Abstract
Thyroid hormone receptors (TRs) are members of the nuclear hormone receptor (NR) superfamily and regulate development, growth, and metabolism. Upon binding thyroid hormone, TR undergoes a conformational change that allows the release of corepressors and the recruitment of coactivators, which in turn regulate target gene transcription. Although a number of TR antagonists have been developed, most are analogs of the endogenous hormone that inhibit ligand binding. In a screen for inhibitors that block the association of TRβ with steroid receptor coactivator 2 (SRC2), we identified a novel methylsulfonylnitrobenzoate (MSNB)-containing series that blocks this interaction at micromolar concentrations. Here we have studied a series of MSNB analogs and characterized their structure activity relationships. MSNB members do not displace thyroid hormone T3 but instead act by direct displacement of SRC2. MSNB series members are selective for the TR over the androgen, vitamin D, and PPARγ NR members, and they antagonize thyroid hormone-activated transcription action in cells. The methylsulfonylnitro group is essential for TRβ antagonism. Side-chain alkylamine substituents showed better inhibitory activity than arylamine substituents. Mass spectrum analysis suggested that MSNB inhibitors bind irreversibly to Cys-298 within the AF-2 cleft of TRβ to disrupt SRC2 association.
Keywords: Coactivator Transcription, Nuclear Receptors, Protein-Protein Interactions, Thyroid, Transcription Factors
Introduction
The nuclear hormone receptors (NR)2 are transcription factors that are a major focus for drug development in the pharmaceutical industry for diverse diseases including metabolic disease, immunology, reproductive health, and cancer (1–3). Most NR modulators are ligand analogs; tamoxifen (4) and faslodex (5) bind the estrogen receptor (ER), hydroxyflutamide (6) and bicalutamide (7, 8) target the androgen receptor (AR), and rosiglitazone (9, 10) and pioglitazone (11, 12) bind the peroxisome proliferator-activated receptor γ (PPARγ). The thyroid hormone receptors (TR) belong to the NR superfamily and regulate development, growth, and metabolism (13, 14). TR has two isoforms, TRα and TRβ, encoded by distinct genes, each with alternatively spliced isoforms (15). Although these isoforms are widely expressed, they are expressed in specific patterns that vary between tissues and developmental stages (16, 17). TRα or TRβ knock-out mice display unique phenotypes, suggesting that the different TR isoforms have unique regulatory roles (18, 19). Thyroid hormone (triiodothyronine (T3)) induces the majority of transcriptional responses mediated by TR in vivo (19).
TR contains three functional domains: an amino-terminal transcription activation domain (AF-1); a central DNA binding domain (DBD); and a carboxyl-terminal ligand binding domain (LBD) that contains a T3-inducible coactivator binding domain, AF-2 (20). TR normally functions as a heterodimer with the retinoid X receptor, which is constitutively bound to thyroid-responsive elements (TRE) in the genome. In the absence of T3, TR is associated with corepressors via the AF-2 domain to cause suppression of basal transcription at TREs. Upon binding of T3, TR undergoes a conformational change that releases corepressor proteins and recruits coactivator proteins, such as the p160 steroid receptor coactivators (SRC) to activate gene transcription from the TRE (21, 22). Members of the SRC family include SRC1 (NcoA1), SRC2 (GRIP1/TIF2), and SRC3 (AIB1/TRAM1/RAC3/ACTR) (23). These proteins contain several functional domains including the nuclear receptor interaction domain and two activation domains that interact with other coregulatory proteins, CBP/p300 and CARM-1/PRMT1.
The coactivators have variable numbers of a conserved LXXLL motif (called collectively NR boxes) within their NR interaction domain that mediates binding to TR (24, 25). The NR boxes interact with hydrophobic amino acid residues located on helixes 3, 4, 5, and 12 of the AF-2 region of the TR LBD (26). Scanning surface mutagenesis reveals that six residues (Val-284, Lys-288, Ile-302, Lys-306, Leu-454, and Glu-457) are important for the coactivator interaction (27). This distinct feature makes the AF-2 domain an ideal target for developing inhibitors of TR that do not competitively inhibit endogenous hormone binding.
Although a number of small molecule modulators of TR have been developed recently, including agonists such as GC-1, TRIAC, reverse T3, and KB-141 and also antagonists such as NH-3 (28, 29), most modulates target the ligand binding pocket in the LBD. We have previously reported the discovery and characterization of a β-aminoketone series that disrupts the TR-coactivator interaction without affecting T3 binding (30–32). These time-dependent, irreversible inhibitors work by generating an enone in situ followed by a reaction between the electrophilic enone and a nucleophilic cysteine in the coactivator binding pocket. The x-ray structure of a β-aminoketone-derived enone bound to TRβ supports this hypothesis (33). TRβ is unique among the nuclear receptors in having four cysteine residues (Cys-294, Cys-298, Cys-308, and Cys-309) located in or near the coactivator binding site. Active site mutagenesis and mass spectroscopy revealed that the enones derived from this β-aminoketone series selectively attack Cys-298. Our efforts to improve the pharmacological profile of the original hit compound improved potency, reduced cytotoxicity, and eliminated hERG (human ether-a-go-go-related gene) activity that hinders use in vivo (34). We recently identified a new TRβ-SRC2 inhibitor from a quantitative high throughput screen (qHTS) using a fluorescence polarization assay (78). Here we have characterized a new class of thyroid hormone receptor-coactivator antagonists that contain a methylsulfonylnitrobenzoate (MSNB) core.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Labeling
SRC2-2 peptide was synthesized and purified by reverse phase HPLC in the Hartwell Center (St. Jude Children's Research Hospital). Texas Red- or fluorescein-maleimide (Molecular Probes) fluoroprobes were conjugated to the amino-terminal cysteine of SRC2-2 peptide as described (35).
Compound Transfer
Compounds were transferred to assay plates by a pin tool equipped with 100 H pins (V&P Scientific).
Fluorescence Polarization Assay
For the TRβ and Texas Red-SRC2-2 assays, all liquid handling was performed on a Biomek FX (Beckman Coulter). Compounds were serially diluted from 10,000 to 5 μm in DMSO into a 384-well plate (Costar). Using a pin tool, 260 nl of each compound was transferred to 20 μl of assay buffer (20 mm Tris (pH 7.4), 100 mm NaCl, 1 mm EDTA, 1 mm DTT, 10% glycerol, 0.01% Nonidet P-40, 1 μm T3, 0.6 μm hTR-LBD, 20 nm Texas Red-labeled SRC2-2 peptide, and 4% DMSO) in a black 384-well assay plate (Corning Inc.). After a 3-h equilibration, fluorescence polarization was measured using an EnVision (PerkinElmer Life Sciences) plate reader. Two independent experiments were carried out in triplicate out for each compound. β-Aminoketone SJ-1 ([3-dibutylamino]-1-(4-hexylphenyl)propan-1-one (DHPPA)), a known thyroid hormone receptor antagonist (33), was used as a positive control. For fluorescence polarization assays using other NRs, see supplemental materials.
Hormone Displacement Assay
The assay was performed as described previously (36). See supplemental materials.
AlphaScreen Assay
Using a pin tool, 260 nl of compound was added to 15 μl of assay buffer (25 mm HEPES, 100 mm NaCl, 1 mm DTT, 0.1% BSA, 0.01% Nonidet P-40, 100 nm TRβ, and 100 nm SRC2-2-PEG 2-biotin) in a white 384-well Optiplate (PerkinElmer Life Sciences), and the samples were equilibrated for 1 h. TRβ antibody (6.3 μg/ml, Santa Cruz Biotechnology, sc-32754) was incubated with 40 μg/ml protein A-acceptor beads, and 5 μl was added to each well. After 30 min, 5 μl of streptavidin donor beads was added, and after 90 min, luminescence was measured by an EnVision (PerkinElmer Life Sciences) plate reader. Two independent experiments were carried out in triplicate for each compound.
Transcription Assay
HEK293 (ATCC) cells were cultured in DMEM containing 10% FBS and maintained in 5% CO2 at 37 °C. T3 (30 nm) was used as a positive control in all assays. HEK293 cells were plated at 8 × 106 cells/dish (∼40–60% confluence) in 100-mm culture dishes in 10 ml of DMEM/F-12 (1:1 mixture, Hyclone Laboratories) containing 2.5 mm l-glutamine and 10% heat-inactivated charcoal-stripped serum (Hyclone Laboratories). After a 6-h incubation, 460 μl of transfection mixture containing 5 μg of CMV-TRβ plasmid, 15 μg of DR4 (AGGTCAcaggAGGTCA)-TRE-firefly luciferase reporter plasmid, 1.25 μg of TK-Renilla luciferase control reporter plasmid (Promega), and 64 μl of FuGENE 6 (Roche Applied Science) was added, and the cells were incubated overnight. Cells were trypsinized and added to 96-well plates (Corning) at 4 × 104 cells/well in 75 μl of DMEM/F-12 medium. Six hours after plating, serially diluted compounds in 25 μl of DMEM/F-12 medium were added to the cell culture medium. After incubation for 18 h, Dual-Glo (Promega) detection reagent was added, and luminescence was measured using an EnVision (PerkinElmer Life Sciences) plate reader. TRE-mediated luciferase activity was normalized by Renilla luciferase activity. The inhibition data were normalized to basal expression (treated with DMSO only) and fully induced expression (treated with T3 solution in DMSO). Two experiments were carried out in triplicate for each compound.
Cytotoxicity Assay
Raji, HepG2, BJ, and HEK293 (ATCC) cells were grown to 80% confluence, collected, and plated at 700–1000 cells/well in 25 μl of medium/well in 384-well plates (Costar 3712). Compounds were diluted and transferred to cells as described above, and the plates were incubated for 72 h at 37 °C in 5% CO2. CellTiter-Glo (Promega) detection reagent was added following the manufacturer's instructions, and luminescence was measured using an EnVision (PerkinElmer Life Sciences) plate reader.
RNA Extraction and RT-PCR
HepG2 cells were split into 6-well plates at a density of 1 × 106 cells/well−1 in DMEM/F-12 medium with 10% charcoal stripped serum. Twenty-four hours later, the cells were treated with T3 or a combination of T3 with compound 1, compound 12, or NH-3. 24 h after treatment, cells were harvested for RNA using RNA Stat-60 (AMS Biotechnology) following the manufacturer's instructions. The resulting RNA was treated with DNase I (Invitrogen, catalog No. 18068-015) to remove contaminating genomic DNA. Then RNA was cleaned up using Qiagen RNeasy Mini (Qiagen) following the manufacturer's instructions. Equal quantities of RNA were then reverse-transcribed using Superscript III (Invitrogen) following the manufacturer's instructions. The resulting cDNA was diluted 1:50 in nuclease-free water and used in real-time PCR reactions with the QuantiFast master mix (Qiagen) in a 7900 HT RT-PCR instrument (Applied Biosystems Inc.). The following primers were used with 18S primer mix (Qiagen, catalog No. QT00199367) and dehydrogenase kinase 4 (PDK4) primer mix (Qiagen, catalog No. QT00003325): phosphoenolpyruvate carboxykinase (PEPCK) ACGGATTCACCCTACGTGGT (forward) and CCCCACAGAATGGAGGCATTT (reverse). The expression of target genes was normalized to the expression of the 18S subunit of the ribosome. The PCR quantization was carried out using the ΔΔCt method, and data were expressed as -fold change relative to DMSO-treated controls.
Solubility
The solubility assay was carried out on a Biomek FX laboratory automation work station (Beckman Coulter). Ten μl of compound stock was added to 190 μl of 1-propanol to make a reference stock plate. Five μl from this reference stock plate was mixed with 70 μl of 1-propanol and 75 μl of PBS (pH 7.4) to make the reference plate; the UV spectrum (250–500 nm) of the reference plate was measured using a SPECTRAmax Plus plate reader (Molecular Devices). Six μl of 10 mm test compound stock was added to 600 μl of PBS in a 96-well storage plate and mixed. The storage plate was sealed and incubated at room temperature for 18 h. The suspension was then filtered through a 96-well filter plate (pION Inc.). Seventy-five μl of filtrate was mixed with 75 μl of 1-propanol to make the sample plate for UV spectroscopic analysis. A single experiment was performed in triplicate for each compound. Solubility was calculated using μSOL Evolution software based on the area under the curve (AUC) of the UV spectrum of the sample plate and the reference plate.
Permeability Assay
The parallel artificial membrane permeability assay (PAMPA) was carried out on a Biomek FX laboratory automation work station (Beckman Coulter). Three μl of test compound stock (10 mm in DMSO) was mixed with 600 μl of system solution buffer (pH 7.4 or 4) (pION Inc.) to dilute the test compound. 150 μl of diluted test compound in system solution buffer was transferred to a UV plate (pION Inc.), and the UV spectrum was measured on a SPECTRAmax Plus plate reader (Molecular Devices) to establish a reference plate. The membrane on a preloaded PAMPA sandwich (pION Inc.) was painted with 4 μl of gastrointestinal tract lipid (pION Inc.). The acceptor chamber was then filled with 200 μl of acceptor solution buffer (pION Inc.), and the donor chamber was filled with 180 μl of test compound diluted in system solution buffer. The PAMPA sandwich (donor and acceptor chambers) was assembled, placed on the Gut-Box (pION Inc.) and stirred for 30 min. The aqueous boundary layer was set to 40 μm for stirring, and the UV spectra (250–500 nm) of the donor and the acceptor chambers were read. A single experiment was performed in triplicate for each compound. The permeability coefficient was calculated using PAMPA Evolution 96 Command software (pION Inc.) based on the area under the curve of the reference, donor, and acceptor plates.
Reversibility Assay
TRβ LBD (1 μm) was incubated with DMSO, SRC2-2 peptide (100 μm), or compound 12 (100 μm) in 1 ml for 3 h in assay buffer. Samples were then dialyzed in 4 liters of assay buffer overnight using 3000 molecular weight cut-off Slide-A-Lyzer MINI dialysis units (Pierce). The protein samples were concentrated by using a 10,000 g spin filter column (Amicon Ultra, Millipore) and then quantified by Bradford analysis. The protein samples were serially diluted from 4 to 0.002 μm in assay buffer in 96-well plates. Then 10 μl of diluted protein was added to 10 μl of 40 nm SRC2 fluorescence probe in a 384-well plate. After a 3-h equilibration, the fluorescence polarization was measured using an EnVision plate reader as described above.
Intact Mass Analysis
TRβ LBD (5 μm) was incubated with compound 13 (25 μm) for 2 h in assay buffer. Samples were precipitated with cold ethanol at −20 °C for 24 h and then air-dried. The samples were desalted using a reverse phase C4 or C8 ZipTips (Millipore) and eluted with aqueous 50% acetonitrile, 2% formic acid. This eluent was ionized by static nanospray using EconoTips (New Objective) on an LCT Premier XE mass spectrometer (Waters Corp.) using positive detection mode. The resultant charge envelope was deconvoluted using a MaxEnt 1 algorithm of MassLynx version 4.0 SP4 software. A mass error of 1 Da for every 10,000 Da is expected when using this mass spectrometer.
Trypsin Digest Peptide Analysis
TRβ LBD (5 μm) was incubated with compound 13 (25 μm) for 2 h in assay buffer. Samples were precipitated with cold ethanol at −20 °C for 24 h and then air-dried. The precipitates were reduced for 1 h at 37 °C with DTT (10 mm) and alkylated for 45 min at ambient temperature with 25 mm iodoacetamide in 100 mm ammonium bicarbonate buffer (pH 8). The protein was digested with trypsin (Promega) for 12 h at 37 °C. The resulting peptide mixture was acidified to pH 3.5 with formic acid and fractionated by nanoflow reverse phase ultra high-pressure liquid chromatography on a nanoACQUITY Ultra Performance LC system (Waters Corp.). Tryptic peptides were loaded onto a Symmetry C18 precolumn (180-μm inner diameter × 20 mm, 5-μm particles; Waters Corp.) which was connected through a zero dead volume union to a BEH C18 analytical column (75-μm inner diameter × 100 mm, 1.7-μm particles; Waters Corp.). Tryptic peptides were eluted over an 86-min gradient (0–70% B in 70 min and 70–100% B in 86 min, where B = 70% acetonitrile, 0.2% formic acid, and 2 mm EDTA) at a flow rate of 250 nl/min and introduced online into a linear ion trap mass spectrometer (Thermo Electron Corp.) using electrospray ionization. Data-dependent scanning was incorporated to select abundant precursor ions for fragmentation by acquisition of a full scan mass spectrum followed by MS/MS on the 10 most abundant ions (one microscan/spectra; precursor m/z ± 1.5 Da, 35% collision energy, 30-ms ion activation, 35-s dynamic exclusion, and repeat count 2).
Data Analysis
Curves were fitted to titration-response data using GraphPad Prism 4.03 (GraphPad Software). IC50 values were obtained by fitting the data to the following equation: (sigmoidal dose response (variable slope)): y = bottom + (top − bottom)/(1 + 10∧((logIC50 − x)*Hill slope)), where x is the logarithm of concentration and y is the response.
For chemical reagents and synthesis and protein expression and purification, see supplemental materials.
RESULTS
A high throughput screen to detect novel inhibitors of the interaction of SRC2-2 with TRβ was carried out using a qHTS variant of the fluorescence polarization screen disclosed previously. The details of the qHTS and initial triage of hits have been reported.
Inhibition of TRβ-SRC2 Interaction by MSNB Series of Compounds
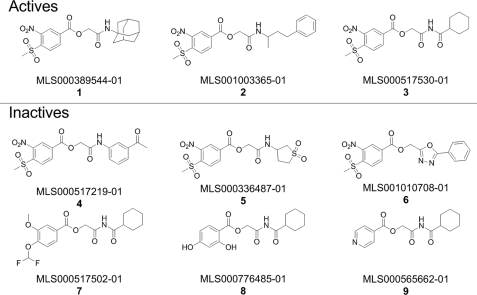
Nine MSNB analogs were identified among the compounds evaluated in the primary screen using fluorescence polarization assays3 of which three inhibited SRC2-2 peptide binding to TRβ LBD and six were inactive (Fig. 1). The MSNB scaffold contains two distinct functional groups. It has two electron withdrawing groups (methylsulfonyl and nitro) on the phenyl ring and an ester linkage with the amide group. This MSNB scaffold has only rarely been disclosed in drug discovery (37, 38). In addition, a search of the PubChem BioAssay database showed that MSNB analogs are active against few targets (PubChem AID 1339 for a ras GTPase assay and AIDs 1304, 1359, and 1861 for a neuropeptide Y receptor type 1 assay), despite having been repeatedly screened in the Molecular Libraries Program. All active compounds identified in the primary screen contained the methylsulfonyl and nitro groups on the phenyl ring and either an amide (CO-NH, 1 and 2) or an imide (CO-NH-CO, 3) bond in the structure. Aniline (4)-, cyclosulfonylamine (5)-, and oxadiazole (6)-substituted MSNB analogs showed no inhibitory activity. Replacement of the methylsulfonyl and nitro groups (7–9) also resulted in a loss of activity.
FIGURE 1.
MSNB analogs tested in the primary screen. Shown is the structure of active and inactive MSNB analogs contained in the set tested during the qHTS using the fluorescence polarization assay. Activity was defined as the ability to provide a saturated antagonistic dose response in the primary screen without significant effects on intrinsic fluorescence.
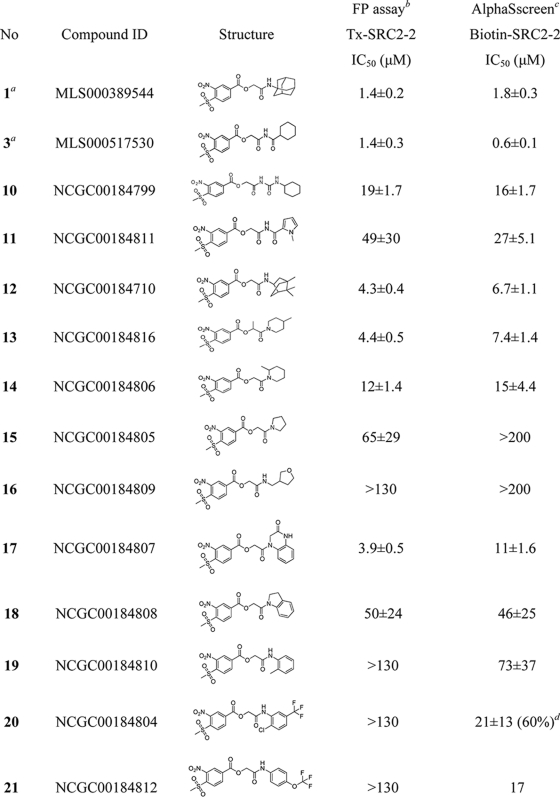
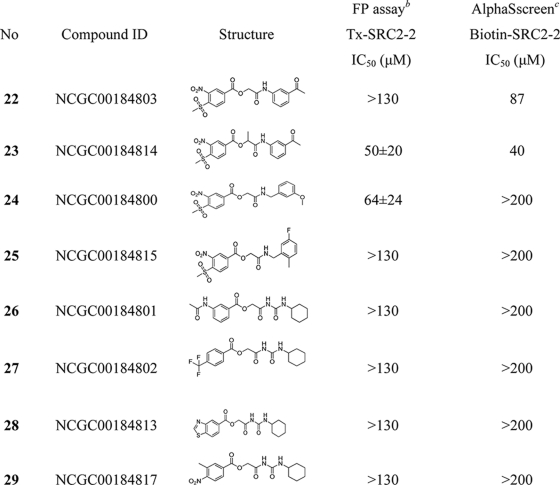
With these basic elements of the structure-activity relationship (SAR) determined from the primary quantitative screening data, 20 MSNB analogs were purchased to further characterize this series. Compounds were serially diluted 3-fold from 130 μm to 6.6 nm and evaluated in fluorescence polarization (FP) assays that measure the interaction of the TRβ LBD with an SRC2-2 fluoroprobe (30). Two fluoroprobes, labeled with Texas Red or fluorescein, were utilized in separate assays to identify false positives arising from compound interference. Compounds 1 and 3, which were identified in the high throughput screen,3 were evaluated as independent samples, and both showed IC50 values near 1 μm as detected by both fluoroprobes (Table 1 and supplemental Table 1). Within this series, the nitro and methylsulfonyl groups were essential for the inhibition of the TRβ-SRC2-2 peptide interaction. Analogs lacking either functionality (compounds 26–29) failed to inhibit the interaction. This suggests that the electrostatic or resonance properties of the ring are essential for activity. Likewise, the amide groups located distally from the MSNB ring were important for activity. For example, the carbamoylamido (10; IC50 = 19.2 μm) and pyrole-imido (11; IC50 = 49.4 μm) compounds showed weak inhibitory activity compared with the original actives (1 and 3). Bicycloheptan (12) and piperidine analogs (13 and 14) showed moderate inhibition (IC50 = 4.3, 4.4, and 11.6 μm, respectively), whereas amines such as pyrrolidine (15) and tetrahydrofuranylmethylamine (16) displayed weak or no inhibition (IC50 = 64.9 and >130 μm, respectively). Anilidoamides (19–23) and benzylamides (24 and 25) showed weak or no activity except for an oxoquinoxaline (17), which inhibited well (IC50 = 3.9 μm). Secondary aniline compounds 17 and 18 were generally active, whereas most primary aniline and benzylamine compounds were inactive. An α-methyl-substituted compound (23) showed slightly increased activity compared with the unsubstituted form (22). The SAR analysis suggested that sterically hindered amides give more favorable interactions with TRβ, which is consistent with the presence of two deep hydrophobic pockets in the targeted binding site (33).
TABLE 1.
Structures and potencies of MSNB analogs in TRB-SRC2 interaction assays
IC50 values were determined using data from two independent experiments in triplicate.
a Primary HTS hit compounds.
b Competitive fluorescence polarization assay in the presence of TRβ-LBD (0.6 μm), T3 (1 μm), and Texas Red-labeled SRC2–2 peptide (20 nm). The data were recorded after a 3-h incubation.
c Competitive AlphaScreen assay in the presence of TRβ-LBD (0.1 μm), T3 (1 μm), and biotin-labeled SRC2–2 peptide (30 nm).
d Percent efficacy is indicated within parentheses.
Biochemical Characterization of MSNB Inhibitors
The association of TRβ with SRC2-2 is ligand-dependent (39). Potentially, MSNB series members could inhibit this interaction by blocking the binding of T3 to TRβ. To test this idea, two MSNB actives (compounds 13 and 17) were evaluated in a ([125I]T3) competition assay using the TRβ LBD (36). Although unlabeled T3 inhibited [125I]T3 binding to the TRβ LBD with an IC50 value of 53 nm, compounds 13 and 17 showed no inhibition at concentrations up 100 μm (supplemental Fig. 1A). Thus, these MSNB inhibitors do not appear to act as T3 competitive antagonists.
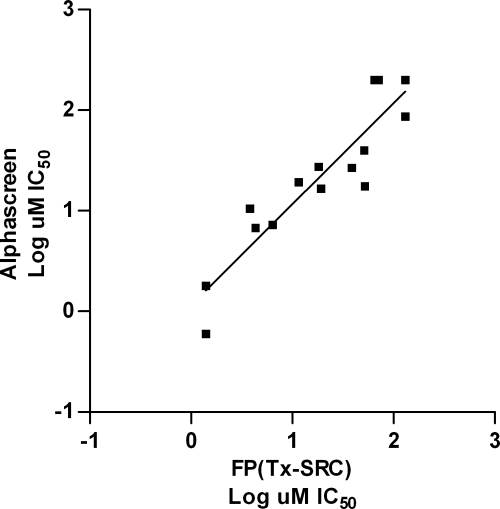
To confirm that MSNB compounds were blocking the interaction of TRβ LBD with SRC2-2, a second interaction assay using AlphaScreen methodology was employed. AlphaScreen, which is widely used for identifying protein-nucleotide, protein-protein, and protein-small molecule interactions (40–42), generates a luminescent signal that is not susceptible to the same perturbations as fluorescence-based assays. In this assay, biotinylated SRC2-2 peptide was bound to streptavidin-conjugated donor beads, and the TRβ LBD was bound to protein A-acceptor beads via an anti-TRβ antibody (supplemental Fig. 1B). When donor and acceptor beads are brought into proximity by the interaction between TRβ and SRC2-2 peptide, 680 nm light excitation produces a chemiluminescent signal. Optimization experiments indicated that 30 nm biotin-SRC2-2 peptide was the minimal concentration to yield a maximal luminescent signal that was completely competed with unlabeled peptide (supplemental Fig. 1C). Selected MSNB inhibitors were tested in the assay over a range of concentrations to determine IC50 values for each (Table 1). The IC50 values determined in the AlphaScreen assay were similar to those measured by the FP assay. A correlation plot comparing the log IC50 values determined by both assays showed very good agreement (Fig. 2, r2 = 0.84, slope = 1). However, aniline-substituted compounds (19–23) that were inactive in the FP assay showed moderate inhibitory activity (IC50 range of 17 to 87 μm) in the AlphaScreen assay. Most likely, this difference arose from the improved dynamic range of the AlphaScreen assay. Analogs (26–29), lacking either the nitro or methyl sulfonyl group, were inactive in both assays. Therefore, these groups are key elements of the active pharmacophore.
FIGURE 2.
Correlation of coactivator antagonism activity determined from FP and AlphaScreen assays. The values of each axis show log IC50 values from an FP assay (x axis) using the Texas Red-SRC2-2 peptide and an AlphaScreen assay (y axis). There is a strong correlation between the two assays.
MSNB Analogs Inhibit TRβ-controlled Transcription in Cells
MSNB series members were tested for their ability to inhibit TRβ-mediated transcription in cells. HEK293 cells were co-transfected with a CMV-TRβ expression vector, a Photonis luciferase reporter fused to a thyroid response element and a Renilla luciferase reporter, used as a control for transfection efficiency. Cells were treated with 30 nm T3 and different concentrations of compounds and incubated for 18 h, and transcriptional reporter activities were measured (Fig. 3A). MSNB analog 1 showed a concentration-dependent decrease of TRβ-mediated activity over the tested concentration range of 2.5 to 20 μm, whereas compound 3 inhibited activity by 50% only at 20 μm. Compound 6 also showed a concentration-dependent decrease in activity, although this effect was slight. Cell viability was monitored in parallel and indicated little to no cell death at the tested concentrations (Fig. 3B). Thus, the biochemical activity of the compounds was recapitulated in cellular models, albeit with ∼10-fold lower potency.
FIGURE 3.
Percent inhibition of T3-induced TRE-luciferase activity in HEK293 cells. A, T3-mediated transcription is reduced by MSNB analogs. HEK293 cells were transfected with CMV-TRβ- and DR4-TRE-driven luciferase expression vectors. The data were normalized to Renilla luciferase activity. B, cell viability based on Renilla luciferase activity. Error bars represent S.D. of two independent experiments performed in triplicate. **, p < 0.01; ***, p < 0.005.
Regulation of TR-targeted Gene Transcription by MSNB Analogs
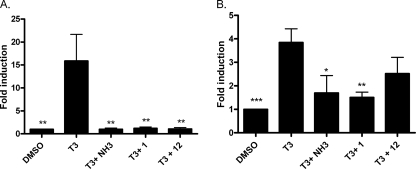
To further validate transcriptional inhibition of T3-driven genes by MSNB compounds, we performed RT-PCR on two well accepted thyroid-responsive genes: PDK4 and PEPCK, which are known to be T3-responsive in HepG2 cells (43, 44) (Fig. 4). Cells were co-treated with T3 (100 nm) and compounds 1 and 12 of NH-3, a known T3 antagonist. (45, 46) (20 μm for MSNBs and 10 μm for NH-3) for 24 h. mRNA was isolated, and real-time PCR experiments were carried out on the diluted cDNA prepared from each mRNA sample. The T3-induced PDK4 gene was fully inhibited by all compounds, whereas the PEPCK gene was moderately inhibited by NH-3 and compound 1, consistent with prior studies of NH-3 and the relative potencies of compounds (47).
FIGURE 4.
Regulation of T3-controlled genes by treatment with MSNBs in HepG2 cells. The cells were exposed to drugs (100 nm T3 and 10 μm NH-3 or 20 μm MSNB) for 20 h. RT-PCR was carried out to determine transcription levels of the PDK and PEPCK genes. The ΔΔCt method was used to calculate -fold induction of expression. A, PDK4 gene expression induced by the treatment of T3 was reduced by NH-3 and MSNBs. B, PEPCK gene expression was only modestly effected by NH-3 and compound 1. Error bars represent S.E. of two independent experiments performed in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
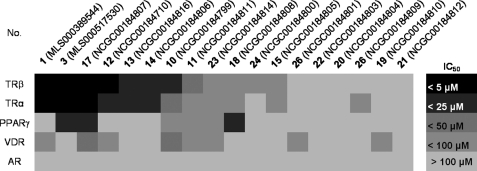
Selectivity of MSNB Analogs against Other Nuclear Hormone Receptors
We examined the specificity of MSNB series members by testing the following NR-coactivator interactions in FP assays: TRα with SRC2-2, PPARγ with vitamin D receptor-interacting protein-2 (DRIP-2), vitamin D receptor (VDR) with SRC2–3, and AR with SRC2–3. The coactivators were selected on the basis of previously published work mapping the preferred interaction partners (48, 49) A set of 18 compounds was surveyed, and the results are depicted in Fig. 5. In general, most compounds exhibited similar antagonistic potency toward both TRα and TRβ. This set of compounds had no effect on AR and only weak effects on PPARγ and VDR. However, for PPARγ, compounds 3, 17, and 18 showed moderate activity (IC50 = 22, 15, and 20.μm, respectively) with compound 18 more potent against PPARγ than against TRβ.
FIGURE 5.
Biochemical selectivity of MSNB analogs in inhibiting coregulator binding to other NR family members. The activity heat map shows IC50 values for the inhibition of coregulatory peptide binding to TRβ, TRα, PPARγ, VDR, and AR. Compounds are ordered by potency against TRβ. The coactivator and NR interactions were: SRC2-2 with TRα, DRIP-2 with PPARγ, and SRC2–3 with VDR or AR.
These MSNB analogs might inhibit the interaction of PPARγ with DRIP-2 by displacing the ligand binding upon which coactivator interaction is dependent (50). To test this idea, a scintillation proximity assay was used to detect the displacement of a radiolabeled ligand analog, [3H]rosiglitazone, from the ligand binding pocket of PPARγ (36). In this assay, the known irreversible ligand competitor GW9662 (51) and unlabeled rosiglitazone inhibited radioligand binding with IC50 values of 0.13 and 0.48 μm, respectively (Table 2). Similarly, MSNB analogs 17 competed [3H]rosiglitazone binding with IC50 values of 1.7 μm. Thus MSNBs active against PPAR act at the ligand binding site and not at the coactivator binding site.
TABLE 2.
Activity profile of FPA and SPA for PPARγ
IC50 values were determined using data from two independent experiments in triplicate. ND, not determined.
| Compound | FPA: IC50vs. DRIP-2 | SPA: IC50vs. [3H]rosiglitazone |
|---|---|---|
| GW9662 | ND | 0.13 ± 0.01 |
| Rosiglitazone | 0.8 ± 0.2 | 0.48 ± 0.04 |
| 17 | 15 ± 1.7 | 1.7 ± 0.3 |
Assessment of MSNB Cytotoxicity and Pharmacological Properties
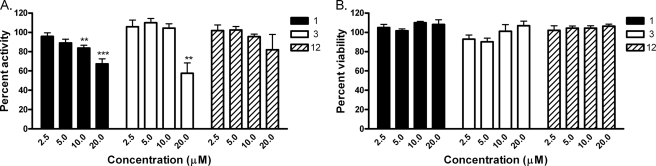
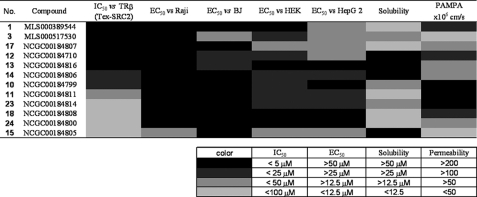
The cytotoxicity of selected inhibitors was assessed in four human cell lines: Raji, a Burkett's lymphoma-derived line; BJ, an immortalized foreskin fibroblast-derived line; HEK293, an embryonic kidney fibroblast-derived line; and HepG2, a hepatocellular carcinoma-derived line. Compounds were serially diluted and incubated with each cell line for 72 h, after which cellular ATP levels were measured using a luminescence-based detection reagent. Curve fits of the titration-response data were used to generate EC50 values (Fig. 6). In Raji and BJ cells, most compounds exhibited weak cytotoxicity with EC50 values significantly above 20 μm. Compared with BJ and Raji cells, HEK293 and HepG2 cells were more sensitive to MSNB analogs, although many showed weak cytotoxic effects (EC50 > 20 μm). Notable exceptions were compounds 15 and 17 (16 and 22 μm for HEK293, respectively) and 1, 15, and 17 (16, 17, and 16 μm for HepG2, respectively). Overall, the cytotoxicity EC50 values of MSNB series members were at least 2–3-fold higher than the IC50 values for the TRβ-SRC2 interaction measured in the FP assay.
FIGURE 6.
Cytotoxicity, solubility, and permeability of MSNB analogs. The heat map shows IC50 values for the TRβ-SRC2-2 FP assay, cell cytotoxicity (Raji, BJ, HEK293, and HepG2), solubility, and permeability. Cytotoxicity potencies were determined by the measurement of total ATP content using CellTiter-Glo® (Promega) after incubation with compound for 72 h. Solubility was measured using the Millipore method in PBS (pH 7.4). Permeability was measured using PAMPA (pH 7.4). The solubility and permeability assay conditions reflect the conditions required for activity in cell-based assays. Compounds are ordered by potency of antagonism of the TRβ and SRC2-2 interaction. The numerical values underlying this heat map are presented in supplemental Table 1.
The MSNB compounds showed good solubility and permeability properties. Compound solubility was determined in PBS buffer containing 1% DMSO, reflecting the conditions of the biochemical assays. All of the tested MSNB analogs showed moderate solubility (2–80 μm), with most active compounds freely soluble at concentrations well above their potency. In general, all aliphatic amines, except for the adamantyl amine (1), were more soluble than aromatic amines. Compound permeability across a membrane bilayer was measured by conducting a PAMPA at pH 7.4, reflecting the conditions of the cell-based assays. All of the compounds showed acceptable to good permeability (>40 × 106 cm/s) except for 15 and 16 (<10 × 106 cm/s).
Mechanism of Inhibition by MSNB Analogs
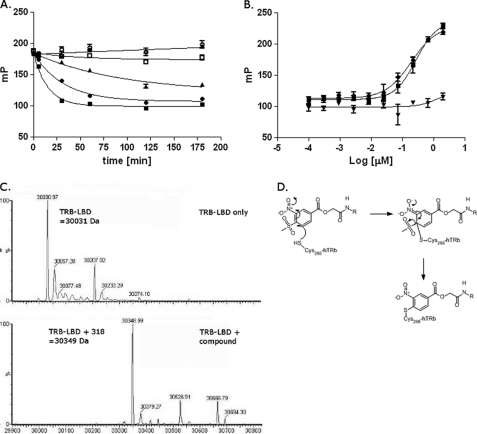
The β-aminoketone inhibitor series blocks TRβ-SRC2 interaction by irreversibly binding to the activator interaction domain via covalent coupling with Cys-298 (30, 33). The knowledge of this mechanism of TRβ inhibition and the similarity of the MSNB series to the GW series of irreversible PPARδ ligand antagonists led us to test whether the MSNB series were irreversible inhibitors as well. We selected compound 12 for these studies because it possessed good solubility (53 μm) in the assay buffers. First, we examined the concentration and time dependence of inhibition by compound 12 in the TRβ-SRC2 FP assay. Time-dependent inhibition was observed with superstoichiometric to stoichiometric concentrations of 12 (35–1.3 μm, Fig. 7A). As the concentrations of 12 were lowered, the rate of inhibition was reduced, and at substoichiometric (0.4 μm) concentrations, no significant inhibition was observed. Such time- and stoichiometry-dependent inhibition is a classical behavior of irreversible inhibitors.
FIGURE 7.
Mechanism of action of the MSNB analogs. A, time dependence of inhibition of TRβ LBD-SRC2 interaction by compound 12 at different concentrations (■, 37.7 μm; ●, 11.6 μm; ▴, 3.9 μm; □, 1.3 μm; ○, 0.4 μm; △, 0.05 μm) as measured by the FP assay using Texas Red-SRC2-2 peptide. B, measurement of TRβ LBD-SRC2 interaction following treatment with DMSO (■), SRC2-2 peptide (●), or compound 12 (▴) as measured by competitive FP assay with Texas Red-SRC2-2 peptide. C, mass spectrometry analysis of TRβ LBD alone (upper panel) or incubated with compound 13 (lower panel). D, proposed mechanism of inhibition of TRβ LBD-SRC2 interaction by MSNB analogs.
To confirm further that MSNB analogs are irreversible inhibitors, we incubated compounds with TRβ and following removal of unbound compounds, we tested whether the recovered TRβ could associate with SRC2-2 peptide. TRβ was pretreated with 100 μm 12, 100 μm SRC2-2 peptide, or DMSO for 3 h, dialyzed overnight to remove reversibly bound molecule or peptide, and then tested for inhibition of SRC2-2 fluoroprobe binding using the FP assay. Following dialysis, TRβ treated with either DMSO or unlabeled SRC2-2 bound the fluoroprobe in the FP assay, and this activity titrated with decreasing TRβ concentrations (Fig. 7B). In contrast, 12 prevented fluoroprobe binding at all TRβ concentrations tested, indicating that 12 bound irreversibly to TRβ and was not removed by dialysis. These results indicate that the MSNB series are irreversible inhibitors of TRβ.
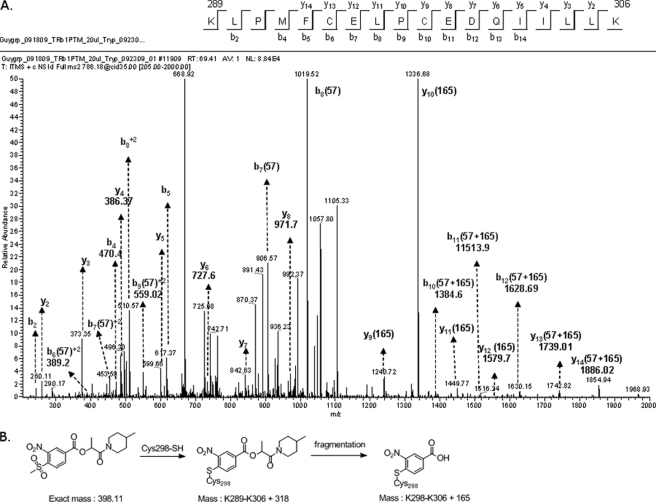
MSNB analogs might covalently attach to TRβ by the ester or aromatic sulfonyl group of MSNB providing an electrophile to react with nucleophilic residues of TRβ such as cysteine or lysine. To determine whether covalent adducts formed between MSNB and TRβ, mass spectroscopy was performed on the TRβ LBD incubated with or without 13. Spectral analysis of the TRβ LBD alone indicated a peak of 30,031 Da corresponding to the predicted mass of the TRβ LBD (Fig. 7C). However, the mass spectrum of TRβ LBD incubated with 13 showed an absence of the 30,031-Da peak and the appearance of a new peak of 30,349 Da, indicative of TRβ LBD plus a 318-Da adduct. The size of the new peak was consistent with a desulfonylated form of 13 but not a thioester adduct (30,258 Da, indicative of TRβ LBD plus 227 Da). Based upon prior work (33), the most likely mechanism is one in which the thiol group of a cysteine residue in the TRβ LBD undergoes a nucleophilic aromatic addition with 13 to displace the methylsufonyl group and generate the modified adduct (Fig. 7D). Similar reactions have been observed in other settings (52, 53).
Identification of the MSNB Covalent Binding Site on TRβ
β-Aminoketone analogs have been shown to covalently attach to one of several cysteine residues within the coactivator binding pocket of TRβ (33). To unambiguously identify the MSNB binding site in TRβ, mass spectrometry analysis was performed on tryptic digests of TRβ incubated with 13. After incubation with or without 13, TRβ was treated with iodoacetamide to alkylate unmodified cysteine residues. The protein was then digested with trypsin, and the resulting peptide mixture was analyzed by nanoscale liquid chromatography followed by tandem mass spectrometry. Analysis of the resulting spectrum indicated alkylation of 13 in a peptide spanning residues 289–306 (KLPMFCELPCEDQIILLK) (Fig. 8A). Fragmentation spectra were obtained for both [M+2]2+ and [M+3]3+ precursor ion forms, and comparison of ions from the unmodified and modified forms of this peptide suggested that Cys-298 was the site of modification. A lower than expected molecular mass shift (+165 Da) was obtained for the fragmented peptides (+318 Da) suggesting that the small molecule itself fragments under these conditions (Fig. 8B). No modifications were observed in other peptides. Thus it appears that the MSNB series acts by irreversible alkylation of the same cysteine targeted by the β-aminoketones (33).
FIGURE 8.
Mass spectrometric confirmation of Cys-298 as the site of MSNB modification. A, fragmentation spectrum collected for a peptide spanning Lys-289 to Lys-306 containing a covalent adduct with compound 13 at Cys-298 is shown. The peptide ([M+H]+:2355) was observed as doubly and triply charged species (m/z = 1178.83, z + 2; m/z = 786.00, z + 3). The peptide showed an increase of +57 D relative to the unmodified material following carbamoylation with iodoacetamide. Cys-298 was the only residue modified by 13. B, structure of compound 13 alone following adduct formation with Cys-298 and after subsequent fragmentation.
Essential Elements of MSNB Inhibition of TRβ Coactivator Interaction
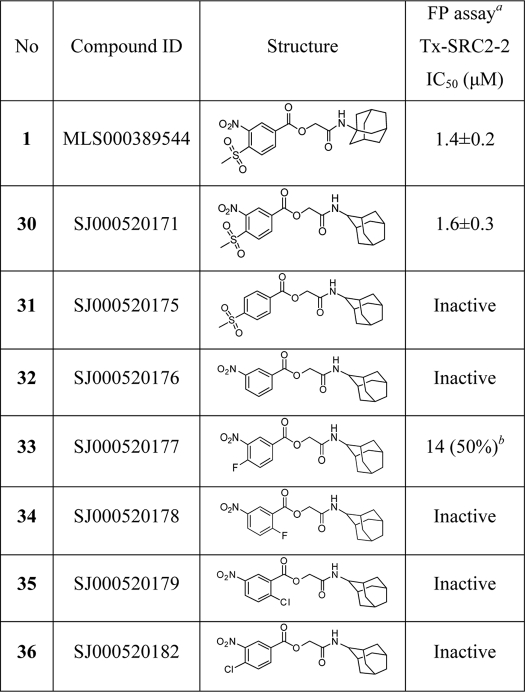
The mass spectroscopy studies demonstrated that MSNB alkylates cysteine 298 of TRβ through a substitution reaction with the aromatic carbon substituted with the methylsulfonyl group. While a methylsufonyl group is not generally considered a good leaving group in aromatic substitution reactions, one recent study reported an irreversible PPARδ antagonist that contains an alkylsulfonylpyridine structure (52). To ascertain the potential of this pharmacophore in the TRβ system, a series of analogs (30–36) was synthesized and tested in the TRβ-SRC2 FP assay (Table 3). The 2-adamantyl-substituted compound 30 was equipotent to the original active 1. However, the para-methylsulfonylbenzoate 31 and meta-nitrobenzoate 32 were both inactive, indicating that either the methylsulfonyl or nitro group alone is insufficient for activity. Halonitrobenzenes are well known as electrophiles that can modify cysteine residues (51, 54) Therefore we synthesized several halonitrobenzoate compounds and tested their activity. Most (34–36) were inactive, but 33 showed weak activity with IC50 = 14 μm and 50% efficacy. These results indicated that inhibitory activity does not track simply with the leaving group ability or electophilicity of the ring. Rather, the reaction appears to require specific activation afforded by the methylsulfonyl group.
TABLE 3.
Primary structure activity relationships for MSNB analogs 30–36
IC50 values were determined using data from two independent experiments in triplicate.
a Competitive fluorescence polarization assay in the presence of TRβ-LBD (0.6 μm), T3 (1 μm), and Texas Red-labeled SRC2–2 peptide (20 nm). The data were recorded after a 3-h incubation.
b Percent efficacy is indicated within parentheses.
DISCUSSION
NR signaling involves ligand-triggered protein-protein interaction to assemble a multiprotein complex that regulates transcription in response to hormone binding (2, 55, 56). Structurally, NRs interact with coactivators through similar surfaces, with a shallow hydrophobic groove on the surface binding to an amphipathic α-helical motif on the coactivator, burying the three leucines into the hydrophobic surface of the NR box (26, 27). Although x-ray cocrystal studies of LXXLL-containing peptides and NRs provide insights into the possible modes of binding (57, 58), it is not clear how the selectivity of NR-coactivator interactions is achieved. Several approaches have been studied to elucidate specific interactions (57, 59). Several selective nuclear receptor modulators have been developed that directly inhibit the binding interaction between receptors and coactivators without affecting ligand binding. Examples include pyrimidines for ERα (60, 61), hydroxytamoxifen for ERβ (62, 63), ketoconazoles for PXR (64, 65), and β-aminoketones for TR (30, 32, 34). However, these inhibitors exhibit either poor potency in cellular models or intolerable toxicity in vivo. For these reasons we sought to identify new chemotypes with fewer liabilities for use in animal models.
Here we have characterized a new chemotype containing an MSNB core that selectively disrupts the interaction between TRβ and SRC2. Active MSNB analogs contain two strong electron withdrawing groups (methylsulfonyl and nitro) on a phenyl ring that can act as an electrophile for nucleophilic aromatic substitution reactions with cysteine residues and an ester linkage with amide group. SAR studies demonstrated that both the methylsulfonyl and nitro groups on the aromatic ring of MSNB are critical for activity. Analogs lacking either functional group were inactive. Mechanism of action studies support the SAR findings indicating that MSNB analogs are covalent inhibitors of TRβ. MSNB analogs were time-dependent inhibitors and required stoichiometric amounts to inhibit the interaction of TRβ with SRC2. In addition, analysis of TRβ recovered from dialysis revealed that MSNB-treated TRβ was irreversibly inhibited. Intact mass analysis also showed that MSNB analogs undergo nucleophilic aromatic substitution with desulfonylation and alkylate a specific cysteine residue, resulting in an increase of mass of TRβ LBD (+318 Da).
The AF-2 pocket of TRβ has four cysteine residues (Cys-294, Cys-298, Cys-308, and Cys-309) not present in any other NR, and Cys-298 and Cys-309 are solvent-exposed (33). Mass spectroscopic analysis indicated that the major target of MSNB is Cys-298, which lies close to the AF-2 cleft. MSNB analogs do not appear to react with Cys-309; in FP assays employing either the wild-type or C309A mutant form of TRβ LBD, MSNB analogs inhibited both equally well (data not shown). In contrast, the irreversible β-aminoketone inhibitor is 50-fold less potent, in inhibiting mutant C309A TRβ LBD compared with the wild type (32), suggesting that Cys-309 is targeted by this inhibitor. In addition, MSNB does not bind to other nucleophilic residues, whereas β-aminoketone modified Lys-211 (located before helix 1) and Cys-388 (located on helix 9) at very low stoichiometry (33). These results indicate that MSNB is more specific to Cys-298 than the previously reported β-aminoketones.
Follow-up studies with structurally modified halonitrobenzoates 30–36 suggested that the ortho-oriented methylsulfonylnitrobenzoate structure is essential for activity. These findings imply that there is a very precise interaction among the inhibitor, the binding site, and the cysteine acceptor that leads to the alkylation event. Perturbation of the binding or activation of the electrophile is sufficient to disrupt inhibition. It is possible that one of several protonated lysine residues in the vicinity of the active binding site activates the sulfonyl group by forming a hydrogen bond. The amide group located distally from the MSNB ring also influences activity. Sterically hindered nitrogen in the amide bond improves inhibitory activity, suggesting that a hydrophilic nitrogen atom is not favorable in the binding pocket. This result is also consistent with β-aminoketones, where compounds containing hydrophilic heteroatoms within the hydrophobic side chain are inactive at any position other than those immediately adjacent to the phenyl ring (34). The TRβ AF-2 pocket contains a narrow hydrophobic passageway with Cys-309 at the bottom, Glu-457 and Lys-306, at the rim and Cys-298 and Lys-288 at the flank (33). Based on the predicted binding mode, the amide group is likely important in threading through the narrow hydrophobic cleft between the two leucine binding subsites within the binding pocket.
We reported previously the equilibrium affinity of the thyroid receptor to a library of potential coregulator peptides including the LXXLL motif (48). This study demonstrated that coregulator peptides bind to TRβ LBD with different affinities (Kd). SRC2-2 binds to TR in a saturable, dose-dependent manner with a measured Kd of 0.7 μm, the most tightly binding of all coregulator peptides studied. In addition, TRAP220 bound but with weaker affinity (Kd = 2.7 μm). We employed the TRβ LBD FP assay with other coactivator peptides such as TRAP220 and RAP250 peptide to determine whether MSNBs could also inhibit those coactivators binding to TR. MSNBs could inhibit the interaction between TRβ and the TRAP220 and RAP250 coactivator peptides, as well as SRC2-2 peptide (data not shown). Thus, the MSNBs broadly block the interaction of the receptor with coregulator peptides.
To investigate the cellular activity of MSNB analogs, we assayed several compounds to determine whether they blocked the TRE response in a luciferase reporter gene assay and in RT-PCR experiments with bona fide TR-regulated genes. The reporter gene assay showed that MSNB analogs inhibited T3 response gene expression driven by the artificial DR4 promoter. In addition, RT-PCR experiments indicated that MSNB can inhibit T3-mediated induction of target genes to a degree similar to the well characterized ligand antagonist NH-3. Together, these data provide support that MSNB analogs effectively block TR-mediated gene transcription.
Later steps in the process of ligand-dependent signaling are driven by a highly conserved interaction between the NRs and coregulators. The conservation of these signaling mechanisms and the nature of the coregulator binding pocket have hindered development of selective NR modulators targeted to this interface. However, the TR has four cysteine residues in the coactivator interaction site, whereas other NRs like AR, VDR, and PPARγ do not. Thus, these distinct cysteine residues might play a critical role in finding selective TR-coactivator modulators. MSNBs are irreversible and are selective for TR over other NR family members, showing no effect on AR and weak effects on VDR and PPAR. Interestingly, several compounds were active against PPARγ and VDR, which have a strongly activated cysteine residue (Cys-285 and Cys-284, respectively) in the ligand binding pocket (66, 67). A ligand displacement assay using [3H]rosiglitazone indicated that compound 17 inhibited hormone binding to the receptor, suggesting that 17 may act as a PPARγ antagonist. In addition, the MSNB series bears structural similarity to a previously reported irreversible ligand antagonist for PPARδ (52). Given these results, we suggest that all MSNB analogs that are active on PPARγ are acting through this alternative mechanism.
We have screened well over 500,000 small molecules in two independent HTS campaigns to identify TR-SRC2 inhibitors. The two screens identified two structurally distinct inhibitor series, β-aminoketones and MSNBs, both of which utilize irreversible mechanisms and target cysteine residues in the coactivator binding pocket. The two series have similar physicochemical properties, and each contains an electrophilic head group and hydrophobic tail group, although they have distinct primary structures. Thus, two inhibitors show a similar mechanism of action in inhibiting the TR-SRC2 interaction. Despite the steric and electrostatic similarities of the TR coactivator binding site with those of other NRs, both inhibitor series are quite selective for TR because of the unique presence of cysteine residues in the TR pocket.
Although these two inhibitor series have a similar physicochemical shape and mechanism, MSNB analogs have several advantages in the development of TR-coactivator inhibitors for use in vivo. First, MSNB members are predicted to lack the cardiac activity exhibited by β-aminoketones, especially by inhibiting the hERG potassium channel. The β-aminoketones have been marketed as drugs such as dyclonine (local anesthetic), hexacaine (antiarrhythmic), and pipoctanone (antihypertensive). Although the mode of action has not been fully elucidated, experimental evidence supports their binding to the inner pore of voltage-gated sodium channels (68–70). The ion channel activity of β-aminoketones is related to the basic tertiary amine moiety (71, 72). Although we successfully reduced the hERG activity of this series by the manipulation of the amine pKa (34), they still exhibit ion channel binding. In contrast, MSNB analogs do not contain basic tertiary amines and therefore are not expected to bind voltage-gated potassium channels. Second, the MSNBs are more stable than the β-aminoketones at physiological pH. Mannich bases are usually stable under acidic conditions but are cleaved into a reactive α,β-unsaturated ketone at basic pH (73, 74). The released α,β-unsaturated ketone can contribute to nonspecific protein binding and general toxicity (75–77). Overall, these advantages suggest that MSNB series members will be more useful for inhibiting TR-coactivator interactions in vivo.
In summary, we have discovered a novel class of TRβ antagonists, the methylsulfonylnitrobenzoates, which act as covalent, irreversible inhibitors. Although the mechanism of activity is similar to the previously reported series of β-aminoketones, the MSNB analogs lack several of the liabilities of the earlier series, especially ion channel activity and poor stability. Therefore, this scaffold provides a good foundation for developing a probe of TR function with a novel cellular mechanism. We are currently investigating the pharmacological properties of this series both ex vivo and in vivo.
Acknowledgments
We thank Sam Michael for automation assistance; Paul Shinn and Danielle Van Leer for compound management; Bill Leister, Chris LeClair, and Jeremy Smith for analytical chemistry; Rajarshi Guha for informatics; and the Hartwell Center (St. Jude Children's Research Hospital) for mass spectrometry analysis of protein samples.
This work was supported, in whole or in part, by National Institutes of Health Grant DK58080, the NIH Roadmap for Medical Research, and the National Institutes of Health NHGRI Intramural Research Program. This work was also supported by the American Lebanese Syrian Associated Charities (ALSAC) and the St. Jude Children's Research Hospital (SJCRH).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Scheme 1, Table 1, and other supplemental material.
- NR
- nuclear hormone receptor
- ER
- estrogen receptor
- AR
- androgen receptor
- PPARγ
- peroxisome proliferator-activated receptor γ
- TR
- thyroid hormone receptor
- T3
- triiodothyronine
- LBD
- ligand binding domain
- TRE
- thyroid-responsive element
- SRC
- steroid receptor coactivator
- MSNB
- methylsulfonylnitrobenzoate
- DMSO
- dimethyl sulfoxide
- h
- human
- PAMPA
- parallel artificial membrane permeability assay
- SAR
- structure-activity relationship
- FP
- fluorescence polarization
- PDK
- pyruvate dehydrogenase kinase
- PEPCK
- phosphoenolpyruvate carboxykinase
- VDR
- vitamin D receptor
- HTS
- high throughput screen
- qHTS
- quantitative high throughput screen.
REFERENCES
- 1. Escriva H., Bertrand S., Laudet V. (2004) Essays Biochem. 40, 11–26 [DOI] [PubMed] [Google Scholar]
- 2. Moore J. T., Collins J. L., Pearce K. H. (2006) ChemMedChem 1, 504–523 [DOI] [PubMed] [Google Scholar]
- 3. Chambon P. (2005) Mol. Endocrinol. 19, 1418–1428 [DOI] [PubMed] [Google Scholar]
- 4. Jordan V. C. (2006) Br. J. Pharmacol. 147, Suppl. 1, S269–S276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kansra S., Yamagata S., Sneade L., Foster L., Ben-Jonathan N. (2005) Mol. Cell. Endocrinol. 239, 27–36 [DOI] [PubMed] [Google Scholar]
- 6. Neri R., Florance K., Koziol P., Van Cleave S. (1972) Endocrinology 91, 427–437 [DOI] [PubMed] [Google Scholar]
- 7. Fradet Y. (2004) Expert Rev. Anticancer Ther. 4, 37–48 [DOI] [PubMed] [Google Scholar]
- 8. Schellhammer P. F., Davis J. W. (2004) Clin. Prostate Cancer 2, 213–219 [DOI] [PubMed] [Google Scholar]
- 9. Mohanty P., Aljada A., Ghanim H., Hofmeyer D., Tripathy D., Syed T., Al-Haddad W., Dhindsa S., Dandona P. (2004) J. Clin. Endocrinol. Metab. 89, 2728–2735 [DOI] [PubMed] [Google Scholar]
- 10. Stumvoll M., Häring H. U. (2002) Ann. Med. 34, 217–224 [PubMed] [Google Scholar]
- 11. Gillies P. S., Dunn C. J. (2000) Drugs 60, 333–343; discussion 344–335 [DOI] [PubMed] [Google Scholar]
- 12. Lawrence J. M., Reckless J. P. (2000) Int. J. Clin. Pract. 54, 614–618 [PubMed] [Google Scholar]
- 13. Cheng S. Y., Leonard J. L., Davis P. J. (2010) Endocr. Rev. 31, 139–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kress E., Samarut J., Plateroti M. (2009) Mol. Cell. Endocrinol. 313, 36–49 [DOI] [PubMed] [Google Scholar]
- 15. Williams G. R. (2000) Mol. Cell. Biol. 20, 8329–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oppenheimer J. H., Schwartz H. L. (1997) Endocr. Rev. 18, 462–475 [DOI] [PubMed] [Google Scholar]
- 17. Yaoita Y., Brown D. D. (1990) Genes Dev. 4, 1917–1924 [DOI] [PubMed] [Google Scholar]
- 18. Brent G. A. (2000) Rev. Endocr. Metab. Disord. 1, 27–33 [DOI] [PubMed] [Google Scholar]
- 19. Harvey C. B., Williams G. R. (2002) Thyroid 12, 441–446 [DOI] [PubMed] [Google Scholar]
- 20. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alonso M., Goodwin C., Liao X., Ortiga-Carvalho T., Machado D. S., Wondisford F. E., Refetoff S., Weiss R. E. (2009) Endocrinology 150, 3927–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paul B. D., Buchholz D. R., Fu L., Shi Y. B. (2007) J. Biol. Chem. 282, 7472–7481 [DOI] [PubMed] [Google Scholar]
- 23. Xu J., Li Q. (2003) Mol. Endocrinol. 17, 1681–1692 [DOI] [PubMed] [Google Scholar]
- 24. Savkur R. S., Burris T. P. (2004) J. Pept. Res. 63, 207–212 [DOI] [PubMed] [Google Scholar]
- 25. Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. (1998) Mol. Endocrinol. 12, 302–313 [DOI] [PubMed] [Google Scholar]
- 26. Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. (1998) Genes Dev. 12, 3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng W., Ribeiro R. C., Wagner R. L., Nguyen H., Apriletti J. W., Fletterick R. J., Baxter J. D., Kushner P. J., West B. L. (1998) Science 280, 1747–1749 [DOI] [PubMed] [Google Scholar]
- 28. Webb P., Nguyen N. H., Chiellini G., Yoshihara H. A., Cunha Lima S. T., Apriletti J. W., Ribeiro R. C., Marimuthu A., West B. L., Goede P., Mellstrom K., Nilsson S., Kushner P. J., Fletterick R. J., Scanlan T. S., Baxter J. D. (2002) J. Steroid Biochem. Mol. Biol. 83, 59–73 [DOI] [PubMed] [Google Scholar]
- 29. Grover G. J., Egan D. M., Sleph P. G., Beehler B. C., Chiellini G., Nguyen N. H., Baxter J. D., Scanlan T. S. (2004) Endocrinology 145, 1656–1661 [DOI] [PubMed] [Google Scholar]
- 30. Arnold L. A., Estébanez-Perpiñá E., Togashi M., Jouravel N., Shelat A., McReynolds A. C., Mar E., Nguyen P., Baxter J. D., Fletterick R. J., Webb P., Guy R. K. (2005) J. Biol. Chem. 280, 43048–43055 [DOI] [PubMed] [Google Scholar]
- 31. Arnold L. A., Estébanez-Perpiñá E., Togashi M., Shelat A., Ocasio C. A., McReynolds A. C., Nguyen P., Baxter J. D., Fletterick R. J., Webb P., Guy R. K. (2006) Sci. STKE 2006, pl3. [DOI] [PubMed] [Google Scholar]
- 32. Arnold L. A., Kosinski A., Estébanez-Perpiñá E., Fletterick R. J., Guy R. K. (2007) J. Med. Chem. 50, 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Estébanez-Perpiñá E., Arnold L. A., Jouravel N., Togashi M., Blethrow J., Mar E., Nguyen P., Phillips K. J., Baxter J. D., Webb P., Guy R. K., Fletterick R. J. (2007) Mol. Endocrinol. 21, 2919–2928 [DOI] [PubMed] [Google Scholar]
- 34. Hwang J. Y., Arnold L. A., Zhu F., Kosinski A., Mangano T. J., Setola V., Roth B. L., Guy R. K. (2009) J. Med. Chem. 52, 3892–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geistlinger T. R., Guy R. K. (2003) Methods Enzymol. 364, 223–246 [DOI] [PubMed] [Google Scholar]
- 36. Féau C., Arnold L. A., Kosinski A., Guy R. K. (2009) J. Biomol. Screen. 14, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng C. Q., Zheng X. S., Holt L. A., Hoong L. K., Somers P. K., Hill R. R., Saxena U. (2001) Bioorg. Med. Chem. Lett. 11, 1823–1827 [DOI] [PubMed] [Google Scholar]
- 38. Coleman M. D., Hadley S., Perris A. D., Jorga K., Seydel J. K. (2002) Environ. Toxicol. Pharmacol. 12, 7–13 [DOI] [PubMed] [Google Scholar]
- 39. Jeyakumar M., Tanen M. R., Bagchi M. K. (1997) Mol. Endocrinol. 11, 755–767 [DOI] [PubMed] [Google Scholar]
- 40. Mills N. L., Shelat A. A., Guy R. K. (2007) J. Biomol. Screen. 12, 946–955 [DOI] [PubMed] [Google Scholar]
- 41. Rouleau N., Turcotte S., Mondou M. H., Roby P., Bossé R. (2003) J. Biomol. Screen. 8, 191–197 [DOI] [PubMed] [Google Scholar]
- 42. Glickman J. F., Wu X., Mercuri R., Illy C., Bowen B. R., He Y., Sills M. (2002) J. Biomol. Screen. 7, 3–10 [DOI] [PubMed] [Google Scholar]
- 43. Attia R. R., Connnaughton S., Boone L. R., Wang F., Elam M. B., Ness G. C., Cook G. A., Park E. A. (2010) J. Biol. Chem. 285, 2375–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park E. A., Song S., Olive M., Roesler W. J. (1997) Biochem. J. 322, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen N. H., Apriletti J. W., Cunha Lima S. T., Webb P., Baxter J. D., Scanlan T. S. (2002) J. Med. Chem. 45, 3310–3320 [DOI] [PubMed] [Google Scholar]
- 46. Shah V., Nguyen P., Nguyen N. H., Togashi M., Scanlan T. S., Baxter J. D., Webb P. (2008) Mol. Cell. Endocrinol. 296, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim W., Nguyen N. H., Yang H. Y., Scanlan T. S., Furlow J. D. (2002) J. Biol. Chem. 277, 35664–35670 [DOI] [PubMed] [Google Scholar]
- 48. Moore J. M., Galicia S. J., McReynolds A. C., Nguyen N. H., Scanlan T. S., Guy R. K. (2004) J. Biol. Chem. 279, 27584–27590 [DOI] [PubMed] [Google Scholar]
- 49. Féau C., Arnold L. A., Kosinski A., Zhu F., Connelly M., Guy R. K. (2009) ACS Chem. Biol. 4, 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y., Kovach A., Suino-Powell K., Martynowski D., Xu H. E. (2008) J. Biol. Chem. 283, 19132–19139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leesnitzer L. M., Parks D. J., Bledsoe R. K., Cobb J. E., Collins J. L., Consler T. G., Davis R. G., Hull-Ryde E. A., Lenhard J. M., Patel L., Plunket K. D., Shenk J. L., Stimmel J. B., Therapontos C., Willson T. M., Blanchard S. G. (2002) Biochemistry 41, 6640–6650 [DOI] [PubMed] [Google Scholar]
- 52. Shearer B. G., Wiethe R. W., Ashe A., Billin A. N., Way J. M., Stanley T. B., Wagner C. D., Xu R. X., Leesnitzer L. M., Merrihew R. V., Shearer T. W., Jeune M. R., Ulrich J. C., Willson T. M. (2010) J. Med. Chem. 53, 1857–1861 [DOI] [PubMed] [Google Scholar]
- 53. Babaoglu K., Simeonov A., Irwin J. J., Nelson M. E., Feng B., Thomas C. J., Cancian L., Costi M. P., Maltby D. A., Jadhav A., Inglese J., Austin C. P., Shoichet B. K. (2008) J. Med. Chem. 51, 2502–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elbrecht A., Chen Y., Adams A., Berger J., Griffin P., Klatt T., Zhang B., Menke J., Zhou G., Smith R. G., Moller D. E. (1999) J. Biol. Chem. 274, 7913–7922 [DOI] [PubMed] [Google Scholar]
- 55. Gronemeyer H., Gustafsson J. A., Laudet V. (2004) Nat. Rev. Drug Discov. 3, 950–964 [DOI] [PubMed] [Google Scholar]
- 56. Chen T. (2008) Curr. Opin. Chem. Biol. 12, 418–426 [DOI] [PubMed] [Google Scholar]
- 57. Chang C., Norris J. D., Grøn H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. (1999) Mol. Cell. Biol. 19, 8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. (1998) Genes Dev. 12, 3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Valadares N. F., Polikarpov I., Garratt R. C. (2008) J. Steroid Biochem. Mol. Biol. 112, 205–212 [DOI] [PubMed] [Google Scholar]
- 60. Rodriguez A. L., Tamrazi A., Collins M. L., Katzenellenbogen J. A. (2004) J. Med. Chem. 47, 600–611 [DOI] [PubMed] [Google Scholar]
- 61. Parent A. A., Gunther J. R., Katzenellenbogen J. A. (2008) J. Med. Chem. 51, 6512–6530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Y., Chirgadze N. Y., Briggs S. L., Khan S., Jensen E. V., Burris T. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9908–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kojetin D. J., Burris T. P., Jensen E. V., Khan S. A. (2008) Endocr. Relat. Cancer 15, 851–870 [DOI] [PubMed] [Google Scholar]
- 64. Huang H., Wang H., Sinz M., Zoeckler M., Staudinger J., Redinbo M. R., Teotico D. G., Locker J., Kalpana G. V., Mani S. (2007) Oncogene 26, 258–268 [DOI] [PubMed] [Google Scholar]
- 65. Wang H., Huang H., Li H., Teotico D. G., Sinz M., Baker S. D., Staudinger J., Kalpana G., Redinbo M. R., Mani S. (2007) Clin. Cancer Res. 13, 2488–2495 [DOI] [PubMed] [Google Scholar]
- 66. Gampe R. T., Jr., Montana V. G., Lambert M. H., Miller A. B., Bledsoe R. K., Milburn M. V., Kliewer S. A., Willson T. M., Xu H. E. (2000) Mol. Cell 5, 545–555 [DOI] [PubMed] [Google Scholar]
- 67. Vanhooke J. L., Benning M. M., Bauer C. B., Pike J. W., DeLuca H. F. (2004) Biochemistry 43, 4101–4110 [DOI] [PubMed] [Google Scholar]
- 68. Armstrong C. M., Hille B. (1998) Neuron 20, 371–380 [DOI] [PubMed] [Google Scholar]
- 69. Hille B., Armstrong C. M., MacKinnon R. (1999) Nat. Med. 5, 1105–1109 [DOI] [PubMed] [Google Scholar]
- 70. Bruhova I., Tikhonov D. B., Zhorov B. S. (2008) Mol. Pharmacol. 74, 1033–1045 [DOI] [PubMed] [Google Scholar]
- 71. Jamieson C., Moir E. M., Rankovic Z., Wishart G. (2006) J. Med. Chem. 49, 5029–5046 [DOI] [PubMed] [Google Scholar]
- 72. Armstrong C. M. (2003) Sci. STKE 2003, re10. [DOI] [PubMed] [Google Scholar]
- 73. Gilmer J. F., Simplício A. L., Clancy J. M. (2005) Eur. J. Pharm. Sci. 24, 315–323 [DOI] [PubMed] [Google Scholar]
- 74. Simplício A. L., Clancy J. M., Gilmer J. F. (2007) Int. J. Pharm. 336, 208–214 [DOI] [PubMed] [Google Scholar]
- 75. Zollner H. (1973) Biochem. Pharmacol. 22, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 76. Morgan D. L., Ward S. M., Wilson R. E., Price H. C., O'Connor R. W., Seely J. C., Cunningham M. L. (2001) Inhal. Toxicol. 13, 633–658 [DOI] [PubMed] [Google Scholar]
- 77. Gul M., Gul H. I., Das U., Hanninen O. (2005) Arzneimittelforschung 55, 332–337 [DOI] [PubMed] [Google Scholar]
- 78. Johnson R. L., Hwang J. Y., Arnold L. A., Huang R., Wichterman J., Augustinaite I., Austin C. P., Inglese J., Guy R. K., Huang W. (2011) J. Biomol. Screen., in press [DOI] [PMC free article] [PubMed] [Google Scholar]