Abstract
Lactating mothers secrete milk sialyloligosaccharides (MSOs) that function as anti-adhesives once provided to the neonate. Particular infant-associated commensals, such as Bifidobacterium longum subsp. infantis, consume neutral milk oligosaccharides, although their ability to utilize acidic oligosaccharides has not been assessed. Temporal glycoprofiling of acidic HMO consumed during fermentation demonstrated a single composition, with several isomers, corresponding to sialylated lacto-N-tetraose. To utilize MSO, B. longum subsp. infantis deploys a sialidase that cleaves α2–6 and α2–3 linkages. NanH2, encoded within the HMO catabolic cluster is up-regulated during HMO fermentation and is active on sialylated lacto-N-tetraose. These results demonstrate that commensal microorganisms do utilize MSO, a substrate that may be enriched in the distal gastrointestinal tract.
Keywords: Bacterial Metabolism, Evolution, Mass Spectrometry (MS), Oligosaccharide, Protein Chemistry, Sialic Acid, Commensal Glycobiology, Human Milk Oligosaccharides
Introduction
Soluble oligosaccharides often exceed protein concentrations in human milk, although their biological role remains poorly defined. These heterogeneous carbohydrates consist of glucose, galactose, N-acetylglucosamine, and frequently fucose and/or N-acetyl-d-neuraminic acid (Neu5Ac or sialic acid) residues via several potential glycosidic linkages. The human milk oligosaccharide (HMO)4 core is typically elongated from a lactose reducing end (Galβ1–4Glc) with iterative Galβ1–3/4GlcNAc units to compose linear or branched oligosaccharides with a degree of polymerization ≥ 4. α1–2/3/4 Fucosylation adds structural complexity and may shield the HMO backbone from exo-glycosidase digestion. Similarly, enzymatic degradation of acidic HMOs or milk sialyloligosaccharides (MSOs) requires cleavage of terminal α2–6- and α2–3-linked sialyl moieties (1). Human milk is a rich source of sialylated glycans, previously determined to be over 40 structures (of over 200 total HMOs) representing nearly 16% of soluble oligosaccharide abundances (2, 3).
In contrast to lactose, the structural organization of HMOs resist host digestive enzymes, thus are introduced intact to microbial communities established along the nursing infant gastrointestinal tract (GIT) (4). Accordingly, HMOs are potent molecular arbiters at the mammalian-microbial interface as they modulate epithelial surface glycan expression and may modulate systemic immune responses (5). Moreover, HMOs limit pathogen colonization by mimicking vulnerable host epitopes thus competing for microbial adhesins. A well-studied example is the inhibition of campylobacter-induced diarrhea in infants by α1–2 fucosylated HMO (6, 7). Likewise, sialylated glycans are bound by Helicobacter pylori and are known to mitigate Streptococcus pneumoniae adherence to the nasopharyngeal mucosa (8–10). In addition, sialylated milk glycoproteins confer further protection by neutralizing infectious particles such as rotavirus (11).
With a few exceptions, the prominence of sialic acids to eukaryotic biology emerged with the deuterostomes and thus microbial interactions denote an evolved commensal, pathogenic, or saprotrophic relationship with animals (12–14). Therefore milk laden with sialylconjugates may enrich for microbes that deploy sialidases (EC 3.2.1.18) to metabolize otherwise inaccessible or perhaps deleterious sialosides passing through the nursing neonate. This is consistent with the frequent overrepresentation of infant-type bifidobacteria in this habitat, many of which exhibit sialidase activity (15). Benefiting the infant, pioneering bifidobacteria sequester colonization sites from pathogens and participate in host metabolism by supplying acetate and lactate, and promoting butyrogenesis by heterologous genera (16–18).
Our recent work detailed the composition and distribution of oligosaccharides secreted into milk (2), demonstrated that specific bifidobacteria exploit HMO as a growth substrate (2, 19), precisely identified preferred neutral oligosaccharides (15, 20) and reconstructed HMO metabolic pathways encoded by B. longum subsp. infantis ATCC15697 (21). In addition, we have observed that this infant-associated commensal degrades milk sialic acids and metabolizes Neu5Ac via its fructose-6-phosphate phosphoketolase pathway (19, 21).
Despite this physiological and genomic evidence, the extent to which B. longum subsp. infantis or other microbes consume MSO is uncertain and is the subject of this study reported herein. Accordingly, MSO utilization and preferred acidic oligosaccharides were determined by monitoring temporal fluxes within a pool of fermented HMO. Moreover, two candidate B. longum subsp. infantis sialidases, one or both of which would be rate-limiting in MSO catabolism were characterized.
EXPERIMENTAL PROCEDURES
Sequence Analyses
Genome and gene sequence analysis was conducted using The Integrated Microbial Genomes (IMG) system (22), MetaCyc (23), VectorNTI (Invitrogen), InterProScan (24), SUPERFAMILY (25), and other standard bioinformatic tools as needed. Sialidase sequences from GIT inhabiting bacteria were retrieved from IMG corresponding to clusters of orthologous groups of proteins (COG4409). Multiple sequence alignment was performed with MUSCLE and manually curated. The phylogeny of 40 sialidase enzyme sequences was inferred by Maximum Likelihood using the JTT matrix-based model (26, 27). 500 replicates were bootstrapped to assess statistical confidence in inferred phylogenetic relationships (28). Distances reflect the number of amino acid substitutions per position. All gaps and incomplete positions were excluded from the final analysis that yielded 274 aligned positions.
Bacterial Growth and DNA Extraction
B. longum subsp. infantis ATCC15697 cultures were routinely propagated in Man-Rogosa-Sharpe (MRS) broth (Becton Dickinson, Franklin Lakes, NJ) supplemented with 0.05% (w/v) l-cysteine at 37 °C under anaerobic conditions. Bacterial DNA was extracted with the MasterPure Gram-positive DNA Purification Kit (Epicenter, Madison, WI) for cloning. Transformed Escherichia coli strains (Top10 and BL21; Invitrogen, Carlsbad, CA) were propagated in Luria Broth under selective conditions (100 μg/ml carbenicillin). Plasmids were extracted with Qiagen (Germantown, MD) plasmid mini-prep kits according to the manufacturer's protocols.
Sialidase Cloning, Heterologous Expression, and Purification
Sialidase genes Blon_0646 (nanH1) and Blon_2348 (nanH2) were PCR amplified from the ATCC15697 chromosome using a high-fidelity polymerase with the following primer pairs: nanH1: (5′-CACCATGGCAGCATCCAACCGATC & 5′-GTGCGTTTCGGCCGCGCCGAA) and nanH2: (5′-CACCATGACGGAGAACGGGATGATG & 5′-GCACCCTCCCTCACCAGACAG). PCR products were cleaned using the Qiagen Qiaquick cleanup kit, quantified, and cloned into a pET101-D expression vector with the ChampionTM pET101 Directional TOPO® Expression kit (Invitrogen) to yield a polyhistidine fusion protein when expressed. Vectors were propagated in E. coli TOP10 and were transformed into E. coli BL21(DE3) for heterologous expression. The NanH1 expression strain was grown for 4 h subsequent to achieving A600 0.5–0.6 at 30 °C, 250 RPM prior to cell harvest. His-tagged NanH2 overproduction was induced with the addition of 1 mm IPTG at A600 0.5–0.6 at 37 °C, 250 RPM and grown for an additional 4 h. Cells were harvested by centrifugation, 5 min of incubation on ice and washed 2× with sterile PBS. Cells were disrupted by chemical lysis using Pierce B-PER (Thermo Scientific, Waltham, MA) according to the manufacturer's directions. Sialidase activity was qualitatively confirmed from the soluble cell-free protein with the BVBlue Diagnostic Assay (Genzyme Diagnostics, Cambridge, MA). The soluble cell-free extract was centrifuged from debris and purified by immobilized metal affinity chromatography with the Pierce HisPur cobalt spin columns (Thermo Scientific) according to the manufacturer's instructions. Purified His-tagged proteins were eluted with 150 mm imidazole, pH 7.4. Sialidases were inspected by SDS-PAGE to conform to expected molecular weight and quantified by bicinchoninic acid (BCA) assay (29).
Sialidase pH Profile
Typical enzymatic assays were performed in duplicate in a 20-μl reaction mixture containing a buffer (200 mm) with a pH in the range of 4.0–9.0, Neu5Acα2–3LacβMU, or Neu5Acα2–6LacβMU (1 mm), and NanH1 (Blon_0646, 3.9 μg) or NanH2 (Blon_2348, 24 ng). The buffers used were: NaOAc-HOAc for pH 4.0–6.0 and Tris-HCl for pH 7.0–9.0. Reactions were allowed to proceed for 15 min at 37 °C and were stopped by adding ice-cold 25% acetonitrile to yield a 10-fold dilution. The samples were analyzed by a Shimadzu LC-2010A system equipped with a membrane on-line degasser, a temperature control unit, and a fluorescence detector. A reverse phase Premier C18 column (250 × 4.6 mm I.D., 5 mm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 25% acetonitrile. The fluorescent compounds LacβMU and Neu5Acα2–3LacβMU (or Neu5Acα2–6LacβMU) were detected with excitation at 325 nm and emission at 372 nm.
Sialidase Kinetics via HPLC Assays
The enzymatic assays were carried out for at least two individual experiments. Each experiment was performed in duplicate in a total volume of 20 μl in a NaOAc/HOAc buffer (200 mm, pH 5.0) containing Neu5Acα2–3LacβMU or Neu5Acα2–6LacβMU and a recombinant sialidase (3.9 μg of NanH1 or 24 ng of NanH2). Apparent kinetic parameters were obtained by varying the Neu5Acα2–3LacβMU or Neu5Acα2–6LacβMU concentration from 0.1–40.0 mm (0.1, 0.2, 0.4, 1, 2, 4, 10, 20, and 40 mm). Reactions were allowed to proceed for 10 min at 37 °C. Apparent kinetic parameters were obtained by fitting the data (the average values of duplicate assay results) into the Michaelis-Menten equation using Grafit 5.0.
Sialidase Substrate Specificity Assays
The assays were carried out at 37 °C in duplicate in 384-well plates (Fisher Scientific) in a total volume of 20 μl containing NaOAc/HOAc buffer (100 mm, pH 5.0), NanH1 (3.9 μg), or NanH2 (12 ng), a sialoside substrate (0.3 mm), and β-galactosidase from Aspergillus oryzae (12 μg, 126 mU) (Sigma-Aldrich). The amount of the β-galactosidase required to completely hydrolyze the GalβpNP within the time frame of the assay was predetermined and confirmed by assays using GalβpNP (0.3 mm). The reactions were carried out for 30 min and were stopped by adding CAPS buffer (40 μl, 0.5 m, pH 10.5). The amount of the para-nitrophenolate formed was determined by measuring the A405 nm of the reaction mixtures using a microtiter plate reader. Reactions of GalβpNP (0.3 mm) and β-galactosidase (12 μg, 126 mU) were used as controls. All substrates used in this assay were synthesized previously as reported (43).
Bacterial Fermentation of HMO
Milk oligosaccharides were purified from pooled human milk as previously described (2). Isolated B. longum subsp. infantis ATCC15697 colonies were cultured in MRS broth and incubated overnight at 37οC in an anaerobic growth chamber (Coy Laboratory Products). The resultant cultures were inoculated at 1% into 200 μl of reconstituted MRS containing 2% HMO instead of glucose as the sole carbohydrate (MRSHMO) and overlaid with 40 μl of sterile mineral oil in a 96-well microtiter plate. Cell growth was monitored in real time by assessing A600 nm using a BioTek PowerWave 340 plate reader (BioTek, Winoosky, VT) every 30 min preceded by 15 s shaking at variable speed. At least two biological replicates were performed for each physiological collection at approximately A600 nm = 0.2, 0.3, 0.6, 0.75, and 1.0. Once harvested, culture supernatants were centrifuged at 2000 × g for 30 min, boiled for 5 min, and filtered through a 0.22-μm pore membrane (Millipore) prior to temporary storage at −80 °C.
Sample Preparation for MS Analysis
Aliquots of supernatant were chemically reduced and spiked with an aliquot of deuterated growth medium that served as an internal standard and allowed for consumption analysis(2,30). Following reduction, all samples were desalted using porous graphitized carbon Top Tip solid phase extraction cartridges (Glygen, Columbia, MD) and the HMOs were eluted from the cartridge using a 20% CH3CN solution (2). The desalted samples were dried in a centrifugal evaporator to dryness and were then reconstituted in 18 mΩ deionized water to a final concentration of 1000 ppm based on the internal standard.
Mass Spectrometry
Working samples were prepared for mass spectral analysis by dilution of each 1000 ppm sample to 10 ppm using a solution of H2O:MeOH (1:1, v/v) + 5 mm ammonium acetate. All mass spectrometry experiments were performed on an IonSpec 9.4 T QFT Fourier transform ion cyclotron resonance mass spectrometer (FT-ICR MS) (Lake Forest, CA) equipped with an Advion Nanomate (Ithaca, NY). Control and programming of the QFT FT-ICR MS was performed in the IonSpec Omega software (version 9.1.2) while the Nanomate was controlled using the accompanying ChipSoft software (version 6.4.5). Prior to the analysis of consumption samples, the experimental parameters of the QFT FT-ICR MS were optimized for oligosaccharide analysis and mass accuracy using maltooligosaccharides (31). All spectra were collected with a total ion intensity of 150–200 (arbitrary units) and were recorded at a transient length of 1 s and 1024K data points in each transient.
Analysis of Mass Spectra to Determine HMO Consumption
Each recorded transient was shortened from 1024K data points to 256K data points and then internally calibrated to the ions corresponding to the deuterated sialylated HMOs, bringing the measured mass error to less than 10 ppm (32). The masses and their associated intensities were copied as a mass-intensity (MI) table from the Omega software into Microsoft Excel 2007 for further data analysis. The MI table was searched for the monoisotopic and 13C peaks of the sialylated HMOs of interest using a custom Excel macro (33). Briefly this macro compares an experimental MI table to a user-defined list of masses of interest and returns those experimental masses, and their intensities, matching the defined list.
The percent consumption of each sialylated HMO was calculated using the ratio of the intensities of deuterated internal standard to the sample HMO (D/H ratio) for a consumption sample and a control sample. In calculating the D/H ratio for a given HMO the 13C contribution of the HMO must be removed from the intensity of the internal standard HMO. The IonSpec Exact Mass Calculator software (version 9.0.15) was used to calculate the isotopic distribution of the sample HMOs as percentages of the monoisotopic peak. In this manner the contribution of the 13C peak for the sample HMO could be subtracted from the intensity of the internal standard monoisotopic peak. This is shown in Equation 1, in which ID is the measured intensity for the deuterated HMO internal standard, IH is the intensity for the reduced HMO, and k is a waiting factor adjusting for the 13C contribution of the reduced HMO to the intensity of the deuterated HMO (2, 30).
 |
After the D/H ratio for the consumption and control samples has been calculated, the percent consumption can then be calculated using Equation 2.
Sialidase Gene Expression
Isolated ATCC15697 colonies were passaged once in reconstituted MRS supplemented with 0.05% l-cysteine with a sole carbohydrate at 2% prior to 1% inoculation into experimental conditions. All bacteria were cultured anaerobically at 37 °C. Mid-log cells (∼A600 nm = 0.5) were harvested on ice, centrifuged at 4 °C followed by the addition of RNAlater (Ambion Inc.) to the aspirated cell pellet following manufacturer's protocols. Cell pellets stored at −20 °C were resuspended in 250 μl of lysozyme (50 mg/ml) and 120 μl of mutanolysin (1000 units/ml) and incubated at 37 °C for 10 min prior to the introduction lysis/binding solution from Ambion's RNAqeous kit used to extract total RNA according to manufacturer's protocols. RNA was cleaned with a Qiagen RNeasy kit prior to cDNA synthesis using random hexamers and Superscript II reverse-transcriptase (Invitrogen) following DNase treatment. 5′ nuclease assay (TaqMan) primers and probes were designed with the PRIMEREXPRESS software (Applied Biosytems) targeting nanH1 and nanH2 sequences and are listed in supplemental Table S1. RT-qPCR was performed on an ABI 7500 Fast Real-Time PCR System using the fast reagents and protocol as directed and supplied by Applied Biosystems. The sequence encoding a cysteinyl-tRNA synthetase (Blon_0393) was targeted as a reference as it is constitutively expressed under several culture conditions and verified for our ATCC15697 laboratory strain (34). Data were analyzed by the ΔΔCt method to determine relative expression by normalizing target values to the reference. p values were determined by a 2-tailed, type 3 t test.
Sialidase Digestion of Purified MSOs
MSOs were isolated from pooled HMO by solid phase extraction (SPE) and HPLC. A fraction containing two abundant MSO species were used for sialidase digestion. Disialyl-lacto-N-tetraose (DSLNT) possessing both α2–3- and α2–6-sialyl linkages was obtained from Dextra Laboratory (Earley Gate, UK). The two purified MSO and commercially obtained DSLNT were treated with NanH1, NanH2 and a recombinant Clostridium perfringens α(2–3)-neuraminidase (New England Biolab). The digestion protocol has been previously reported and deviates only by omitting dialysis to conserve enzymes prior to digestion (30, 35). Briefly, the reaction consisted of 0.1 m ammonium acetate buffer, MSO, and enzyme in a 2:1:1 ratio incubated in a water bath at 37 °C. MALDI FT-ICR MS was used to monitor the digestion reaction every 30 min.
Determination of Extent of Sialylation in HMO Pool
The extent of sialylation in the HMO pool was determined using a previously described method that utilizes a porous graphitized carbon nano-LC-MS platform for the separation and identification of milk oligosaccharides (36). Briefly, an aliquot of the fermentation medium was spiked with an equal amount of deuterated growth medium as described in the preceeding sections. Six replicate injections of this sample were performed and the generated chromatograms were analyzed to determine the extent of sialylation in the growth medium as percentages of the total ion current.
RESULTS
B. longum subsp. infantis MSO Consumption
Mass-spectrometry (MS) was employed to ascertain the preferred MSO compositions, and at what point in the cell cycle B. longum subsp. infantis ATCC15697 utilize these acidic glycans. Briefly, Fourier transform ion cyclotron resonance MS (FTICR-MS) was used to monitor fluxes in a mixed population of purified HMOs (i.e. <200 neutral and acidic compositions) subjected to bacterial fermentation. Consumption was represented by the ratio of oligosaccharides recovered from the fermentate to a deuterated internal standard generated from the identical, albeit sterile, starting HMO pool. Discrete mass to charge ratios (m/z) corresponds to a characteristic oligosaccharide degree of polymerization (DP) and monosaccharide composition.
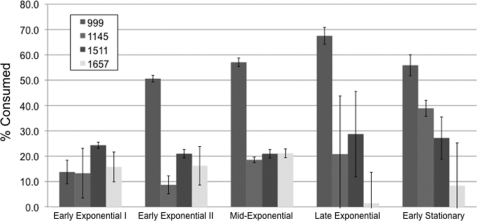
The four most abundant MSO compositions were examined comprising ∼1.7% of the total oligosaccharide content of the HMO pool (supplemental Table S2). Sialyllactose was not detected despite applying identical methods that have previously isolated sialyllactose from bovine milk (37). To resolve MSO preferences and exclude spurious degradation, culture supernatants were sampled at various cell physiological stages as defined by culture optical densities (A600 nm) ∼0.2, 0.3, 0.6, 0.75, and 1.0 (Fig. 1). Strikingly, only m/z 999, corresponding to sialyllacto-N-tetraose (SLNT), was consumed in a temporal-dependent manner with a final consumption >50% of the starting material. Three SLNT isomers have been isolated from human milk and are either α2–3/6 sialylated type I HMO or an α2–6 sialylated type II (supplemental Fig. S1). Mono- and di-fucosylayed sialylated lacto-N-hexaose (m/z 1511 and m/z 1657) were not consumed appreciably over the course of fermentation. Repression of SLNT utilization by neutral milk oligosaccharides was not observed, although this cannot be excluded for the other MSO compositions. Furthermore, MSO containing O-acetylated Neu5Ac was not detected in the MSO pool, despite the potential to hydrolyze acetyl ester derivatives encoded by a putative sialate O-acetylesterase gene (Blon_1907).
FIGURE 1.
Temporal glycoprofile of abundant MSO consumption by B. longum subsp. infantis ATCC15697. MSO compositions are represented by an m/z value signifying a characteristic oligosaccharide composition. Only 999 (SLNT) is consumed appreciably (≥50%) at the initiation of stationary phase.
B. longum subsp. infantis Sialidases
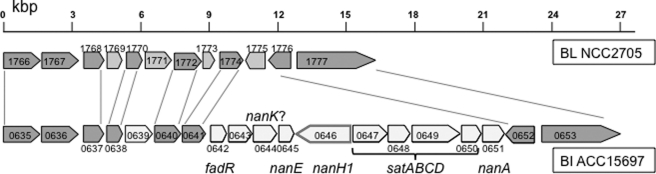
Microbes deploy exo-glycolytic sialidases (EC 3.2.1.18) to utilize terminally sialylated glycans. We have identified two-candidate sialidase sequences encoded within the B. longum subsp. infantis ATCC15697 chromosome, Blon_0646 and Blon_2348 (21). These predictions were verified, and additional sequences excluded, by scanning the ATCC15697 genome for conserved sialidase motifs. The first sialidase gene, nanH1 (Blon_0646) appears in a gene cluster dedicated to sialic acid catabolism including a N-acetyl neuraminic acid lyase (nanA1), a putative kinase and an epimerase (nanE) (Fig. 2). This encoded pathway provides N-acetyl glucosamine-6-P for further processing by NagA (encoded by Blon_0882; EC 3.2.1.49) and NagB (encoded by Blon_0881; EC 3.5.99.6) into fructose-6-P to enter central metabolism as previously observed in ATCC15697 Neu5Ac metabolism (supplemental Fig. S2 and supplemental Table S3) (21). In addition, a putative sialic acid-specific ABC transporter is encoded proximally (Blon_0647- Blon_0650) which shares sequence similarity to the satABCD sialic acid transport system first described in Haemophilus ducreyi (38).
FIGURE 2.
B. longum subsp. infantis ATCC15697 sialic acid utilization cluster. The analogous loci in the closely related B. longum subsp. longum NCC2705 is included for comparative purposes. Dark gray arrows depict genes conserved in B. longum subsp. infantis ATCC15697 (BI ATCC15697) and B. longum subsp. longum NCC2705 (BL NCC2705). White arrows denote genes unique to ATCC15697, including sialic acid catabolism, with light gray arrows marking those genes specific to NCC2705. Loci are preceded by BL and Blon_ for NCC2705 and ATCC15697, respectively.
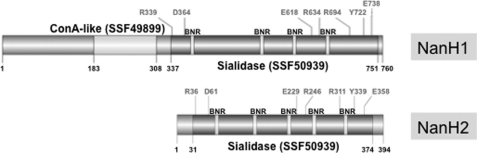
The nanH1 locus encodes an 83-kDa protein with a 414 amino acid sialidase domain (SSF50939) and a concanavalin A-like lectin domain (SSF49899) (Fig. 3). Similar ConA domains are found in other actinobacterial sialidases and may facilitate substrate recognition and binding. The deduced NanH1 enzyme sequence exhibited all seven expected catalytic residues, and four Asp-box repeats ((S/T) XDXGXR(W/F)) inherent to bacterial sialidases (39, 40). However, an identifiable export signal, transmembrane domain or cell wall anchor motif were lacking for both sialidase sequences, suggesting likely intracellular localization. Although sialidase activity has been observed in the cell wall extracts of fractionated cells.5 Similarly, the recently sequenced Bifidobacterium breve DSM20213 draft genome includes a predicted intracellular sialidase homolog (BIFBRE_01961) exhibiting 55% amino acid identity to nanH1 (41).
FIGURE 3.
Domain structure of the two ATCC15697 sialidase enzymes. Catalytic residues appear above the protein, with domain boundaries demarked below. SCOP Superfamily sialidase domains (SSF50939) are depicted encompassing several bacterial neuraminidase repeats (BNR) (i.e. Asp-boxes).
In contrast, nanH2 (Blon_2348) is co-localized with a putative N-acetylneuraminate lyase (nanA2) to the B. longum subsp. infantis HMO cluster, and in the absence of other Neu5Ac catabolic genes, aside from a potential lyase found immediately adjacent (Blon_2349). The smaller protein (42 kDa) encoded by nanH2 consists of a single sialidase domain. Like NanH1, all 7 catalytic residues were readily identifiable in this predicted intracellular sialidase that possesses 5 Asp box motifs. Interestingly, the two nanH2 orthologs presented by the Bifidobacterium bifidum NCIMB41171 draft genome appears to be associated with the cell envelope similar to its extracellular fucosidases (42). The lack of sialic acid catabolic genes evident in the NCIMB41171 genome contrasts with other infant-associated bifidobacteria, B. longum subsp. infantis ATCC15697 and B. breve DSM20213 included, as well as the human gut commensal Bifidobacterium gallicum DSM20093 (Table 1). The presence or absence of Neu5Ac genes was consistent with utilization phenotypes previously observed in similar B. bifidum and B. breve strains (19).
TABLE 1.
Apparent kinetic parameters of NanH1 and NanH2 when Neu5Acα2–3LacβMU or Neu5Acα2–6LacβMU was used as the substrate
| Enzymes | NanH1 | NanH2 | ||
|---|---|---|---|---|
| Substrates | Neu5Acα2–3LacβMU | Neu5Acα2–6LacβMU | Neu5Acα2–3LacβMU | Neu5Acα2–6LacβMU |
| Km (mm) | 7.6 ± 0.3 | 3.7 ± 0.1 | 7.6 ± 0.2 | 3.8 ± 0.1 |
| Vmax (mm s−1) | (2.1 ± 0.2) × 10−3 | (1.8 ± 0.1) × 10−3 | (3.6 ± 0.1) × 10−3 | (3.7 ± 0.3) × 10−3 |
| kcat (s−1) | 1.0 ± 0.1 | 0.8 ± 0.1 | (1.4 ± 0.1) × 102 | (1.4 ± 0.1) × 102 |
| kcat/Km (s−1 mm−1) | 0.13 | 0.22 | 18 | 37 |
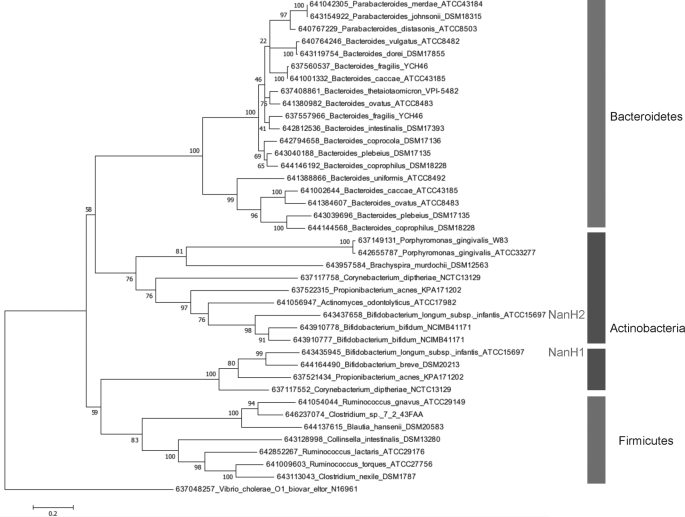
As expected, the inferred phylogeny derived from representative gut bacterial sialidases bifurcates into Bacteroidetes and Gram-positive protein sequences (Fig. 4). Interestingly, actinobacterial sialidases are paraphyletic, as they unambiguously assembled into discrete clades. In instances that an Actinobacterium species encodes two sialidases, the proteins phylogenetically segregated into each clade suggestive of differential function for these lineages and/or lateral gene transfer. The exception to this was B. bifidum NCIMB41171 that encodes two sialidase paralogs in tandem on the contig on which they appear. Major branches of the phylogeny reconstructed by Maximum Likelihood received significant support and were confirmed by a similar topology yielded from maximum parsimony.
FIGURE 4.
Phylogenetic relationship of sialidases encoded by gut bacteria. Branch lengths are in the same units (number of amino acid substitutions per site) as those of the evolutionary distances used to construct the tree. The organism and loci are listed in which these sialidases are found.
To functionally characterize the two putative ATCC15697 sialidases, and characterize their activity toward purified MSOs, these sequences were cloned and expressed in a heterologous host. Purified polyhistidine-tagged proteins of the expected molecular weight were verified as sialidases in a qualitative assay of their activity.
Sialidase Gene Expression
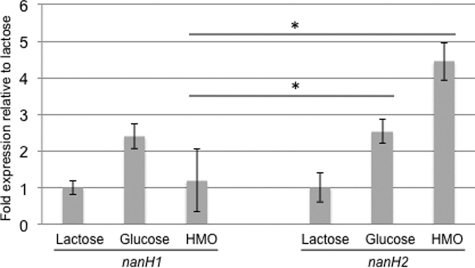
A RT-qPCR 5′ nuclease assay was developed to evaluate expression of the two B. longum subsp. infantis ATCC15697 sialidase genes. Interestingly, when actively utilizing HMO, nanH2 was up-regulated ∼4-fold relative to expression levels on lactose as the sole fermentation substrate (Fig. 5). Proteomics has previously shown that the HMO cluster-encoded nanH2 was expressed during growth on this substrate (21). In contrast, nanH1 expression was not induced to the same extent as determined by RT-qPCR. Growth on glucose resulted in a 2-fold induction of both nanH1 or nanH2 expression. This is consistent with a previously observed preference for lactose over glucose that may repress sialidases and other carbohydrate-active enzymes.
FIGURE 5.
ATCC15697 sialidase gene expression during carbohydrate fermentation. Gene expression is calculated relative to levels while growing on lactose as a sole carbon source. Bars represent averages from three independent experiments ± standard error. *, p ≤ 0.05.
Substrate Specificity and Kinetics of B. longum subsp. infantis Sialidases
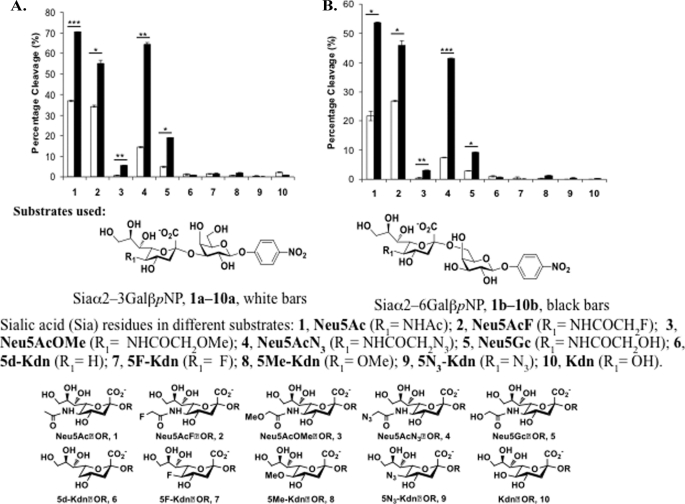
A library consisting of para-nitrophenol (pNP)-tagged sialylgalactosides was screened to characterize NanH1 and NanH2 substrate specificities. This sialosides library was synthesized in a one-pot, three-enzyme reaction to yield sialogalactosides with different sialyl linkages, and various sialic acid forms. The substrate specificity study was conducted in a high throughput screen format using a 384-well plate (43, 44). Briefly, removal of the terminal appropriate sialic acid form by sialidase activity exposes a terminal galactose residue that can be cleaved by excess β-galactosidase in the reaction mixture to release para-nitrophenol. Upon adjusting the pH of the reaction mixture to >9.5, the majority of the para-nitrophenol produced will be converted to para-nitrophenolate that could be quantified by a plate reader at A405 nm.
The two purified sialidases cleaved both α2–3- and α2–6-linked sialosides, with a consistent preference for the α2–6 linkage (Fig. 6). Both sialidases do not have significant activity (<5%) in cleaving 2-keto-3-deoxy-d-glycero-d-galacto-nonulosonic acid (Kdn) or its derivatives in either α2–3- or α2–6-linkage. NanH1 generally exhibited lower activity on all assayed substrates. Interestingly, both sialidases cleave α2–3- and α2–6-linked Neu5Gc from compounds Neu5Gcα2–3GalβpNP 5a and Neu5Gcα2–6GalβpNP 5b, but a 5C glycolyl reduces their activities. Surprisingly, an azido group addition on the N-acetyl group of N-acetylneuraminic acid (Neu5Ac) does not change the activity of either sialidases when Neu5AcN3α2–6GalβpNP 4b is used as a substrate, but the activity of both sialidases decreases when Neu5AcN3α2–3GalβpNP 4b is used as a substrate. In addition, a fluorine substitution of one of the hydrogens in the N-acetyl group of Neu5Ac in both α2–3- and α2–6-linked sialosides Neu5AcFα2–3/6GalβpNP 2a and 2b does not affect the activity of both sialidases. In contrast, the addition of an O-methyl group at the N-acetyl of Neu5Ac in both α2–3- and α2–6-linked sialosides Neu5AcOMeα2–3/6GalβpNP 3a and 3b decreases the activity of both sialidases significantly (Fig. 6).
FIGURE 6.
B. longum subsp. infantis ATCC15697 sialidase substrate specificity assay. NanH1 (A) and NanH2 (B) were assayed for activity on a library of sialosides with either α2,3- or α2,6-sialyl linkages depicted in white and black, respectively. The error bars in the graphs represent the standard errors of experimental values obtained from duplicated samples in an individual experiment. Significant differences were determined with a Student's t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Sialidase kinetics were characterized using both α2–3- and α2–6-linked sialyllactosyl 4-methylumbelliferol (MU) (Neu5Acα2–3/6Lacβ-Mu). Interestingly, both sialidases had similar affinities toward these two substrates with similar Km values (supplemental Fig. S4). NanH1 was approximately twice as efficient at hydrolyzing α2–6-linked Neu5Ac (0.22 s−1 mm−1) as α2–3 sialyl linkages (0.13 s−1 mm−1) because of greater affinity for this substrate (Table 1). The same preference for α2–6 linkages was exhibited by NanH2, though with a considerably higher efficiency reflecting a 175-fold greater turnover (kcat) rate than NanH1. NanH2 hydrolyzed α2–3 linkages more efficiently than NanH1 as well. Interestingly, both sialidases had similar affinities toward the two α linkages as approximated by Km. Furthermore, both sialidases were more active under acidic conditions (pH 4.5–6.0), a pH range typical of several bacterial sialidases (Fig. 4).
B. longum subsp. infantis NanH1 and NanH2 Sialidase Activity on Purified MSO
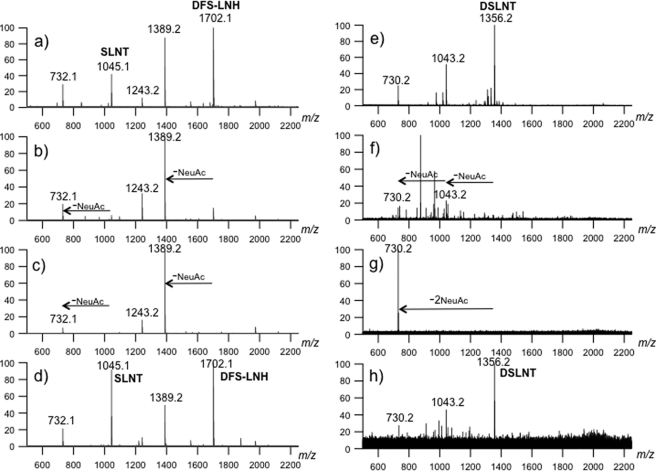
To link exo-sialidase function as the committed step in MSO utilization, NanH1 and NanH2 were assayed for hydrolysis of purified MSO substrates by mass spectrometry. Accordingly, single isomers of SLNT and di-fucosylayed sialylated lacto-N-hexaose-like oligosaccharide (DFS-LNH) were isolated from pooled HMO by solid phase extraction prior to liquid chromatography (Fig. 7a). These two MSO species, as well as commercially obtained α2–3- and α2–6-linked disialyl-lacto-N-tetraose (DSLNT) (Fig. 7e) were incubated with both recombinant ATCC15697 sialidases and a control α(2–3)-neuraminidase. All three purified MSO species were rapidly digested by NanH2 and the control α(2–3)-neuraminidase (Fig. 7, b, c and f, g). Significantly contrasting with this, NanH1 did not hydrolyze any of the three purified sialylated milk oligosaccharides (Fig. 7, d and h) following an extended incubation of 1 h.
FIGURE 7.
Digestion of MSO species with 3 recombinant sialidases for 30 min. SLNT and DFSLNH were purified from a pool of HMO and DSLNT was obtained from a commercial source. HPLC fraction before digestion of purified HMO (i.e. SLNT and DFSLNH) (a); digestion of purified HMO with an α2–3-neuraminidase (b); digestion of purified HMO with NanH2 (c); digestion of purified HMO with NanH1 (d); commercially obtained DSLNT prior to digestion (e); DSLNT digestion with an α2–3-neuraminidase (f); DSLNT digestion with NanH2 (g); DSLNT digestion with NanH1 (h).
DISCUSSION
György et al. (1) reported that sialyl moieties protect HMO from bacterial consumption. Their HMO-utilizing B. bifidum strain (strict α2–3 sialidase activity) did not ferment sialylated milk oligosaccharides without an in vitro hydrolysis of α2–6 linkages predominant in milk glycans. Therefore it is significant that B. longum subsp. infantis ATCC15697 utilizes SLNT and produces the efficient NanH2 α2–6 sialidase active on MSO linkages. This functionally links substrates incorporated into milk with the 43-kbp gene cluster previously predicted to enable HMO catabolism (21). To this end, HMO (neutral and/or acidic) fermentation likely initiates a signal cascade to up-regulate nanH2 expression to utilize MSO.
Long regarded as a virulence factor, sialic acid catabolism confers an in vivo competitive advantage regarded to be critical to pathogenesis, as recently described in a murine Vibrio cholerae infection model (45). While saccharolytic commensals pervade the distal infant GIT, utilization of sialylated glycans delivered to their niche through nursing was previously unknown, and may be a significant factor involved in colonization and persistence. That SLNT is metabolized in the presence of highly preferred neutral HMOs signifies that they are of an equivalent value to B. longum subsp. infantis thus tantalizingly suggestive of a lifestyle predicated on the acquisition of MSO.
The progressive disappearance of SLNT during HMO fermentation is a consequence of ATCC15697 metabolism and is not confounded by extracellular degradation. If spurious digestion had occurred, all four monitored MSO compositions would have been eliminated to the same extent as SLNT, likely approaching 100% disappearance as observed in the in vitro assays on purified MSO. This would be similar to the nonspecific glycoprofile of the prominent GIT commensal Bacteroides fragilis that employs several extracellular glycoside hydrolases to utilize HMO (46). Moreover, an increase in the abundance of an LNT-like tetrasaccharide, concomitant with SLNT desialylation was not observed (data not shown). Rather, both LNT and free sialic acids disappear during pooled HMO fermentation, with the latter previously demonstrated to enter the fructose-6-phosphate phosphoketolase pathway when utilized as a sole carbon source (19, 21).
Bacteria competent for sialic acid metabolism derive energy and biomass from Neu5Ac or sequester it to be deployed as extracellular decoration, cloaked from immunogenic responses (reviewed in Refs. 13, 47). Recently, B. fragilis was characterized with a peculiar Neu5Ac catabolism in which N-acetylmannosamine epimerase (pfam07221) converts manNAc to N-acetylglucosamine to be subsequently phosphorylated (48). This contrasts with the canonical pathway in which ManNAc is phopshorylated prior to epimerization (49). While the ATCC15697 chromosome does not encode an epimerase with significant identity, ROK kinases appear in the sialic acid utilization cluster (Blon_0644) and upstream of nagA and nagB (Blon_0880) clustering with N-acylglucosamine 2-epimerase (pfam07221; Blon_0875). These kinases, and putative epimerase, may function in a similar manner to the B. fragilis system, mitigating the lack of a clearly identifiable nanK gene within the ATCC15697 chromosome (50).
In contrast to NanH2, a linkage between NanH1 and MSO utilization is less evident, though consistent with the physiology of other bifidobacterial species. Specifically, B. breve strains tested to date do not subsist on HMO/MSO despite the presence of a nanH1 homolog within its genome. B. bifidum strains, in contrast, utilize HMO and degrade MSO at its extracellular surface, ostensibly accomplished with secreted NanH2 homologs (19, 42). NanH1 may be active on other sialylated glycans encountered in the infant gut, be it delivered by milk or endogenously present. Possible NanH1 substrates include various milk glycopeptides, monosialoganglioside, or colostrum disialoganglioside, or sialylated mucins.
In any event, the importance of translocation to MSO metabolism is accentuated by intracellular localization of B. longum subsp. infantis sialidase activity. Whereas bacterial sialic acid transporters have been elucidated (i.e. SiaT (51), NanT (52), SatABCD (38)), transport of acidic oligosaccharides remains unexplored. The multitude of transporters encoded within the ATCC15697 genome present several attractive candidates, including several transport genes within the HMO cluster or others unlinked in cis. Interestingly, the ATCC15697 locus comprised of a predicted epimerase and ROK genes upstream of nagAB are proximal to a solute binding protein, which binds LNT-like carbohydrates, thus potentially involved in SLNT transport.5
Until now, direct microbial interactions with milk sialyloligosaccharides have been solely characterized in terms of innate immune function. Inherently shielded by their acidic moiety, MSOs are frequently fucosylated to present a multi-layered barrier to microorganisms seeking to metabolize HMO core components (e.g. LNT), an insurmountable prospect in the absence of fucosidase and sialidase activities. Recalcitrant MSOs are therefore likely concentrated in the distal GIT, potentially more so than neutral species that are consumed by a more diverse assortment of microbial scavengers. Thus the ability to access these highly restricted carbohydrates may contribute to the establishment of bifidobacteria in the nursing infant gut. If digestible by the infant, HMOs would be a calorically dense nutrient for the infant, irrespective of sialylation. As such, HMOs are either appropriated for other early developmental functions (e.g. immune or neural function) or do not increase progeny fitness, with the latter in need of reconciliation with the considerable maternal resources expended in their synthesis. At any rate, the provision of HMO to the neonate has evolved either reciprocally or has arisen unilaterally in the bifidobacteria likely to the benefit of the host.
Acknowledgments
We thank Eric Vimr for helpful discussions and Karen Kalenetra for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health NICHD Awards 5R01HD059127 and 1R01HD061923 and by National Institutes of Health-NIGMS 3R01GM076360. This work was also supported by grant support from the University of California Discovery Grant Program, Dairy Management Inc., the California Dairy Research Foundation, and the USDA NRI-CSREES Award 2008-35200-18776.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
D. A. Mills, unpublished data.
- HMO
- human milk oligosaccharide
- MSO
- milk sialyloligosaccharide
- SLNT
- sialylated lacto-N-tetraose
- MI
- mass-intensity
- DSLNT
- disialyl-lacto-N-tetraose
- FTICR-MS
- Fourier transform ion cyclotron resonance MS
- ConA
- concanavalin A
- pNP
- para-nitrophenol
- DFS-LNH
- di-fucosylayed sialylated lacto-N-hexaose-like oligosaccharide
- SPE
- solid phase extraction.
REFERENCES
- 1. György P., Jeanloz R. W., von Nicolai H., Zilliken F. (1974) Eur. J. Biochem. 43, 29–33 [DOI] [PubMed] [Google Scholar]
- 2. Ward R. E., Niñonuevo M., Mills D. A., Lebrilla C. B., German J. B. (2006) Appl. Environ. Microbiol. 72, 4497–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bao Y., Zhu L., Newburg D. S. (2007) Anal. Biochem. 370, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Engfer M. B., Stahl B., Finke B., Sawatzki G., Daniel H. (2000) Am. J. Clin. Nutr. 71, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 5. Bode L. (2006) J. Nutr. 136, 2127–2130 [DOI] [PubMed] [Google Scholar]
- 6. Morrow A. L., Ruiz-Palacios G. M., Altaye M., Jiang X., Guerrero M. L., Meinzen-Derr J. K., Farkas T., Chaturvedi P., Pickering L. K., Newburg D. S. (2004) J. Pediatr. 145, 297–303 [DOI] [PubMed] [Google Scholar]
- 7. Ruiz-Palacios G. M., Calva J. J., Pickering L. K., Lopez-Vidal Y., Volkow P., Pezzarossi H., West M. S. (1990) J. Pediatr. 116, 707–713 [DOI] [PubMed] [Google Scholar]
- 8. Barthelson R., Mobasseri A., Zopf D., Simon P. (1998) Infect. Immun. 66, 1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mahdavi J., Sondén B., Hurtig M., Olfat F. O., Forsberg L., Roche N., Angstrom J., Larsson T., Teneberg S., Karlsson K. A., Altraja S., Wadström T., Kersulyte D., Berg D. E., Dubois A., Petersson C., Magnusson K. E., Norberg T., Lindh F., Lundskog B. B., Arnqvist A., Hammarström L., Borén T. (2002) Science 297, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon P. M., Goode P. L., Mobasseri A., Zopf D. (1997) Infect. Immun. 65, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yolken R. H., Peterson J. A., Vonderfecht S. L., Fouts E. T., Midthun K., Newburg D. S. (1992) J. Clin. Invest. 90, 1984–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schauer R. (2000) Glycoconj. J. 17, 485–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vimr E. R., Kalivoda K. A., Deszo E. L., Steenbergen S. M. (2004) Microbiol. Mol. Biol. Rev. 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewis A. L., Desa N., Hansen E. E., Knirel Y. A., Gordon J. I., Gagneux P., Nizet V., Varki A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13552–13557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. LoCascio R. G., Ninonuevo M. R., Freeman S. L., Sela D. A., Grimm R., Lebrilla C. B., Mills D. A., German J. B. (2007) J. Agric. Food Chem. 55, 8914–8919 [DOI] [PubMed] [Google Scholar]
- 16. Le Blay G., Michel C., Blottière H. M., Cherbut C. (1999) J. Nutr. 129, 2231–2235 [DOI] [PubMed] [Google Scholar]
- 17. Falony G., Vlachou A., Verbrugghe K., De Vuyst L. (2006) Appl. Environ. Microbiol. 72, 7835–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liévin V., Peiffer I., Hudault S., Rochat F., Brassart D., Neeser J. R., Servin A. L. (2000) Gut. 47, 646–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward R. E., Niñonuevo M., Mills D. A., Lebrilla C. B., German J. B. (2007) Mol. Nutr. Food Res. 51, 1398–1405 [DOI] [PubMed] [Google Scholar]
- 20. LoCascio R. G., Niñonuevo M., Kronewitter S. R., Freeman S. L., German J. B., Lebrilla C. B., Mills D. A. (2009) Microb. Biotechnol. 2, 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sela D. A., Chapman J., Adeuya A., Kim J. H., Chen F., Whitehead T. R., Lapidus A., Rokhsar D. S., Lebrilla C. B., German J. B., Price N. P., Richardson P. M., Mills D. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Markowitz V. M., Kyrpides N. C. (2007) Methods Mol. Biol. 395, 35–56 [DOI] [PubMed] [Google Scholar]
- 23. Caspi R., Foerster H., Fulcher C. A., Kaipa P., Krummenacker M., Latendresse M., Paley S., Rhee S. Y., Shearer A. G., Tissier C., Walk T. C., Zhang P., Karp P. D. (2008) Nucleic Acids Res. 36, D623–D631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R. (2005) Nucleic Acids Res. 33, W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilson D., Madera M., Vogel C., Chothia C., Gough J. (2007) Nucleic Acids Res. 35, D308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K., Dudley J., Nei M., Kumar S. (2007) Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 27. Jones D. T., Taylor W. R., Thornton J. M. (1992) Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 28. Felsenstein J. (1985) Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 29. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 30. Xie Y., Liu J., Zhang J., Hedrick J. L., Lebrilla C. B. (2004) Anal. Chem. 2004, 5186–5197 [DOI] [PubMed] [Google Scholar]
- 31. Clowers B. H., Dodds E. D., Seipert R. R., Lebrilla C. B. (2008) Anal. Biochem. 381, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dodds E. D., Tassone F., Hagerman P. J., Lebrilla C. B. (2009) Anal. Chem. 81, 5533–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dodds E. D., German J. B., Lebrilla C. B. (2007) Anal. Chem. 79, 9547–9556 [DOI] [PubMed] [Google Scholar]
- 34. Parche S., Beleut M., Rezzonico E., Jacobs D., Arigoni F., Titgemeyer F., Jankovic I. (2006) J. Bacteriol. 188, 1260–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J. H., Lindsay L. L., Hedrick J. L., Lebrilla C. B. (2004) Anal. Chem. 76, 5990–6001 [DOI] [PubMed] [Google Scholar]
- 36. Niñonuevo M., An H., Yin H., Killeen K., Grimm R., Ward R., German B., Lebrilla C. (2005) Electrophoresis 26, 3641–3649 [DOI] [PubMed] [Google Scholar]
- 37. Tao N., DePeters E. J., Freeman S., German J. B., Grimm R., Lebrilla C. B. (2008) J. Dairy Sci. 91, 3768–3778 [DOI] [PubMed] [Google Scholar]
- 38. Post D. M., Mungur R., Gibson B. W., Munson R. S., Jr. (2005) Infect. Immun. 73, 6727–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor G., Dineley L., Glowka M., Laver G. (1992) J. Mol. Biol. 225, 1135–1136 [DOI] [PubMed] [Google Scholar]
- 40. Gaskell A., Crennell S., Taylor G. (1995) Structure 3, 1197–1205 [DOI] [PubMed] [Google Scholar]
- 41. Mavromatis K., Chu K., Ivanova N., Hooper S. D., Markowitz V. M., Kyrpides N. C. (2009) PLoS One 4, e7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ashida H., Miyake A., Kiyohara M., Wada J., Yoshida E., Kumagai H., Katayama T., Yamamoto K. (2009) Glycobiology 19, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 43. Cao H., Li Y., Lau K., Muthana S., Yu H., Cheng J., Chokhawala H. A., Sugiarto G., Zhang L., Chen X. (2009) Org. Biomol. Chem. 7, 5137–5145 [DOI] [PubMed] [Google Scholar]
- 44. Chokhawala H. A., Yu H., Chen X. (2007) Chembiochem. 8, 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Almagro-Moreno S., Boyd E. F. (2009) Infect. Immun. 77, 3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marcobal A., Barboza M., Froehlich J. W., Block D. E., German J. B., Lebrilla C. B., Mills D. A. (2010) J. Agric. Food Chem. 58, 5334–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Severi E., Hood D. W., Thomas G. H. (2007) Microbiology 153, 2817–2822 [DOI] [PubMed] [Google Scholar]
- 48. Brigham C., Caughlan R., Gallegos R., Dallas M. B., Godoy V. G., Malamy M. H. (2009) J. Bacteriol. 191, 3629–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vimr E. R., Troy F. A. (1985) J. Bacteriol. 164, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brigham C. J., Malamy M. H. (2005) J. Bacteriol. 187, 890–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Allen S., Zaleski A., Johnston J. W., Gibson B. W., Apicella M. A. (2005) Infect. Immun. 73, 5291–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martinez J., Steenbergen S., Vimr E. (1995) J. Bacteriol. 177, 6005–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deleted in proof.