Abstract
Sertoli cells (SCs) are the central, essential coordinators of spermatogenesis, without which germ cell development cannot occur. We previously showed that Dicer, an RNaseIII endonuclease required for microRNA (miRNA) biogenesis, is absolutely essential for Sertoli cells to mature, survive, and ultimately sustain germ cell development. Here, using isotope-coded protein labeling, a technique for protein relative quantification by mass spectrometry, we investigated the impact of Sertoli cell-Dicer and subsequent miRNA loss on the testicular proteome. We found that, a large proportion of proteins (50 out of 130) are up-regulated by more that 1.3-fold in testes lacking Sertoli cell-Dicer, yet that this protein up-regulation is mild, never exceeding a 2-fold change, and is not preceeded by alterations of the corresponding mRNAs. Of note, the expression levels of six proteins of interest were further validated using the Absolute Quantification (AQUA) peptide technology. Furthermore, through 3′UTR luciferase assays we identified one up-regulated protein, SOD-1, a Cu/Zn superoxide dismutase whose overexpression has been linked to enhanced cell death through apoptosis, as a likely direct target of three Sertoli cell-expressed miRNAs, miR-125a-3p, miR-872 and miR-24. Altogether, our study, which is one of the few in vivo analyses of miRNA effects on protein output, suggests that, at least in our system, miRNAs play a significant role in translation control.
In all sexually reproducing organisms, germ cells (GCs)1, in contrast to somatic cells, are the only cells that can give rise to a new organism; GCs give rise to the gametes—egg in females and sperm in males. Spermatogenesis refers to the development of mature haploid sperm from diploid spermatogonial cells within the testis of the male reproductive tract. It is typically divided in three strictly regulated phases, the mitotic, the meiotic, and the phase of spermiogenesis, which culminates with spermiation, the release of spermatozoa in the testicular seminiferous tubule's lumen. Spermatogenesis ensures continuous gamete production and occurs throughout adulthood in consecutive waves within the seminiferous tubules of the testis (reviewed in (1)). Apart from the GCs, which undergo spermatogenesis, the supporting cells of the testis called Sertoli cells (SCs), play a central role in the coordination of this process (for review see (2, 3)). SCs structurally and nutritionally support GCs and secrete factors that control, among other events, the survival and progression of GCs through the sequential steps of spermatogenesis (for example see (4–6)).
Post-transcriptional control plays an essential role in the regulation of spermatogenesis. During GC development, transcription and translation are un-coupled: transcription occurs massively following meiosis, with postmeiotic transcripts accumulating in large amounts, becoming deadenylated and stored in a repressed, dormant form in the spermatid cytoplasm for 4–5 days, whereas translation occurs at later stages (7). In addition to this “classic” mechanism, a novel system of post-transcriptional control mediated by microRNAs (miRNAs) is lately emerging with an important role during spermatogenesis ((8–10), and reviewed in (11)). miRNAs are endogenous, single-stranded, noncoding RNAs of ∼22 nucleotides that act as post-transcriptional regulators of gene expression. They are generated through a multistep enzymatic process that involves the function of Dicer (Dcr), an RNaseIII endonuclease essential for the production of mature miRNAs (reviewed in (12)). miRNAs bind most frequently to the 3′UTR (3′ untranslated region) of target mRNAs, although recent studies show that some can also bind within the coding sequence (CDS) of mRNAs (reviewed in (13)), and depending on sequence complementarity, induce either mRNA degradation or translational repression of their target (for review see (14)). Importantly though, it has been reported that in some cases, miRNAs can also promote gene expression (15, 16), thus broadening even more their range of effects.
Although miRNA effects at the mRNA level have been frequently evaluated (for example (17–19)), their impact on protein output, which is thought to be the primary effect of animal miRNAs, has been, technically, more difficult to assess. One study used stable isotope labeling by amino acids in cell culture (SILAC) technology to investigate the effect of a single miRNA on protein output and reported that miR-1 can regulate a substantial percentage of the HeLa proteome (20). Only recently though two groups performed a large-scale protein analysis that unraveled the impact of miRNAs on protein output; both concluded that, in addition to down-regulating mRNA levels, a single miRNA can repress the production of hundreds of proteins, but that this repression is relatively mild (21, 22).
We previously generated a mouse model in which Dcr -and miRNAs- are eliminated uniquely in the SCs of the testis (10). We found that this ablation leads to complete infertility because of severe spermatogenic defects and gradual testicular degeneration; importantly, significant transcriptome (mRNA) down-regulation of genes such as Gdnf, KitL, Man2a2, and Wt1, all with essential roles during spermatogenesis, was detected upon SC-Dcr loss (10). Here, in order to investigate the impact of SC-Dcr loss at the proteome level, we performed ICPL (isotope-coded protein label) analysis (23), which allowed us, by means of MS, to relatively quantify proteins whose expression was altered between SC-Dcr-depleted (Dcrfx/fx;MisCre, hereafter referred to as mutant) and wild-type (Dcrfx/fx, hereafter referred to as control) testes. We found that more than a third of 130 quantified testicular proteins are up-regulated in mutant testes, yet at a relatively mild level, and that, importantly, this up-regulation does not reflect detectable changes in their respective mRNA levels. Of note, protein absolute quantification was achieved in independent experiments using the AQUA (Absolute QUAntification) peptide strategy (24) on a selected set of proteins and thus validated the results obtained through ICPL analysis. In addition, we identified Sod-1, a gene up-regulated at the protein level, as a direct in vitro post-transcriptional target of three SC-expressed miRNAs, miR-125a-3p, miR-872, and miR-24, we hypothesize that its up-regulation upon SC-Dcr and miRNA loss could account, partially, for the observed testicular degeneration. Globally, our findings further reinforce the current notion of animal miRNAs exerting one of their primary negative effects at the translational level, but most importantly, open new perspectives in studying the testicular proteome and its relation to the miRNA machinery.
EXPERIMENTAL PROCEDURES
Affymetrix Microarray Analysis
Microarray analysis is described in (10). All microarray data are available through ArrayExpress (http://www.ebi.ac.uk/arrayexpress/, accession number: E-TABM-426).
Protein Extraction and ICPL Labeling
Performing differential proteomics analysis using extremely small micro-dissected tissue samples is indeed a challenge. Considering in addition, the cost of knockout animals, experiments were only performed once. Protein extracts were prepared from 20 pairs of control and 20 pairs of mutant P0 (postnatal day 0) testes; for additional information on the generation of the Dcrfx/fx;MisCre mouse strain, refer to (10). Tissues were homogenized by sonication on dry ice in a lysis buffer (6 m guanidine HCl, pH 8.5, tissue/buffer: 1/1.5(w/v)) and were then placed for 1 h at 4 °C before being centrifuged (15,000 × g, 30min, 4 °C). The resulting supernatants were then ultracentrifuged (105,000 × g, 1 h, 4 °C). Protein concentration of the resulting supernatants was measured with a bicinchoninic acid assay (Sigma-Aldrich) and was adjusted to 5 mg/ml by addition of lysis buffer. Disulfide bonds were reduced with 0.2 m tris(2-carboxyethyl)phosphine and then alkylated with 0.4 mm iodoacetamide. For each sample, 100 μg of proteins were labeled using the ICPL-kit (Serva Electrophoresis, Heidelberg, Germany) according to the manufacturer's instructions. Briefly, free amino groups (lysine residues and N-terminal NH2) of proteins from control and mutant extracts were labeled at room temperature for 2 h with the light (12C- nicotinoyloxysuccinimide) and the heavy (13C- nicotinoyloxysuccinimide) ICPL reagents, respectively. Following quenching excess reagent with 6 m hydroxylamine, the two labeled samples were mixed, purified by acetone-precipitation (−20 °C, overnight), and subsequently dissolved in 20 mm HEPES. Labeled proteins (50 μg) were separated by SDS-PAGE on a 12% precast gel (Gebagel, Gene Bio-Applications, Interchim, Montluçon, France). The gel was subsequently stained with Coomasie blue R-350 using the EZBlue gel staining reagent (Sigma-Aldrich, Saint-Quentin Fallavier, France). The entire gel lane was cut into 20 bands, which were washed with different acetonitrile (ACN)/100 mm NH4HCO3 solutions. In-gel digestion was performed overnight at 37 °C with modified trypsin (Promega, Charbonnières-les-Bains, France). Proteolytic peptides were then extracted from the gel by sequential incubation in the following solutions: ACN/H2O/TFA, 70:30:0.1 (v/v/v), 100% ACN and ACN/H2O/TFA, 70:30:0.1 (v/v/v), and extracts were eventually concentrated by evaporation to a final volume of 30 μl.
Nano-LC-MS-MS Analysis
Proteolytic mixtures were separated on a nano-high performance liquid chromatography system (Ultimate 3000, Dionex, Jouy-en-Josas, France), with an injection volume of 22 μl: first, they were concentrated on a C18-PepMap trapping reverse phase column (5 μm, 300 Å/300 μm i.d. × 5 mm, Dionex), and were then eluted with a 75-min, 2–90% ACN gradient in 0.05% formic acid, at a flow rate of 250 nL/min. The nano-LC apparatus was coupled on-line with an Esquire HCT Ultra PTM Discovery mass spectrometer (Bruker Daltonik, GmbH, Bremen, Germany), equipped with a nanoflow electrospray ionization (ESI) source and an ion trap analyzer (ITMS). The mass spectrometer was operated in the positive ionization mode. The EsquireControlTM software (Bruker Daltonik, GmbH) automatically alternated MS and MS-MS acquisitions and was tuned to preferentially subject ICPL labeled peptides to MS-MS acquisitions.
Protein Identification and Relative Quantification
The DataAnalysis 3.4 software (Bruker Daltonik, GmbH) was used to create the peak lists from raw data. For each acquisition, a maximum of 500 compounds were detected with an intensity threshold of 250,000 and the charge state of precursor ions was automatically determined by resolved-isotope deconvolution. The Biotools 3.1 software (Bruker Daltonik, GmbH) was used to submit MS/MS data to the Swiss-Prot database (version 47, Mus musculus taxonomy, 568,851 sequence entries) using the Mascot algorithm (Mascot server v2.2; http://www.matrixscience.com). Given that modification of lysine residues by ICPL labeling prevents their cleavage by trypsin, arginine C was selected as enzyme with one allowed missed cleavage. In addition, carbamidomethylation of cysteins was set as fixed modifications, and labeling of lysine residues and of the N-terminal NH2 group of proteins by light or heavy ICPL reagents, as well as methionine oxidation were considered as variable modifications. The mass tolerance for parent and fragment ions was set to 1.2 and 0.5 Da, respectively. Peptide identifications were accepted if the individual ion scores were above 25 (the ion score is −10*log(P), where P is the probability that the observed match is a random event). Protein identifications were accepted if the score indicated identity or extensive homology, i.e. the probability that the identification is a random match was lower than 5%. Matches corresponding to the heavy and the light labeled forms of the same peptide counted for one peptide. Single peptide-based identifications were accepted because missed cleavages of labeled lysine residues leads to a global reduction in the number of peptides produced in comparison to “classical” trypsin digestion and to the formation of relatively long peptides that can single-handedly represent a sufficient percentage of protein sequence coverage.
MS/MS spectra were searched against a randomized sequence (decoy) database using Mascot to determine the false discovery rate defined as the number of validated decoy hits/(number of validated target hits + number of decoy hits)*100. Thus, a satisfactory false discovery rate of 1.15% was obtained for the totality of identifications acquired during ICPL analysis.
Relative protein quantification was obtained using the WarpLC 1.1 software (Bruker Daltonik, GmbH). This automatically calculates the heavy-to-light (H/L) ratios by comparing the relative intensities of the extracted ion chromatograms (EIC) that are reconstituted by extraction of the intensities of m/z ratios corresponding to the labeled peptides observed on MS spectra. For each EIC, the contribution of 1+, 2+, and 3+ charge states of the peptide was considered and smoothing was applied (one smoothing cycle with Gauss algorithm and a smoothing width of 3 s). For each protein, the H/L ratio was calculated by averaging the different H/L ratios obtained for each pair of labeled peptides.
In the present study, the amount of peptides obtained following peptide extraction from both sample pools was not enough to perform technical replicates. Reproducibility and accuracy of ICPL experiments performed by ESI-ITMS were evaluated in five independent technical replicates using a standard mixture of ICPL labeled proteins containing bovine serum albumin with a heavy-to-light ratio of 1:1 (ICPL-kit, Serva Electrophoresis). For bovine serum albumin, an average H/L ratio of 0.94 was obtained, very close to the theoretical value of 1, and corresponding to a variation coefficient of 8%.
AQUA Peptide Analysis
AQUA [13C6, 15N2] K-Lysine-labeled and [13C6, 15N4] R-Arginine-labeled peptides (listed in supplemental Table 1) were synthesized and quality- and quantity-controlled by Sigma-Aldrich. All AQUA peptide standard solutions were prepared from stock solutions at 5 pmol/μl according to the manufacturer's instructions. All samples used for AQUA peptide experiments were systematically prepared in low adsorption tubes (LoBind tubes, Eppendorf, Le Pecq, France) to minimize errors because of peptide adsorption (25). AQUA peptide standard solutions were prepared at 0.1, 0.2, 0.5, 1, 2 and 5 fmol/μl and were analyzed by nano-liquid chromatography (LC)-ESI-ITMS with an injection volume of 10 μl in three analytical replicates for calibration. For each AQUA peptide, the corresponding EIC area was automatically determined by the QuantAnalysis 1.8 software (Bruker Daltonik, GmbH) and plotted against the injected amount to obtain the calibration curves. The linearity of the response was verified for all AQUA peptides with correlation coefficients ranging from 0.988 to 0.996. In addition, the analytical repeatability of measurement of EIC areas corresponding to the different AQUA peptides was evaluated: satisfactory coefficient of variations (CV) ranging from 2% to 9% (n = 6) were obtained for an AQUA peptide concentration of 2 fmol/μl.
Protein extracts used for the ICPL experiment, that is, one sample of 20 pairs of control and one sample of 20 pairs of mutant testes, were also used for AQUA peptide analysis. Proteins from the control and mutant sample were precipitated with acetone overnight at −20 °C and dissolved in Laemmli buffer (Gene Bio-Application). The two samples were then independently separated by SDS-PAGE on a 12% precast gel (Gebagel). Following fixation, washing, and staining, both entire gel lanes were manually cut into 20 pieces. Disulfide bonds were then reduced with dithiotreitol and alkylated with iodoacetamide. Protein in-gel digestion and proteolytic peptide extraction from each gel band was then performed. In order to precisely control the final volume of proteolytic peptides, extracts were completely dried by evaporation and dissolved with 20 μl of H2O/formic acid (95/5, v/v) solution, then with 20 μl of a H2O/ACN/formic acid (95/5/0.2, v/v/v) solution and vigorously sonicated and vortexed. AQUA peptide standards were added in precise amounts to the samples just before nano-LC-MS analysis. For all AQUA peptide analysis, the EsquireControl software was operated in the Multiple Reaction Monitoring mode to specifically subject the labeled (AQUA peptides) and unlabeled (peptides from the sample) peptides to MS/MS fragmentations. Then, for each fragmented peptide, an EIC was reconstituted by extracting the signals corresponding to fragment ions specific to the peptide of interest. Absolute quantification was obtained by comparing the EIC areas of the unlabeled peptide and its corresponding AQUA peptide added in precise amount.
Real-Time Quantitative PCR
Total RNAs from six control and six mutant P0 testes were extracted using the RNeasy Micro Kit (Qiagen, Basel, Switzerland) according to the manufacturer's protocol. For each of the 12 individual samples, 1 μg of total RNA was reverse transcribed with the Superscript II Reverse Transcriptase (Invitrogen, Basel, Switzerland) according to the manufacturer's instructions, and 1/40 of the cDNA was used as template for Real-Time PCR amplification on a Freedom Evo 150 System (Tecan, Männedorf, Switzerland) using the Power SYBR Green PCR master mix (ABI, Foster City, CA). Raw threshold-cycle (Ct) values were obtained with the SDS 2.0 software (ABI). Relative quantities (RQ) were calculated with the formula RQ = E–Ct, using efficiencies (E) calculated with the DART-PCR algorithm, as described (26). Mean quantities were calculated from triplicate PCR reactions for each sample, and were normalized to two similarly measured quantities of Gapdh and Trf1R as described (27). Normalized quantities were averaged for three replicates for each data point and represented as the mean ± S.D. The highest normalized relative quantity was arbitrarily designated as a value of 100.0. Fold changes were calculated from the quotient of means of these normalized quantities and reported as values ± S.E. The statistical significance of fold-changes was determined by an unpaired Student's t test. Primers used are listed in supplemental Table 2.
Spermatogenic Cell Purification
Mature Sertoli and peritubular myoid cells were prepared from 10 C57BL/6J males aged P16, whereas immature Sertoli cells were prepared from 16 C57BL/6J animals aged P6, as previously described (28). Germ cells were prepared using the STAPUT technique according to (29); spermatogonia were prepared from 40 C57BL/6J males aged P6–8, whereas pachytene spermatocytes and spermatids were prepared from six adult (P60) C57BL/6J mice. To verify cell purity, 5 × 105 cells were fixed in phosphate-buffered saline (PBS)/PAF 1% for 10′ at room temperature, washed in PBS and then conserved overnight at 4 °C in PBS/FCS 1%. Cells were then marked with propidium iodide (100 μg/ml) in PBS/0.2% saponin (30′, RT) and were sorted on a FacsCalibur machine (Beckton Dickinson, France), in order to quantify their contamination. Leydig cells were prepared from 16 adult (12-week-old) mice, as previously described (30); their purity was assessed by incubating cells with NAD (in Nitro Blue Tetrazolium, NBT, N-6876, Sigma-Aldrich) for 90' and quantifying the percentage of cells having acquired a violet color, indicative of the presence of 3β-HSD.
microRNA Expression Profiling with Illumina Arrays
Total RNA was isolated with Trizol (Invitrogen, Basel, Switzerland) and quality controlled for RNA integrity by capillary electrophoresis on an Agilent 2100 Bioanalyzer. miRNA profiling was performed according to the manufacturer's protocol using the Illumina MicroRNA Expression Profiling Mouse Panel (Illumina, Hayward, CA), which contains 656 assays for miRNAs described in miRBase v12. Briefly, for each sample, 500 ng of total RNA was polyadenylated and converted into cDNA using an oligo dT-Reverse PCR primer. miRNA-specific oligos (extended with specific address sequences and Forward PCR primer sequences) were then hybridized to cDNAs. Following extension using DNA polymerase, products were PCR-amplified using Cy3-labeled forward and unlabeled reverse primers, then purified and eventually hybridized onto a Sentrix Array Matrix overnight. The Sentrix Array Matrix was washed and scanned on a BeadArrays reader. Data were normalized and analyzed using the Illumina Beadstudio 3.1.3 (background correction and quantile normalization without scaling). Expression profiles for each sample were imported into GeneSpringGX 7.3.1 (Agilent Technologies) and MatLab (MathWorks, Inc) and further analyzed in order to identify differentially expressed miRNAs. MicroRNAs were considered as being expressed when the expression was above 1000 (arbitrary units).
MicroRNA Target Recognition Analysis
Target sites were initially identified by the presence of miRNA seeds (the minimum sequence of nucleotides required for successful miRNA binding on its target, see supplemental Fig. 1A), and their biological relevance was estimated using the following three models: The first model relies on the thermodynamics of the miRNA-mRNA interactions. The energy balance of these interactions (ΔΔG) was computed with a method similar to that of (31): It includes the free energy gain resulting from the formation of the miRNA-target duplex (ΔG duplex) and the free energy required for the unfolding of the target site and of at least 10 nucleotides upstream and 15 downstream of the target site (ΔG open). The second model relies on sequence features as described in (32). The third model relies on conservation of the seed sequences among placental species: the Phastcons scores of the seed sequence bases, provided by the UCSC genome browser (33), were summed (the sum allows to include the effect of the seed length). For seed identification, we used standard parameters, requiring seed length to be 6–8 bases from position 2 of the miRNA, and not allowing mismatches except for a single G:U wobble in 7-mers and two G:U wobbles in 8-mers. Of note, for each model, the first quartile of the ranked predictions was considered as biologically significant in our target site enrichment analysis. A schematic representation of the described strategy is shown in supplemental Fig. 1B.
In vitro Luciferase Assays
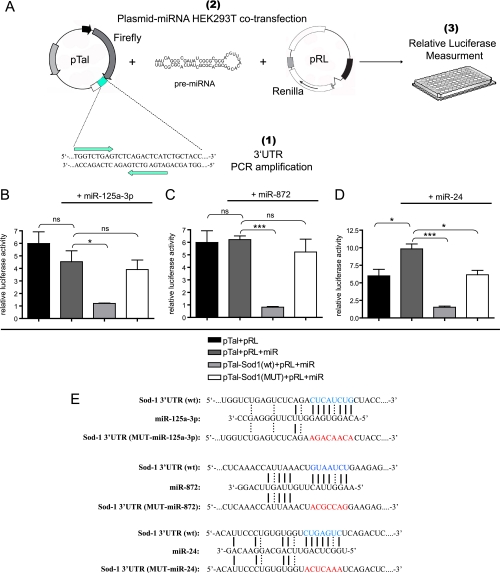
The 3′UTR of Sod-1 was PCR-amplified from genomic DNA using the following oligos: F:5′-ATATGGTCTAGAACATTCCCTGTGTGGTCTGAG-3′, R:5′-ATATGGCCGGCCGTCACACAGTTACAA-3′, and was subcloned in a TOPOII vector (Invitrogen, Basel, Switzerland). The insert was then digested out and directionally inserted downstream of the Firefly luciferase coding sequence in the XbaI and FseI sites of the pTal-Luc vector (Clontech, Sait-Germain-en-Laye, France). Mutated Sod-1 3′UTR constructs were generated with the QuikChange II Site-Directed Mutagenesis kit (Stratagene, Agilent Technologies, Schweiz AG), as described in the manufacturer's protocol, using oligos carrying a fully mutated seed sequence. The day before transfection, 104 HEK293T cells/well were seeded in 96-well plates; transfection was performed with (i) 100 ng of the pTal-Luc-Sod1–3′UTR (wild-type or mutant) Firefly plasmid, (ii) 5 ng of the transfection control pRL-SV40 Renilla luciferase plasmid (Promega AG, Dübendorf)) and (iii) 10 nm of the pre-miR-125a-3p (#PM12378), pre-miR-872 (#PM12800), or pre-miR-24 (#PM10737) (Ambion, Applied Biosystems Europe BV), using Lipofectamine 2000 (Invitrogen, Basel, Switzerland) according to the manufacturer's instructions. The Firefly and Renilla luciferase activities were measured 48 h post-transfection using the Dual Luciferase Assay system (Promega AG, Dübendorf) as described in the manufacturer's protocol. All experiments were performed three times, with each experimental condition being performed in four technical replicates. A schematic representation of the Luciferase assays' strategy is shown in Fig. 6A.
Fig. 6.
Testing the binding of the Sod-1 3′UTR by SC-expressed miRNAs. A, The Sod-1 3′UTR was PCR-amplified (1) and cloned just downstream of the Firefly luciferase CDS in the pTal plasmid. The pTal-Sod1–3′UTR plasmid was cotransfected with a pRL transfection control vector expressing the Renilla luciferase CDS and 10 nm of a miRNA precursor in HEK293T cells (2) and the relative luciferase activity was measured (3). Cotransfection of pTal (Firefly) and pRL (Renilla) (black bars) yielded a certain Firefly/Renilla luciferase level; addition of either pre-miR-125a-3p or pre-miR-872 did not alter the relative luciferase levels (B, C, dark gray bars), however that of pre-miR-24 did (see text) (D, dark gray bar); cloning the wt Sod-1 3′UTR in pTal in the presence of either one of three miRNAs caused a significant reduction in the relative luciferase activity (B, C, D, light gray bars), whereas mutation of the seed sequences abolished the miRNA repressive effect (B, C, D, white bars); *p < 0.05, **p < 0.01, ***p < 0.001 versus controls; ns: not significant. (E) Schematic representation of the binding of miR-125a-3p, miR-872 and miR-24 on either the wt or the mutated Sod-1 3′UTR sequence. Wt seeds are marked in blue, mutated seeds in red, Watson-Crick base pairing with a straight line and U-G wobbles with a dotted line.
RESULTS
SC-expressed miRNAs Affect Testicular Transcription
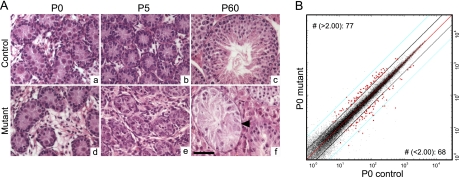
We previously generated a mouse model in which Dicer (Dcr), and subsequently miRNAs, are specifically eliminated in the Sertoli cells (SCs) of the testis (Dcrfx/fx;MisCre), and found that this loss leads to complete infertility (10). We were able to detect already by postnatal day 5 (P5), a delay in SC maturation and an initial increase in SC proliferation followed by highly elevated levels of SC and GC apoptosis, events that ultimately led to a dramatic testicular degeneration during adulthood (Fig. 1A). Importantly, although at birth (P0) no morphological (histological) defects were detected (Fig. 1A), we measured several alterations of the testicular transcriptome. More precisely, we found 77 and 68 genes to be ≥ 2 -fold up- and down-regulated respectively in P0 testes lacking Dcr in SCs (Fig. 1B). Deregulated genes included among others Gdnf, Kitl, Serpin5a, Sox9, Wt1, and Cldn11, all of which have key roles during spermatogenesis. However, the in vivo effect of SC-miRNA depletion on protein output was not addressed.
Fig. 1.
Morphological abnormalities appear by postnatal (P) day 5, yet mRNA transcripts are affected upon SC-Dcr loss already by P0. A, Hematoxylin and eosin-stained paraffin sections from P0 (a, d), P5 (b, e) and P60 (c, f) testis sections of control (Dcrfx/fx) (a, b, c) and mutant (Dcrfx/fx;MisCre) (d, e, f) mice; note the dramatic spermatogenic defects (arrowhead points to a tube containing only SCs) observed in adult P60 mutant testes. Scale bar: 50 μm. B, Scatterplot depicting genes showing at P0 differential expression between control and mutant whole testes. Each dot (black or red) represents a gene; genes represented as red dots are those which are either up-regulated (77 genes) or down-regulated (68 genes) >2-fold in mutant testes. Diagonal black bars represent a 2-fold threshold.
Sertoli-cell Loss of Dicer Causes Significant Proteome Alterations
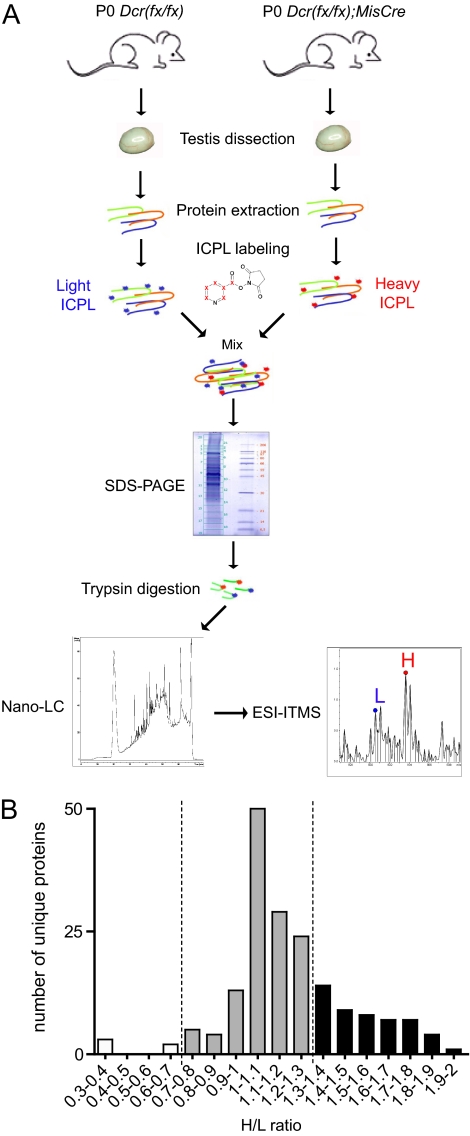
Here, to assess the impact of SC-miRNA loss on the testicular proteome, we performed relative quantification of proteins on P0 whole testis protein extracts of control and mutant mice using ICPL: Proteins extracted from P0 control and mutant testes were labeled with the light (L) and heavy (H) ICPL reagent respectively, mixed, prefractionated by gel electrophoresis, excised, and trypsin digested. The obtained peptide mixtures were analyzed by nano-ESI-ITMS for protein identification and relative quantification (Fig. 2A).
Fig. 2.
SC-loss of Dcr causes significant proteome alterations at birth. A, Schematic representation of the experimental design employed for relative quantification of proteins using ICPL. B, Shown here are proteins up-regulated (black bars), unaffected (gray bars) or down-regulated (white bars) upon SC-Dcr loss. H/L (mutant/control) ratios between 0.7 and 1.3 are indicative of no difference between control and mutant testes.
By querying the Swiss-Prot database with the Mascot algorithm, we obtained 240 protein identifications showing a score superior to the identity or the extensive homology threshold. These 240 identifications actually corresponded to 168 proteins, each associated with a nonredundant EntrezGene (EG) identifier. Out of these 168 nonredundant proteins, a mutant/control (H/L) protein ratio was calculated for 130 of them (all 130 proteins are listed in supplementary Table 3, and all peptide sequences for the identified proteins are listed in supplementary Table 4); for the remaining 38, this was not possible because of the absence of detected labeled peptides from either the control or the mutant sample. The minimum variation of H/L ratios associated to significant variation of protein expression was determined similarly to (34). The average of the CV (coefficient of variation) obtained for H/L ratios of all proteins for which at least two peptides were quantified was 11.1%. We thus considered that a variation of 30% (>2 CV) was significant. This significant variation of 30% largely overcomes technical variability in our experiments that was demonstrated to be 8%. Of these 130 quantified proteins, 50 were up-regulated (H/L ratio ≥1.3), whereas only 3 were down-regulated (H/L ≤0.7) in mutant testes (Table I). The remaining 77 showed no significant difference in abundance between control and mutant testes (0.7<H/L<1.3). More precisely, of the 50 up-regulated proteins, 23 showed a mild (1.3–1.5-fold change) up-regulation, and the remaining 27 displayed an H/L ratio between 1.5- and 2- (Fig. 2B and Table I). These findings are in agreement with two recent studies that reported a relatively mild repression of hundreds of proteins upon miRNA overexpression (21, 22).
Table I. List of proteins whose mutant/control (H/L) ratio, as revealed by ICPL analysis, is >1.3 (50 proteins, Sfrs-Stip1) or <0.7 (3 proteins, Hbb-b1, Fasn, Hbb-b2). Also shown are two unaffected (0.7<H/L<1.3) proteins (Gstm1, Prdx1) that were used for AQUA validation analysis.
| Protein Name | Gene Name | H/L (ICPL)a | H/L (AQUA)b | mRNA (Affy)c | mRNA (qPCR)d |
|---|---|---|---|---|---|
| Splicing factor, arginine/serine-rich 1 | Sfrs | 1.97 | −1.08 | ||
| Heat shock protein HSP 90-alpha | Hsp90aa1 | 1.86 | 1.12 | 0.75 | |
| Heat shock 70 kDa protein 1L | Hspa1l | 1.84 | −1.17 | ||
| Heat shock protein HSP 90-beta | Hsp90ab1 | 1.82 | 1.02 | ||
| 40S ribosomal protein S11 | Rps11 | 1.8 | 1.04 | ||
| Tubulin α-1B chain | Tuba1b | 1.79 | 1.08 | ||
| Annexin A2 | Anxa2 | 1.78 | 1.87 | 1 | ns |
| ADP/ATP translocase 1 | Slc25a4 | 1.77 | 1.13 | ||
| Poly(rC)-binding protein 1 | Pcbp1 | 1.77 | 1.01 | ||
| Profilin-2 | Pfn2 | 1.76 | 1.09 | ||
| Endoplasmin | Hsp90b1 | 1.74 | 1.05 | ||
| Apolipoprotein A-I | Apoa1 | 1.72 | −1.22 | ||
| Vimentin | Vim | 1.69 | 1.93 | 1.03 | 0.75 |
| Superoxide dismutase [Cu-Zn] | Sod1 | 1.68 | 1.44 | −1.04 | ns |
| Elongation factor 2 | Eef2 | 1.66 | 1.06 | ||
| Rho GDP-dissociation inhibitor 1 | Arhgdia | 1.62 | 1.09 | ||
| Lamin-B1 | Lmnb1 | 1.61 | 1.09 | ||
| Actin, cytoplasmic 2 | Actg1 | 1.6 | 1.1 | ||
| Serum albumin | Alb | 1.6 | −1.21 | ||
| Heat shock 70 kDa protein 1B | Hspa1b | 1.58 | – | ||
| 60S ribosomal protein L14 | Rpl14 | 1.57 | −1.15 | 0.75 | |
| 40S ribosomal protein S14 | Rps14 | 1.56 | 1.05 | ||
| Histone H4 | Histh4 | 1.56 | – | ||
| Protein disulfide-isomerase | P4hb | 1.56 | 1.08 | ||
| ADP/ATP translocase 2 | Slc25a5 | 1.55 | −0.99 | ||
| Splicing factor, proline- and glutamine-rich | Sfpq | 1.53 | 1.03 | ns | |
| ATP synthase subunit delta, mitochondrial | Atp5d | 1.52 | 1.78 | 1.02 | ns |
| Ig κchain C region | Igk-C | 1.48 | – | ||
| Tubulin β-5 chain | Tubb5 | 1.47 | 1.11 | ||
| Actin, γ-enteric smooth muscle | Actg2 | 1.46 | 1.13 | ||
| 60S ribosomal protein L10 | Rpl10 | 1.45 | 1.04 | ||
| 60S ribosomal protein L28 | Rpl28 | 1.45 | 1.02 | ||
| Elongation factor 1-α1 | Eef1a1 | 1.42 | 1.07 | ||
| Phosphoglycerate mutase 1 | Pgam1 | 1.42 | 1.01 | ||
| Peptidyl-prolyl cis-trans isomerase A | Ppia | 1.41 | 1.08 | ||
| Nucleophosmin | Npm1 | 1.4 | 1.06 | ||
| Calmodulin | Calm3;Calm1;Calm2 | 1.38 | −1.04;1.07;1.03 | ||
| Heat shock cognate 71 kDa protein | Hspa8 | 1.37 | −1.01 | ||
| 40S ribosomal protein S20 | Rps20 | 1.35 | 1.01 | ||
| 60 kDa heat shock protein, mitochondrial | Hspd1 | 1.35 | 1.12 | ||
| 60S ribosomal protein L18 | Rpl18 | 1.35 | 1.09 | ||
| ATP-citrate synthase | Acly | 1.35 | 1.17 | ||
| 40S ribosomal protein S3 | Rps3 | 1.34 | 1.04 | ||
| ATP synthase subunit β, mitochondrial | Atp5b | 1.34 | 1.01 | ||
| 60S ribosomal protein L13 | Rpl13 | 1.33 | 1.02 | ||
| 40S ribosomal protein S8 | Rps8 | 1.32 | 1.01 | ||
| ATP synthase subunit α, mitochondrial | Atp5a1 | 1.32 | 1.06 | ||
| Heterogeneous nuclear ribonucleoprotein A3 | Hnrnpa3 | 1.32 | 1.11 | ||
| Histone H1.2 | Hist1h1c | 1.3 | 1 | ||
| Stress-induced-phosphoprotein 1 | Stip1 | 1.3 | −0.99 | ||
| Hemoglobin subunit β-1 | Hbb-b1 | 0.36 | 1.06 | ||
| Fatty acid synthase | Fasn | 0.37 | −1.18 | ||
| Hemoglobin subunit β-2 | Hbb-b2 | 0.38 | 1.06 | ||
| Glutathione S-transferase Mu 1 | Gstm1 | 0.85 | 0.86 | −1.38 | 0.5 |
| Peroxiredoxin-1 | Prdx1 | 1.18 | 1.3 | 0.99 |
a Marked here is the H/L protein ratio, as measured through the ICPL analysis.
b Marked here is the H/L ratio of six selected proteins, as measured through AQUA peptide analysis.
c Shown in this column are the mutant/control mRNA ratios revealed by our Affymetrix (Affy) analysis (Ref (10)). Note that no statistically significant difference in abundance of mRNAs is observed between mutants and controls. The mark (–) indicates that the mentioned protein corresponds to multiple EG identifiers and thus, we were not able to sort out the correct corresponding Affymetrix probeset value.
d Marked here is the mutant/control mRNA ratio as measured through quantitative Real-Time PCR (also see Fig. 4). ‘ns’ indicates no significant difference.
Independent Validation of Testicular Protein Levels by AQUA Peptide Analysis
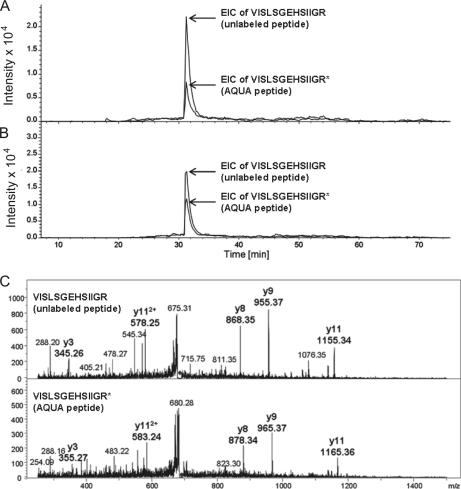
To confirm the differential expression levels we detected through ICPL analysis, we selected six proteins, namely four up-regulated (Vimentin, Atp5d, Anxa2, and Sod1) in mutant testes and two unaffected (Prdx1 and Gstm1), for further validation by means of AQUA peptide analysis, an MS-based technique for the absolute quantification of proteins. AQUA peptides are chemically synthesized isotope-labeled peptides whose sequences correspond to proteolytic peptides of the proteins to be quantified. They were spiked into the sample in known quantities before LC-MS/MS analysis. Absolute quantification was achieved by comparing the signals corresponding to AQUA and proteolytic peptides (an example for SOD-1 is shown in Fig. 3). Absolute quantities determined in mutant and control samples were used to calculate mutant-to-control ratios, which we found to be close to those obtained with relative quantification by ICPL for all six proteins, a result that validated our ICPL results. Note that all AQUA values are indicated in Table I.
Fig. 3.
AQUA peptide analysis validated the ICPL results: shown here is the example of SOD-1. Extracted ion chromatograms (EICs) were reconstituted by extracting the signals of y3, y8, y9, y11, and y112+ sequence ions of the VISLSGEHSIIGR unlabeled peptide and the VISLSGEHSIIGR* AQUA peptide used for the quantification of SOD1 by multiple reaction monitoring in mutant (A) and control (B) testes and their corresponding MS/MS spectra (C). The EICs obtained for AQUA peptides used as internal standards in (A) and (B) correspond to an injected amount of 30 fmol.
Protein Up-regulation Upon Sertoli-cell Loss of Dcr is not Accompanied by mRNA Alterations
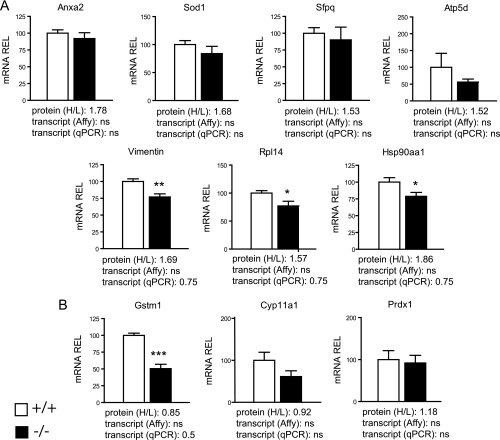
Next, we went on to assess whether the changes we measured in protein output were the result of changes in mRNA expression levels. Forty-seven out of 50 up-regulated proteins showed no difference in their mRNA expression levels between controls and mutants, as evidenced by their expression levels measured on the Affymetrix microarray (for the remaining three proteins, we were not able to sort out the corresponding Affymetrix probeset value, because they match to multiple EG identifiers) (Table I), thus suggesting that they represent genes whose expression is controlled at the translational level. In fact, we further selected seven up-regulated proteins (among them were those that had been validated by AQUA peptide analysis) and performed RealTime qPCR for their respective genes on P0 control and mutant whole testes: these genes showed either no difference (Anxa2, Sod1, Sfpq, and Atp5d), or, interestingly, a ∼25% reduction in their mRNA levels (Vimentin, Rpl14, and Hsp90aa1), whereas their respective proteins were >1.3 times more abundant in mutant testes (Fig. 4A). We also selected three unaffected proteins (Gstm1, Cyp11a1, and Prdx1) to evaluate their mRNA levels, and found that the mRNA expression levels of Cyp11a1 and Prdx1 remained unaffected, whereas that of Gstm1 showed a ∼50% reduction in mutant testes (Fig. 4B). Taken together, these findings demonstrate that loss of Dcr and miRNAs in SCs has a significant impact on testicular proteins, without however affecting the amounts of the corresponding mRNAs.
Fig. 4.
Real-Time qPCR revealed that protein up-regulation upon SC-Dcr loss is not accompanied by alterations at the mRNA level. A, Shown here are four genes (Anxa2, Sod1, Sfpq, and Atp5d) whose mRNA levels are not significantly different between controls and mutants (qPCR and Affy), but whose respective proteins are up-regulated in mutant testes (H/L>1.3), and three genes (Vimentin, Rpl14, Hsp90aa1) whose mRNA levels show a ∼25% reduction (qPCR), but whose respective proteins are up-regulated in mutant testes (H/L>1.3). B, Gstm1, Cyp11a1 and Prdx1 protein levels were unaffected and so were their transcript levels, except for Gstm1, which showed a 50% reduction in mutant testes. For each gene, the mutant/control (H/L) protein ratio revealed by ICPL analysis, as well as their mutant/control mRNA (transcript) ratio revealed either through microarray (Affy) or qPCR is given beneath the graph. Results are mean ± S.E. (n = 6 animals/group), *p < 0.05, **p < 0.01, ***p < 0.001 versus controls; ns: not significant. Primers used are listed on supplemental Table S2. +/+ and −/− are abbreviations for control Dcrfx/fx, and mutant Dcrfx/fx;MisCre animals respectively. REL: relative expression levels.
Several microRNAs are Expressed in Immature SCs
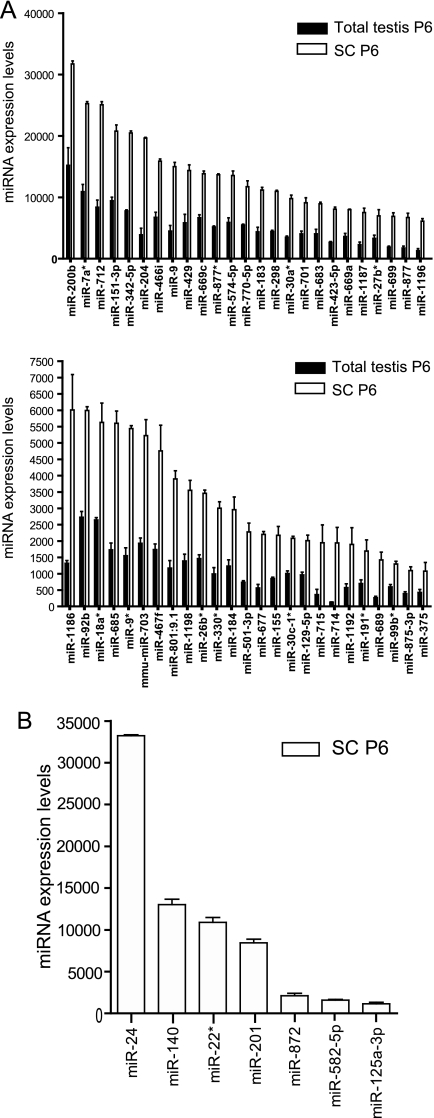
The fact that the protein up-regulation we measured is not accompanied by an mRNA up-regulation in mutant testes strongly suggested that their respective genes are most likely to represent direct SC-miRNA targets regulated at the translational, and not the transcriptional level. To further investigate this hypothesis, we first set out to characterize the miRNA expression profile of SCs using a miRNA microarray. For this purpose, we analyzed the expression of 656 murine miRNAs in purified populations of testicular cells, namely immature P6 and mature P17 SCs, in adult Leydig cells, in different types of germ cells (spermatogonia A, spermatogonia B, and intermediate, pachytene spermatocytes and spermatids), as well as in immature P6 whole testes, using the Illumina microRNA Expression Profiling System. We identified a set of 382 miRNAs expressed in SCs (supplemental Table 5). Of these, we found that 50 are expressed more than two times more abundantly in immature P6 SCs in comparison to P6 whole testes (shown in two graphs in Fig. 5A), a finding that could suggest a potential biological role for these miRNAs in SCs, without however neglecting the potential role of other SC-expressed miRNAs.
Fig. 5.
miRNAs expressed in SCs. A, Shown here in two graphs are the expression profiles of the 50 SC-enriched miRNAs, in descending order of expression in P6 SCs. The entire list of 382 miRNAs expressed in SCs, along with their sequences is provided in supplemental Table 5. B, Shown here are the expression profiles of 7 SC-expressed miRNAs predicted to have a putative binding site in the 3′UTR of Sod-1. Results are represented as mean values of three biological replicates ±s.d. miRNA nomenclature is based on the Sanger miRBase, v12.
The 3′UTRs of Genes Up-regulated Upon SC-Dcr Loss at the Protein Level are Enriched for SC-miRNA Target Sites
Having in hand these SC-expressed miRNAs, and given that miRNAs most frequently bind on regions of an mRNA's 3′UTR, we went on to assess whether the 3′UTRs of transcripts coding for up-regulated proteins are actually enriched for SC-miRNA target sites. First, we considered as a criterion for successful miRNA binding only the presence of a seed (the minimum sequence of nucleotides required for successful miRNA binding on its target) in the transcripts' 3′UTR (supplemental Fig. 1), but found that none of the seed sequences were significantly enriched in genes coding for up-regulated proteins in mutant testes, when compared with the unaffected proteins (One-sided Fisher test p value = 0.40, Table II). However, when we further refined our query to target sites bearing a conserved-in-placental-species seed sequence, we found that genes coding for up-regulated proteins in mutant testes were slightly, yet significantly, enriched in SC-miRNA target sites (One-sided Fisher test p value = 0.0128, Table II). Interestingly, nonsignificant enrichments were observed when considering either energetically favorable miRNA-mRNA duplexes (ΔG duplex, One-sided Fisher test p value = 0.0721, Table II) or a favorable target site sequence context (One-sided Fisher test p value = 0.71, Table II). Altogether, although the low statistical significance of the enrichment is acknowledged, these findings tend to suggest that genes coding for up-regulated proteins upon SC-Dcr loss are likely to represent direct SC-miRNA targets.
Table II. SC-miRNA target site enrichment among genes coding for proteins deregulated upon SC-Dcr loss. Shown here are the % of up-regulated proteins (up) versus those unaffected (equal) upon SC-Dcr loss that were enriched in SC-miRNA target sites, when considering either only the presence of a seed sequence (seeds), an energetically favorable miRNA-mRNA duplex (ΔG duplex), a favorable target site sequence context (Targetscan) or the conservation of the seed sequence (conservation). The numbers in parenthesis indicate the number of genes taken into consideration in each fraction.
| Fractions | Proteome fractions with an H/L threshold of 1.3 |
||
|---|---|---|---|
| Up (59) |
Equal (96) |
||
| % targeted | Enrichment (p value) | % targeted | |
| Seeds | 93.2 | 0.40 | 90.6 |
| ΔG duplex | 77.8 | 0.072 | 65.2 |
| Targetscan | 74.6 | 0.71 | 77.1 |
| Conservation | 78 | 0.013 | 59.4 |
SOD-1, a Protein Up-regulated Upon SC-Dcr Loss, is a Direct Target of Three SC-expressed miRNAs
Next, we selected Sod-1, one of the genes we found to be up-regulated at the protein level, in order to evaluate its direct post-transcriptional targeting by SC-miRNAs. Sod-1 was selected because of its potential biological interest: SOD-1 is a Cu-Zn superoxide dismutase that catalyzes the reaction 2O2− + 2H+ → O2 + H2O2, thus protecting cells from oxidative damage (reviewed in (35)). Perturbation of this reaction's equilibrium may lead to oxidative damage; indeed, it has been shown that overproduction of SOD-1 can cause increased oxidative damage resulting in enhanced cell death by apoptosis (for example see (36, 37)). We thus reckoned that Sod-1 would be an interesting candidate gene for the explanation of the testicular degeneration phenotype we observed. Using the three prediction models described in the Experimental Procedures, we predicted one putative target site for each of seven SC-expressed miRNAs, namely miR-125a-3p, miR-140, miR-24, miR-201, miR-22*, miR-872, and miR-582–5p, on the 3′UTR of Sod-1. Of these, miR-24 showed the strongest expression in P6 SCs, miR-872 much lower, and miR-125a-3p the lowest (Fig. 5B), although the latter was slightly enriched in P6 SCs in comparison to P6 whole testis (data not shown). These three miRNAs were selected for further analysis: The Sod-1 3′UTR was cloned downstream of the Firefly luciferase coding sequence in the pTal plasmid. A dual-luciferase assay was then performed in HEK293T cells by transfecting the pTal plasmid harboring or not the 3′UTR of Sod-1, along with one of the three precursor miRNAs and a transfection control vector expressing the Renilla luciferase coding sequence (pRL) (Fig. 6A). Transfection of the empty pTal and pRL plasmids in the presence of pre-miR-125a-3p or pre-miR-872 did not significantly alter the relative luciferase levels (Figs. 6B and 6C, dark gray bars). However cotransfection with pre-miR-24 caused an increase in the relative luciferase levels (Fig. 6D, dark gray bar), which is most likely due to the fact that the Renilla CDS harbors one putative miR-24 binding site: binding of miR-24 causes a decrease in the Renilla levels, thus an increase in the Firefly/Renilla ratio. When cloning the wild-type (wt) Sod-1 3′UTR downstream of the Firefly luciferase coding sequence, the Firefly/Renilla luciferase levels showed a significant decrease for all three miRNAs (Figs. 6B, 6C and 6D, light gray bars). To further confirm the specificity of these effects, we generated three Sod-1 3′UTR constructs, each harboring a mutated seed sequence for the three SC-expressed miRNAs (Fig. 6E) and repeated the luciferase assays as described above. For all three miRNAs, seed mutation abolished the miRNA repressive effect on the Sod-1 3′UTR (Figs. 6B, 6C, and 6D, white bars), thus strongly suggesting that the 3′UTR of Sod-1 is a direct target of these three SC-expressed miRNAs.
DISCUSSION
Although the primary effect of animal miRNAs is thought to occur at the level of translational repression, most studies until today have measured their effect at the mRNA level, mostly through DNA microarrays. We ourselves measured the impact of an SC-Dcr loss at the mRNA level by performing an Affymetrix microarray on P0 and P5 whole testes and measured several transcriptome alterations occurring in mutant testes (10). The effect of miRNAs on protein output has been more difficult to study in a high-throughput manner, and only recently two groups performed such an analysis, showing that a single miRNA can repress the production of hundreds of proteins, yet at a relatively mild level (21, 22). Additional studies have unraveled similar results, although at a smaller scale (for example see (38, 39)). Here, in an effort to reveal the molecular factors whose deregulation causes infertility in mice lacking SC-Dcr, we have used quantitative mass spectrometry to measure the effect of SC-Dcr and subsequent miRNA loss on the testicular proteome. To our knowledge, this is the first report using ICPL technology to study in vivo differential protein expression in the testis. We report the quantification of 130 nonredundant proteins, of which 50 are up-regulated more than 30% in mutant testes, whereas the remaining are unaffected (77 proteins) or down-regulated (3 proteins). We find that protein up-regulation is mild, that is, never exceeding a 2-fold change, yet the fact that from 53 differentially expressed proteins, the vast majority (50 proteins) is up-regulated in mutant testes further reinforces the notion that translational repression, at least in our system, is one of the primary effects of animal miRNAs.
Indeed, one striking finding of our study is the large proportion of proteins up-regulated upon SC-Dcr loss (50/130 or ∼38%). Certainly, we acknowledge that extrapolation to the whole proteome would be speculative, because our mass spectrometry analysis quantified only a small set of highly expressed proteins, however, it would tend to suggest that SC-miRNAs have a significant impact on testicular translational control. In itself, this is not unexpected: recently, the global impact of miRNAs on protein output was investigated by quantitative mass spectrometry and showed that a single miRNA can directly repress translation of hundreds of genes (21, 22). Taking into account that SCs express hundreds of miRNAs and that in silico analyses have predicted several thousands of protein-coding genes to be potential targets of hundreds of miRNAs, our own findings suggest that miRNAs play a broad role in the fine-tuning of protein synthesis in SCs. They are thereby essential for their survival and maturation and eventually for the entire male reproductive function.
A careful comparison of our own data to those of the two above-mentioned studies, (21, 22) reveals some interesting points worth of discussion. First, the Selbach and Baek papers conclude that only targets translationally repressed by more than a third also display detectable mRNA alterations, whereas those modestly repressed show little or no change at the mRNA level. The protein up-regulation we measure in our system, which falls within a 1.3–2-fold range, is actually not accompanied at all by alterations at the mRNA level. This finding could be interpreted as a more significant miRNA-mediated translational control in SCs than in other systems, although, again, extrapolation to the whole proteome must be done with caution. We should also mention here that, in comparison to our study, the fact that the Selbach and Baek papers report the identification of ∼3000 proteins, is likely because of the following reasons: (a) They used stable isotope labeling with amino acids in cell culture (SILAC) a technique not applicable to tissues, in which trypsin cleavage occurs following both lysine and arginine residues, thus generating numerous peptides and increasing the chances of protein identification and quantification; in our study, ICPL labeling prevents trypsin cleavage following lysine residues, allowing cleavage to occur only next to arginine residues, therefore the number of generated peptides is significantly smaller; (b) The mass spectrometer used in the Selbach and Baek papers was an LTQ-Orbitrap instrument, which is of higher performance and resolution than the one we used.
It would be worth noting at this point that, for purely technical reasons, our starting material for the mass spectrometry analysis was whole testis protein extracts, whereas depletion of miRNAs was performed uniquely in SCs. Thus, the presence of a heterogeneous population of cells in P0 testes probably masks the true impact on the protein output of SCs because of the dilution by proteins originating from other cell populations. The use of purified SCs would certainly further refine our results and thereby unravel novel, or additional, molecular targets that could explain the observed testicular degeneration and eventual infertility caused by the loss of SC-Dcr and miRNAs. In fact, as a first step toward this direction, we used purified wild-type SCs to perform a miRNA expression profiling analysis. This allowed us to unravel several SC-expressed miRNAs that we then used to assess whether the transcripts coding for up-regulated proteins upon SC-Dcr loss are enriched for SC-miRNA target sites. The enrichment was significant only when taking into consideration seed conservation among placental species. However, neither an energetically favorable miRNA-mRNA duplex, nor a favorable target site sequence context yielded a significant SC-miRNA target site enrichment. This could be explained by the fact that the ΔG duplex feature is based on an RNA-only model and that more importantly, the sequence context parameters are evaluated based on RNA-microarrays (32). As described above, the differences in protein level we measured here are because of translational differences and not mRNA alterations. Prediction features were described to be differentially relevant at each step of the RNAi pathway (40), therefore, models for target prediction trained on the mRNA level are expected to be less accurate when no mRNA degradation is involved. This might thus explain our insignificant target site enrichment when considering a favorable sequence context or miRNA-mRNA duplex. In contrast, the conservation feature captures a “blinder” information (i.e. without a regulation model), which allows us to significantly isolate miRNA effects on the measured proteome.
Among the proteins up-regulated in mutant testes, SOD-1 retained our attention. We reckoned that because SOD-1 is a Cu/Zn-superoxide dismutase whose overproduction causes increased oxidative damage resulting in enhanced cell death through apoptosis (for example see (36, 37)), its up-regulation could be detrimental for cell survival, and thereby account, at least partially, for the testicular degeneration we observed upon SC-Dcr loss. By performing an in vitro dual-luciferase assay, we found the 3′UTR of Sod-1 to be directly targeted by three SC-expressed miRNAs: miR-125a-3p, miR-872 and miR-24. Of note, because SOD-1 is present in both SCs and in all types of GCs (41), the effect could be Sertoli-cell autonomous or not. In either case, taking also into consideration the ∼3-fold mRNA up-regulation of Bcl2l11, a facilitator of apoptosis we previously detected at P0 (10), we are tempted to believe that two independent, miRNA-mediated, cell-death molecular mechanisms are at the origin of -at least part of- the observed testicular degeneration. It would certainly be interesting to find out whether the observed SOD-1 increase upon SC-Dcr loss at birth is maintained at later stages of testis development. If this were indeed true, a chronic oxidative damage could most likely explain the almost complete loss of testicular structures upon aging.
An additional interesting issue raised by our findings is whether the effects on protein output of mutant testes are because of direct SC-miRNA-mediated inhibition of protein synthesis, or because of indirect repressive mechanisms. Several indications suggest that a direct effect of miRNAs on target genes may account for most of the proteome alterations. First, we performed all of our analyses at an early stage of testis development (P0), when miRNAs are beginning to be depleted from SCs and when no morphological and histological alterations are yet detected, a fact that would tend to suggest a direct miRNA effect because of a rather restricted time window for secondary, indirect effects to occur. Second, although we did observe transcriptional alterations in mutant testes, the deregulated genes at P0 represent only 0.5% of the total transcriptome (145 deregulated probe sets versus 29.000 probe sets considered to be expressed in our Affymetrix analysis (10)), and most importantly, do not account for any protein deregulation, thus suggesting that, any alterations at the protein level are most likely to represent direct effects in our system. Finally, the fact that the 3′UTR of transcripts coding for up-regulated proteins are enriched, although slightly, for SC-miRNA target sites, and that one of these proteins, SOD-1, is, at least in vitro, directly targeted by three SC-expressed miRNAs, points toward a direct negative miRNA effect on protein synthesis.
Overall, with this study, we unravel a molecular mechanism that could partially explain the observed testicular degeneration caused by SC-Dcr and miRNA loss. Most importantly, we show, for the first time to our knowledge, that miRNAs have quite a significant impact on the testicular protein output and thus further reinforce the current notion of animal miRNAs exerting their primary negative effect at the translational level.
Acknowledgments
We would like to thank Nicolas Veillard for excellent technical assistance; all the members of the Nef Laboratory for critical comments and discussion on the manuscript; all members of the Proteomics Core Facility Biogenouest for valuable assistance during the mass spectrometry experiments.
Footnotes
 This article contains supplemental Fig. S1 and Tables S1 to S5.
This article contains supplemental Fig. S1 and Tables S1 to S5.
1 The abbreviations used are:
- GCs
- germ cells
- DCR
- Dicer
- ITMS
- ion trap mass spectrometry
- ICPL
- isotope-coded protein labeling
- miRNA
- microRNA
- SCs
- Sertoli cells.
REFERENCES
- 1. Cooke H. J., Saunders P. T. (2002) Mouse models of male infertility. Nat. Rev. Genet 3, 790–801 [DOI] [PubMed] [Google Scholar]
- 2. Jegou B. (1992) The Sertoli cell. Clin. Endocrinol Metab. 6, 273–311 [DOI] [PubMed] [Google Scholar]
- 3. Jégou B. (1993) The Sertoli-germ cell communication network in mammals. Int. Rev. Cytol. 147, 25–96 [PubMed] [Google Scholar]
- 4. Chen C., Ouyang W., Grigura V., Zhou Q., Carnes K., Lim H., Zhao G. Q., Arber S., Kurpios N., Murphy T. L., Cheng A. M., Hassell J. A., Chandrashekar V., Hofmann M. C., Hess R. A., Murphy K. M. (2005) ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436, 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J., Pandolfi P. P. (2004) Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Gen. 36, 653–659 [DOI] [PubMed] [Google Scholar]
- 6. Meng X., Lindahl M., Hyvönen M. E., Parvinen M., de Rooij D. G., Hess M. W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M., Pichel J. G., Westphal H., Saarma M., Sariola H. (2000) Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493 [DOI] [PubMed] [Google Scholar]
- 7. Braun R. E. (1998) Post-transcriptional control of gene expression during spermatogenesis. Sem. Cell Develop. Biol. 9, 483–489 [DOI] [PubMed] [Google Scholar]
- 8. Hayashi K., Chuva de Sousa Lopes S. M., Kaneda M., Tang F., Hajkova P., Lao K., O'Carroll D., Das P. P., Tarakhovsky A., Miska E. A., Surani M. A. (2008) MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3, e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maatouk D. M., Loveland K. L., McManus M. T., Moore K., Harfe B. D. (2008) Dicer1 is required for differentiation of the mouse male germline. Biol. Reprod. 79, 696–703 [DOI] [PubMed] [Google Scholar]
- 10. Papaioannou M. D., Pitetti J. L., Ro S., Park C., Aubry F., Schaad O., Vejnar C. E., Kühne F., Descombes P., Zdobnov E. M., McManus M. T., Guillou F., Harfe B. D., Yan W., Jégou B., Nef S. (2009) Sertoli cell Dicer is essential for spermatogenesis in mice. Develop. Biol. 326, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papaioannou M. D., Nef S. (2010) microRNAs in the Testis: Building Up Male Fertility. J. Androl. 31, 26–33 [DOI] [PubMed] [Google Scholar]
- 12. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 13. Rigoutsos I. (2009) New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 69, 3245–3248 [DOI] [PubMed] [Google Scholar]
- 14. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 15. Vasudevan S., Tong Y., Steitz J. A. (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318, 1931–1934 [DOI] [PubMed] [Google Scholar]
- 16. Place R. F., Li L. C., Pookot D., Noonan E. J., Dahiya R. (2008) MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U. S. A. 105, 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A. E. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563 [DOI] [PubMed] [Google Scholar]
- 18. Giraldez A. J., Mishima Y., Rihel J., Grocock R. J., Van Dongen S., Inoue K., Enright A. J., Schier A. F. (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79 [DOI] [PubMed] [Google Scholar]
- 19. Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 20. Vinther J., Hedegaard M. M., Gardner P. P., Andersen J. S., Arctander P. (2006) Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucl. Acids Res. 34, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 23. Schmidt A., Kellermann J., Lottspeich F. (2005) A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 5, 4–15 [DOI] [PubMed] [Google Scholar]
- 24. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brun V., Dupuis A., Adrait A., Marcellin M., Thomas D., Court M., Vandenesch F., Garin J. (2007) Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol. Cell Proteomics 6, 2139–2149 [DOI] [PubMed] [Google Scholar]
- 26. Peirson S. N., Butler J. N., Foster R. G. (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucl. Acids Res. 31, e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toebosch A. M., Robertson D. M., Klaij I. A., de Jong F. H., Grootegoed J. A. (1989) Effects of FSH and testosterone on highly purified rat Sertoli cells: inhibin alpha-subunit mRNA expression and inhibin secretion are enhanced by FSH but not by testosterone. J. Endocrinol. 122, 757–762 [DOI] [PubMed] [Google Scholar]
- 29. Bellvé A. R. (1993) Purification, culture, and fractionation of spermatogenic cells. Meth. Enzymol. 225, 84–113 [DOI] [PubMed] [Google Scholar]
- 30. Manna P. R., Tena-Sempere M., Huhtaniemi I. T. (1999) Molecular mechanisms of thyroid hormone-stimulated steroidogenesis in mouse leydig tumor cells. Involvement of the steroidogenic acute regulatory (StAR) protein. J. Biol. Chem. 274, 5909–5918 [DOI] [PubMed] [Google Scholar]
- 31. Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. (2007) The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 32. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karolchik D., Kuhn R. M., Baertsch R., Barber G. P., Clawson H., Diekhans M., Giardine B., Harte R. A., Hinrichs A. S., Hsu F., Kober K. M., Miller W., Pedersen J. S., Pohl A., Raney B. J., Rhead B., Rosenbloom K. R., Smith K. E., Stanke M., Thakkapallayil A., Trumbower H., Wang T., Zweig A. S., Haussler D., Kent W. J. (2008) The UCSC Genome Browser Database: 2008 update. Nucl. Acids Res. 36, D773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sarioglu H., Brandner S., Jacobsen C., Meindl T., Schmidt A., Kellermann J., Lottspeich F., Andrae U. (2006) Quantitative analysis of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced proteome alterations in 5L rat hepatoma cells using isotope-coded protein labels. Proteomics 6, 2407–2421 [DOI] [PubMed] [Google Scholar]
- 35. Turner T. T., Lysiak J. J. (2008) Oxidative stress: a common factor in testicular dysfunction. J. Androl. 29, 488–498 [DOI] [PubMed] [Google Scholar]
- 36. Peled-Kamar M., Lotem J., Okon E., Sachs L., Groner Y. (1995) Thymic abnormalities and enhanced apoptosis of thymocytes and bone marrow cells in transgenic mice overexpressing Cu/Zn-superoxide dismutase: implications for Down syndrome. The EMBO J. 14, 4985–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanij E., Hatzistavrou T., Hertzog P., Kola I., Wolvetang E. J. (2001) Ets-2 is induced by oxidative stress and sensitizes cells to H(2)O(2)-induced apoptosis: implications for Down's syndrome. Biochem. Biophys. Res. Comm. 287, 1003–1008 [DOI] [PubMed] [Google Scholar]
- 38. Taguchi A., Yanagisawa K., Tanaka M., Cao K., Matsuyama Y., Goto H., Takahashi T. (2008) Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res. 68, 5540–5545 [DOI] [PubMed] [Google Scholar]
- 39. Yang Y., Chaerkady R., Beer M. A., Mendell J. T., Pandey A. (2009) Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 9, 1374–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hausser J., Landthaler M., Jaskiewicz L., Gaidatzis D., Zavolan M. (2009) Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 19, 2009–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gu W., Morales C., Hecht N. B. (1995) In male mouse germ cells, copper-zinc superoxide dismutase utilizes alternative promoters that produce multiple transcripts with different translation potential. J. Biol. Chem. 270, 236–243 [DOI] [PubMed] [Google Scholar]