous studies showing that cellular decisions are encoded in the precise timing of signaling events (31). This principle may allow cells not only to sense environmental changes and respond accordingly, but may also enable cells to deconvolve mixtures of different and even opposing signals to effect an appropriate and uniform outcome.
In summary, the screening and analysis strategies outlined in this study are general and can easily be applied to other high-throughput, antibody-based assay technologies such as immunocytochemistry and flow cytometry. Our results show that lysate microarrays are broadly useful in a variety of cellular contexts, and we find no intrinsic limitations in extending these studies to other areas of cell biology. These characteristics, coupled with the high-throughput nature of this technology, suggest that lysate microarrays will play an important role in systems-level investigations of the cell.
Acknowledgments
We thank Joel Wagner, Jordan Krall, Elsa Beyer and Taran Gujral for helpful suggestions and critical reading of the manuscript. We are grateful to Toni Holway of Aushon Biosystems for microarray printing and assistance.
Footnotes
* This work was supported by an award from the WM Keck Foundation and by grants from the National Institutes of Health (R21 CA126720 and P50 GM068762). M. S. is the recipient of an Alfred and Isabel Bader fellowship and a Jacques-Emile Dubois fellowship. G. M. is a founder, employee, and shareholder of Merrimack Pharmaceuticals; a founder, consultant, and shareholder of Makoto Life Sciences; and a scientific advisory board member of Aushon Biosystems.
 This article contains supplemental Experimental Procedures, supplemental Figures S1–S5, supplemental Tables S1–S4, as well as supplemental Analysis.
This article contains supplemental Experimental Procedures, supplemental Figures S1–S5, supplemental Tables S1–S4, as well as supplemental Analysis.
1 The abbreviations used are:
- PTM
- post-translational modification
- RTK
- receptor tyrosine kinase
- TNFα
- Tumor necrosis factor α.
REFERENCES
- 1. Hale M. B., Nolan G. P. (2006) Phospho-specific flow cytometry: intersection of immunology and biochemistry at the single-cell level. Curr. Opin. Mol. Ther. 8, 215–224 [PubMed] [Google Scholar]
- 2. Kellar K. L., Mahmutovic A. J., Bandyopadhyay K. (2006) Multiplexed microsphere-based flow cytometric immunoassays. Curr. Protoc. Cytom. [DOI] [PubMed] [Google Scholar]
- 3. Wu W., Slåstad H., de, la, Rosa, Carrillo D., Frey T., Tjønnfjord G., Boretti E., Aasheim H. C., Horejsi V., Lund-Johansen F. (2009) Antibody array analysis with label-based detection and resolution of protein size. Mol. Cell Proteomics 8, 245–257 [DOI] [PubMed] [Google Scholar]
- 4. Carpenter A. E. (2007) Image-based chemical screening. Nat. Chem. Biol. 3, 461–465 [DOI] [PubMed] [Google Scholar]
- 5. Ciaccio M. F., Wagner J. P., Chuu C. P., Lauffenburger D. A., Jones R. B. (2010) Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat. Methods 7, 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haab B. B., Dunham M. J., Brown P. O. (2001) Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2, RESEARCH0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou H., Bouwman K., Schotanus M., Verweij C., Marrero J. A., Dillon D., Costa J., Lizardi P., Haab B. B. (2004) Two-color, rolling-circle amplification on antibody microarrays for sensitive, multiplexed serum-protein measurements. Genome Biol. 5, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen S., LaRoche T., Hamelinck D., Bergsma D., Brenner D., Simeone D., Brand R. E., Haab B. B. (2007) Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods 4, 437–444 [DOI] [PubMed] [Google Scholar]
- 9. Gembitsky D. S., Lawlor K., Jacovina A., Yaneva M., Tempst P. (2004) A prototype antibody microarray platform to monitor changes in protein tyrosine phosphorylation. Mol. Cell Proteomics 3, 1102–1118 [DOI] [PubMed] [Google Scholar]
- 10. Knickerbocker T., Chen J. R., Thadhani R., MacBeath G. (2007) An integrated approach to prognosis using protein microarrays and nonparametric methods. Mol. Syst. Biol. 3, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nielsen U. B., Cardone M. H., Sinskey A. J., MacBeath G., Sorger P. K. (2003) Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc. Natl. Acad. Sci. U.S.A. 100, 9330–9335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paweletz C. P., Charboneau L., Bichsel V. E., Simone N. L., Chen T., Gillespie J. W., Emmert-Buck M. R., Roth M. J., Petricoin E. F, III, Liotta L. A. (2001) Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 13. Sevecka M., MacBeath G. (2006) State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat. Methods 3, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chan S. M., Ermann J., Su L., Fathman C. G., Utz P. J. (2004) Protein microarrays for multiplex analysis of signal transduction pathways. Nat. Med. 10, 1390–1396 [DOI] [PubMed] [Google Scholar]
- 15. Spurrier B., Ramalingam S., Nishizuka S. (2008) Reverse-phase protein lysate microarrays for cell signaling analysis. Nat. Protoc. 3, 1796–1808 [DOI] [PubMed] [Google Scholar]
- 16. Spurrier B., Washburn F. L., Asin S., Ramalingam S., Nishizuka S. (2007) Antibody screening database for protein kinetic modeling. Proteomics 7, 3259–3263 [DOI] [PubMed] [Google Scholar]
- 17. Calvert V. S., Tang Y., Boveia V., Wulfkuhle J., Schutz-Geschwender A., Olive D. M., Liotta L. A., Petricoin E. F. (2004) Development of multiplexed protein profiling and detection using near infrared detection of reverse-phase protein microarrays. Clin. Proteomics 1, 81–90 [Google Scholar]
- 18. Vesanto J., Himberg J., Alhoniemi E., Parkankangas J. (2000) SOM toolbox for Matlab 5. Technical Report A57, Helsinki University of Technology, Helsinki, Finland [Google Scholar]
- 19. Ultsch A., Siemon H. (1990) Kohonen's self organizing feature maps for exploratory data analysis. Int. Neural Network conf., pp. 305–308, Dordrecht, Netherlands [Google Scholar]
- 20. Hautaniemi S., Yli-Harja O., Astola J., Kauraniemi P., Kallioniemi A., Wolf M., Ruiz J., Mousses S., Kallioniemi O. P. (2003) Analysis and visualization of gene expression microarray data in human cancer using self-organizing maps. Machine Learning 52, 45–66 [Google Scholar]
- 21. Gordus A., Krall J. A., Beyer E. M., Kaushansky A., Wolf-Yadlin A., Sevecka M., Chang B. H., Rush J., MacBeath G. (2009) Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol. Syst. Biol. 5, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marmor M. D., Skaria K. B., Yarden Y. (2004) Signal transduction and oncogenesis by ErbB/HER receptors. Int. J. Radiat. Oncol. Biol. Phys. 58, 903–913 [DOI] [PubMed] [Google Scholar]
- 23. Schwenk J. M., Lindberg J., Sundberg M., Uhlén M., Nilsson P. (2007) Determination of binding specificities in highly multiplexed bead-based assays for antibody proteomics. Mol. Cell Proteomics 6, 125–132 [DOI] [PubMed] [Google Scholar]
- 24. Kohonen T. (2001) Self-organizing maps, 3rd Ed., Springer, Berlin; New York [Google Scholar]
- 25. Wolf-Yadlin A., Kumar N., Zhang Y., Hautaniemi S., Zaman M., Kim H. D., Grantcharova V., Lauffenburger D. A., White F. M. (2006) Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol. Syst. Biol. 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vesanto J., Alhoniemi E. (2000) Clustering of the self-organizing map. IEEE Trans. Neural Netw 11, 586–600 [DOI] [PubMed] [Google Scholar]
- 27. Thomas G. (2002) The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 35, 305–313 [DOI] [PubMed] [Google Scholar]
- 28. Ruvinsky I., Meyuhas O. (2006) Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31, 342–348 [DOI] [PubMed] [Google Scholar]
- 29. Roux P. P., Blenis J. (2004) ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68, 320–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wada T., Penninger J. M. (2004) Mitogen-activated protein kinases in apoptosis regulation. Oncogene 23, 2838–2849 [DOI] [PubMed] [Google Scholar]
- 31. Marshall C. J. (1995) Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185 [DOI] [PubMed] [Google Scholar]