Abstract
Previous studies of E2F family members have suggested that protein-protein interactions may be the mechanism by which E2F proteins are recruited to specific genomic regions. We have addressed this hypothesis on a genome-wide scale using ChIP-seq analysis of MCF7 cell lines that express tagged wild type and mutant E2F1 proteins. First, we performed ChIP-seq for tagged WT E2F1. Then, we analyzed E2F1 proteins that lacked the N-terminal SP1 and cyclin A binding domains, the C-terminal transactivation and pocket protein binding domains, and the internal marked box domain. Surprisingly, we found that the ChIP-seq patterns of the mutant proteins were identical to that of WT E2F1. However, mutation of the DNA binding domain abrogated all E2F1 binding to the genome. These results suggested that the interaction between the E2F1 DNA binding domain and a consensus motif may be the primary determinant of E2F1 recruitment. To address this possibility, we analyzed the in vivo binding sites for the in vitro-derived consensus E2F1 motif (TTTSSCGC) and also performed de novo motif analysis. We found that only 12% of the ChIP-seq peaks contained the TTTSSCGC motif. De novo motif analysis indicated that most of the in vivo sites lacked the 5′ half of the in vitro-derived consensus, having instead the in vivo consensus of CGCGC. In summary, our findings do not provide support for the model that protein-protein interactions are involved in recruiting E2F1 to the genome, but rather suggest that recognition of a motif found at most human promoters is the critical determinant.
Keywords: Chromatin Immunoprecipitation (ChIP), DNA Binding Protein, E2F Transcription Factor, Transcription Promoter, Transcription Regulation
Introduction
A critical question in gene regulation is how selective sets of transcription factors are specifically recruited to their target sites. For site-specific DNA binding factors, a major component of the genomic recruitment mechanism is the highly specific interaction of the DNA binding protein with its consensus motif. The relatively new technology of ChIP-seq has allowed very precise analyses of sequences involved in recruitment of site-specific DNA binding factors (and proteins complexes associated with DNA binding factors) to specific genomic locations. For example, the in vivo binding sites for transcription factors such as p63, STAT1, and REST show high enrichment for a specific motif. In fact, ∼75% of the peaks identified by ChIP-seq for these factors contain the known consensus motif for that factor within 50 nucleotides of either side of the center of the peak (1). However, there are clear examples of genomic recruitment of site-specific transcription factors being dictated, at least in part, by protein-protein interactions. For example, approximately half of the binding sites for the serum response factor are cell type-specific, and it has been proposed that the cell type-specific binding is due to serum response factor making different protein-protein interactions in different cell types (2). Although tethered recruitment has been proposed as a mechanism by which human transcription factors can be recruited to the genome, very few studies have tested this possibility by analyzing the in vivo binding patterns of transcription factors that have been mutated in their DNA binding and/or protein interaction domains. However, a recent study has shown that the estrogen receptor can be recruited to the genome through both a direct interaction of its DNA binding domain with a well characterized estrogen response element and via tethering mediated by interactions of the estrogen receptor and other DNA binding proteins such as Runx (3).
E2F1 is the founding member of a set of transcription factors that have been implicated in controlling critical cellular (entrance into S phase, regulation of mitosis, apoptosis, DNA repair, and DNA damage checkpoint control) and organismal (regulation of differentiation, development, and tumorigenesis) functions (4–6). There are eight genes for E2F family members encoded in the human genome (see Refs. 5 and 7 for recent reviews of the E2F family), with the highest degree of homology among the E2F family members being in their DNA binding domains (DBDs).3 E2F family members bind poorly in vitro unless they are complexed with a member of the DP family of transcription factors (5, 8–10). However, E2F7 and E2F8 are exceptions to this rule, functioning as homodimers or heterodimers with each other (11–17). The DBD of E2F1, located between amino acids 120–191, consists of a basic helix-loop-helix structure (4), with a fold resembling a winged helix DNA binding motif, as revealed by crystal structure analysis (18). Although the DBD is required for direct binding to DNA, it is not sufficient for in vitro binding. High affinity binding to DNA also requires the contribution of the adjacent hydrophobic heptad repeat leucine zipper domain (amino acids 188–241), which is known to be involved in heterodimerization with the DP family of transcription factors (10, 19–23). A multitude of in vitro DNA-protein interaction studies and promoter reporter assays have identified an E2F consensus motif of TTTSSCGC, where S is either a G or a C (4, 24), which is both necessary and sufficient for E2F binding in vitro (4, 24).
Although the DNA binding domain of E2F1 is clearly critical for in vitro DNA binding (25), it has also been suggested that other site-specific transcription factors may influence the recruitment of E2F family members to in vivo binding sites. For example, using cells stably transfected with wild type (WT) or mutant herpes simplex virus thymidine kinase promoter constructs, Karlseder et al. (26) showed that occupancy of the E2F site in that promoter required the adjacent SP1 consensus site. Furthermore, the N terminus of the E2F1 protein was shown to directly interact with SP1, suggesting that tethering of E2F1 to the genome was mediated by SP1 (27). Several additional studies have investigated a possible partnership between these two transcription factors and confirmed cooperative binding between SP1 and E2F1 at the c-myc, DHFR, and mouse TK promoters (26, 28, 29). Because an SP1 consensus motif has been identified as one of the most common motifs present in human promoters (30), it is possible that tethering of E2F1 to the genome via interaction of its N terminus with SP1 may be an important recruitment mechanism. In addition to the N terminus, other domains of E2F1 have been implicated in protein-protein interactions. For example, previous studies have demonstrated that TFE-3 physically interacts with E2F3 and helps to recruit E2F3 to the ribonucleotide reductase 1, ribonucleotide reductase 2, and DNA polymerase α p68 subunit promoters (31, 32). Similarly, RYBP (Ring1 and YY1 binding protein) was identified as a “bridging” molecule between YY1 and certain E2F family members that can assist in the regulation of the CDC6 promoter (33). Of note, the protein-protein interactions between either TFE-3 or RYBP with E2F proteins were shown to be dependent on the E2F marked box domain (amino acids 243–358). The E2F marked box domain has also been implicated in facilitating DNA binding of E2F proteins via its interaction with DP1, in contributing to E2F-mediated DNA bending (34, 35), and in interactions with other factors such as Jab1 (36). Finally, NF-YA has been shown to be required for adjacent binding of E2F3 to the cdc2 promoter (36), whereas E2F4 binding to the c-myc promoter was shown to depend on simultaneous binding of the SMAD proteins (37). However, in these latter two cases, the domain of E2F required for the interaction has not been delineated.
In addition to interacting with other site-specific DNA binding factors, members of the E2F family have also been shown to interact with components of the general transcriptional machinery and/or other types of co-regulatory proteins. For example, the C-terminal transactivation domain of E2F1 (amino acids 368–437) can interact with the basal transcription factors TFIID, TFIIH, and TBP, as well as with transcription coactivators, including CBP/p300, TRRAP, GCN5, Tip60, and NCOA3 (38–46). Unlike many transcription factors that bind to both promoter and enhancer regions (see Ref. 47 for a review), E2F1 binds almost exclusively to core promoter regions (48–50), and the binding pattern of E2F1 is essentially indistinguishable from that of RNA polymerase II or TAF1 (the largest subunit of TFIID). Therefore, it is quite possible that E2F1 could be tethered to certain promoters via the strong interactions of its C-terminal transactivation domain with general transcription factors. The transactivation domain of E2F1 can also interact with members of the retinoblastoma tumor suppressor protein family. Although retinoblastoma lacks the ability to bind directly to DNA, it does interact with site-specific transcription factors such as AP2 and thus may serve as a bridge that allows AP2 to tether E2F1 to the genome (51, 52). The C-terminal 70 amino acids of E2F1 can also interact with ANCCA (AAA nuclear coregulator cancer-associated protein, also known as ATAD2) (53). In addition to interacting with SP1, the N terminus of E2F can also interact with ANCCA and with cyclin A (53, 54).
Taken together, the many functional studies of the E2F family suggest that protein-protein interactions may play an important role in recruiting E2F1 to the genome. However, most of the above-mentioned studies were performed in vitro or focused on one, or at most a handful, of genomic binding sites. Therefore, we have now used ChIP-seq to test the hypothesis that protein-protein interactions are involved in recruiting E2F1 to target sites in the human genome.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mm glutamine, and 1% penicillin and Streptomycin. All cells were incubated at 37 °C in a humidified 5% CO2 incubator.
Cloning of E2F1 Mutants
All E2F1 mutants were amplified from the pCMV-HA-ER-E2F1wild type plasmid DNA template by PCR with either AccuTaq (Sigma) or Finnzymes Phusion high fidelity polymerase (New England Biolabs, catalog no. F-531) using primers that introduced unique BamHI sites immediately 5′ and 3′ to the coding sequence site of interest, respectively. The resulting E2F1 mutant proteins were introduced into the pCMV HA estrogen receptor (ER) plasmid (a generous gift from Kristian Helin) using the BamHI sites; successful cloning of the various inserts was confirmed by sequencing at the University of California, Davis Sequencing Facility. The pCMV-HA-ER, pCMV-HA-ER-E2F1 wild type, or pCMV-HA-ER-E2F1 (E132) constructs were all generous gifts from Kristian Helin.
Generation of HA-ER-E2F Stable Cell Lines
Stable clones were generated by transfecting MCF7 cells on six-well dishes with 1 μg of either pCMV-HA-ER-E2F1ΔC mutant, the pCMV-HA-ER-E2F1ΔN/C mutant, or the pCMV-HA-ER- E2F1ΔMB mutant using FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's recommendations. Forty-eight hours after transfection, the cells were placed under selection in medium containing 1 mg/ml G418. Individual drug-resistant colonies were isolated and assayed for ectopic HA-ER-E2F fusion protein expression by Western blot analysis of total cellular protein using a 1:1000 dilution of anti-HA.11 (16B12 clone; Covance catalog no. MMS-101P) and a 1:10,000 dilution of anti-nucleoporin p62 (BD Transduction Laboratories, catalog no. N43620) antibodies in 5% milk. To ensure that the stably integrated fusion proteins properly translocated into the nucleus upon stimulation with 4-hydroxytamoxifen (4-OHT), clones determined to have high ectopic expression of each fusion protein were treated with 600 nm of 4-OHT (Sigma) for 30 min and processed for cytoplasmic and nuclear protein extraction. Briefly, subconfluent cells were harvested after 4-OHT treatment by scraping in ice-cold PBS containing 1 mm PMSF and processed using the NE-PER nuclear and cytoplasmic extraction kit from Pierce, according to the manufacturer's instructions. Both the nuclear and cytoplasmic extracts (20 μg) were boiled in 4× SDS sample buffer for 5 min, loaded onto a 10% SDS-polyacrylamide gel, and further processed for Western blot as described previously by Xu et al. (27). Positive MCF7 clones for each fusion protein were expanded in culture and subsequently treated for 30 min with 4-hydroxytamoxifen (Sigma) at a final concentration of 600 nm immediately prior to formaldehyde cross-linking and harvesting for ChIP assays.
ChIP-seq Assays
All cell cultures were cross-linked for 10 min by adding formaldehyde to the growth medium to a final concentration of 1%. Cross-linking was stopped by the addition of glycine to a final concentration of 125 mm, and cells were washed three times with ice-cold PBS prior to harvesting by scraping of the plates. Chromatin was fragmented using the Bioruptor sonicator (Diagenode) for 20 min (15-s pulses and 1-min pauses in between) to produce fragments ∼500 nt in size. ChIP assays were performed using 1 × 108 cells for each ChIP as described at the Farmham Laboratory Protocol Web site. The antibody used was HA.11 (16B12 clone; Covance catalog no. MMS-101P). Immunoprecipitates were collected using the Staph A method. ChIP samples were tested by PCR using positive and negative control primer sets prior to making the library (supplemental Fig. S3). ChIP libraries were created according to Robertson et al. (55). Libraries were run on a 2% agarose gel, and the 150–400 bp, 200–400 bp, or 400–600 bp fraction of the library was extracted and purified; the library with the highest enrichment, as monitored by quantitative PCR, was used for sequencing. The libraries were quantitated using serial dilutions by real-time PCR using primers complementary to the library adapters or by Bioanalyzer analysis. See supplemental File S1 for more details concerning library preparation.
Comparison of E2F1 and H3K4me3 Binding Patterns
H3K4me3 ChIP-seq data from MCF7 cells was provided by the University of Washington ENCODE group led by John Stamatoyannopoulos (a part of the ENCODE Project Consortium). The raw reads were initially mapped as Sequence Alignment/Map format to the human HG19 genome assembly to obtain 30,857,387 uniquely mapped reads. The HG19 version of the mapped reads was converted to the HG18 version using a liftover program. Using our BELT program (56), a set of 34,164 binding sites for H3K4me3 was identified with a false discovery rate of 4.8%. We used the top 20,000 H3K4me3 binding sites to eliminate any false positives at the bottom of the list of ranked peaks. To fairly compare binding patterns between E2F1 and H3K4me3, we also called peaks for the E2F1 ChIP-seq data using BELT. We used the top 10,000 binding sites for E2F1 called by BELT to be consistent with the other analyses of E2F1 binding. We compared the E2F1 and H3K4me3 binding patterns using 100-nt intervals.
RESULTS
E2F1 DNA Binding and Heterodimerization Domains Are Sufficient for Recruitment to all Genomic Binding Sites
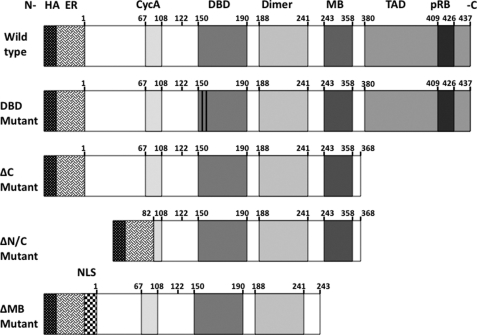
To address the question as to whether protein-protein interactions are involved in recruiting E2F1 to the genome, we first needed to engineer a system that would allow us to perform genome-wide ChIP-seq analysis of mutant proteins. We have shown previously that a continual high level expression of E2F1 is toxic to cells (57). Therefore, it was critical that we use an inducible system to express E2F1 derivatives. Another requirement for our studies was that we needed to distinguish the introduced E2F proteins from the endogenous E2F1 protein in our ChIP-seq experiments. Therefore, we cloned the WT and mutant E2F proteins into an expression construct that provides an N-terminal HA tag, followed by a modified ER ligand binding domain (Fig. 1). The advantages of using this particular expression system for these studies is that the HA tag allows for the specific isolation of these mutants using an HA antibody in subsequent chromatin immunoprecipitation steps, whereas the ER domain allows for the regulated translocation of the E2F1 proteins into the nucleus upon treatment with the anti-estrogen 4-hydroxytamoxifen (4-OHT). We created cell lines stably maintaining the plasmids harboring the constructs shown in Fig. 1, grew the cells to the large number required for ChIP-seq, treated with 4-OHT, and then harvested cells for Western blot analysis and ChIP-seq (see Table 1 for a summary of the ChIP-seq results for WT E2F1 and all of the E2F1 mutants).
FIGURE 1.
Schematic of E2F1 constructs. A schematic of the various E2F1 fusion protein constructs is shown. All constructs contain the influenza HA tag and the estrogen receptor ligand binding domain (ER) immediately preceding the N terminus (indicated as -N) of the E2F1 coding sequence. The amino acid positions of the different domains are indicated above each construct. These domains include the cyclin A binding domain (CycA), the DBD, the heterodimerization domain (Dimer), the marked box domain (MB), and the C-terminal (indicated as -C) transactivation domain (TAD) with the pocket protein binding domain (pRB) embedded within it. The E2F1ΔMB is unique in that it has the SV40 large T antigen nuclear localization domain (NLS) introduced immediately upstream of the E2F1 coding sequence. For the E2F1 DBD mutant, the two vertical bars within the DBD domain represent a two-point amino acid substitution.
TABLE 1.
Summary of ChIP-seq peaks
| Number of peaks | Highest peak height | Median peak height | Lowest peak height | Average peak width | |
|---|---|---|---|---|---|
| WT1 | 10,233 | 221 | 25 | 11 | 452 |
| WT2 | 11,794 | 226 | 33 | 15 | 432 |
| WT merged | 15,944 | 229 | 39 | 14 | 469 |
| 1–368 (ΔC) | 17,944 | 229 | 37 | 12 | 485 |
| 82–368 (ΔNC) | 29,258 | 229 | 32 | 14 | 520 |
| 1–243 (ΔMB) | 28,427 | 214 | 30 | 12 | 419 |
| E132 (ΔDNA) | 508 | 88 | 17 | 10 | 259 |
We have shown previously, using ChIP-chip and NimbleGen high density oligonucleotide arrays (50), that HA tagged WT E2F1 binds to the same targets as does the endogenous E2F1 in MCF7 cells. Therefore, we have used the binding pattern of the HA-tagged WT E2F1 as our standard for comparison to the binding pattern of all mutant E2F1 proteins for our ChIP-seq experiments. We first performed ChIP-seq for the WT E2F1, identified binding sites using the Sole search peak calling program (58) and found that E2F1 binds to ∼10,000 genomic locations using a false discovery rate cut-off of 0.0001. To test the reproducibility of our assay, we grew an independent culture of MCF7 cells, induced nuclear translocation of the WT E2F1, and performed another ChIP-seq experiment. The second replicate (called WT2) produced ∼11,000 peaks. A direct overlap of the top 10,000 peaks identified in the two replicates gave a 78% overlap. Because the smallest peaks identified by ChIP-seq can vary from experiment to experiment (often depending on the number of sequenced tags), the ENCODE Consortium has developed a method for comparing replicates. This method involves comparing the top 40% of the peaks in one replicate to the entire set of peaks from the other replicate. Essentially, this method ensures that the majority of the big peaks are the same from experiment to experiment. Most biological replicates of the same factor show an 80–90% overlap using this method of comparison. Using this “top 40% method” to determine overlap, we found that the two WT E2F1 ChIP-seq experiments gave a 97% overlap; thus, our experiments are very reproducible.
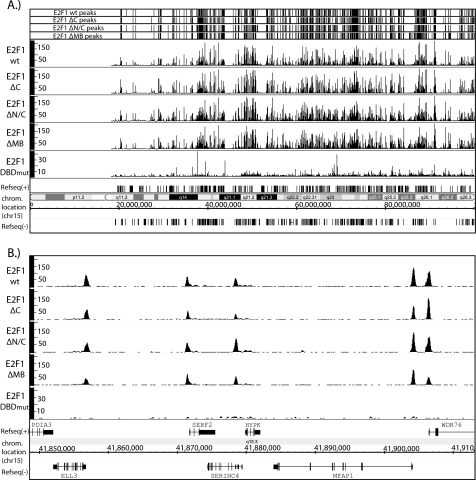
As expected from our previous studies (48, 49, 58), E2F1 binds almost exclusively to core promoter regions of the well characterized set of RefSeq genes (supplemental Fig. S1). As indicated above, the transactivation domain of E2F1 has been shown previously to bind to a variety of transcriptional regulatory proteins, including TBP, TFIIH, CBP, retinoblastoma, CBP/p300, TRAPP, GCN5, Tip60, and ACTR/AIB1 (38–46). Therefore, we tested the hypothesis that interactions between the E2F1 transactivation domain and components of the general transcriptional machinery were involved in recruiting E2F1 to core promoter regions. To do so, we created a stable MCF7 cell line expressing E2F1ΔC, induced nuclear translocation, and performed a ChIP-seq assay. We identified ∼18,000 peaks for E2F1ΔC. As shown in Fig. 2, top panel, the called peaks for the E2F1ΔC are very similar to the called peaks of wt E2F1. In fact, an overlap analysis indicates that the similarity of E2F1ΔC to E2F1 WT1 is essentially the same as the similarity of two biologically independent E2F1 WT ChIP-seq experiments (Table 2). When we performed the top 40% overlap analysis comparing the E2F1ΔC with WT E2F1, we found that all of the top-ranked peaks are the same (99% overlap; Table 2). Closer examination of the binding patterns show that binding of E2F1ΔC is indistinguishable from the binding of WT E2F1 (Fig. 2, middle and bottom panels).
FIGURE 2.
Comparison of the binding patterns of the E2F1 constructs. A, the positions of the top 10,000 ranked peaks are shown for the WT, ΔC, ΔN/C, and ΔMB E2F1 fusion protein ChIP-seq data sets for the entire chromosome 15. The Sequence GRaph visualization files for the WT, ΔC, ΔN/C, ΔMB, and the DBD mutant E2F1 fusion protein ChIP-seq data sets are also shown. The number of tags is shown on the y axis of each track. (Note that a different scale was used for the DBD mutant (DBDmut) than the rest of the E2F1 fusion proteins because fewer unique mapped reads were obtained for this mutant.) The chromosomal coordinates and the location (chrom. location) of the RefSeq genes, transcribed either in the forward (+) or reverse direction (−), are indicated on the x axis. B, a closer view of a ∼60-kb region of chromosome 15, showing a more detailed profile of the peaks for each of the E2F1 fusion proteins. The x and y coordinates are as described in A.
TABLE 2.
Overlap analysis of ChIP-seq peaks
The percent overlap between the top 10,000 ranked peaks for the first E2F1 wild type ChIP-seq replicate versus the second E2F1 wild type ChIP-seq replicate (WT2) or the indicated E2F1 mutants is listed in the left column. The percent overlap between the top 40% of the top ranked peaks for the first E2F1 wild type replicate and all of the top ranked peaks of WT2 or the indicated E2F1 mutants is listed in the right column. All overlaps were performed using the Sole-Search GFF overlap tool.
| Top 10,000 overlap | Top 40% overlap | |
|---|---|---|
| WT2 | 78 | 97 |
| ΔC | 80 | 99 |
| ΔN/C | 76 | 96 |
| ΔMB | 74 | 92 |
We next investigated the role of the N-terminal domain of E2F1. This domain has been implicated previously in interaction with the transcription factor SP1 and with other proteins such as cyclin A. Because SP1 binding motifs are one of the most common motifs in human core promoters (30), it was possible that interaction of E2F1 via its N terminus with SP1 is critical for the genomic recruitment of E2F1 to many of its thousands of target promoters. To test this hypothesis, we deleted the first 82 amino acids from the E2F1ΔC protein, creating E2F1ΔN/C. We then created a stable cell line, induced nuclear translocation of E2F1ΔN/C, performed ChIP assays, created a library, and analyzed the binding pattern by ChIP-seq. Once again, the pattern of peaks called using Sole-search for E2F1ΔN/C is very similar to the pattern of peaks for WT E2F1 (Fig. 2, top panel), the actual binding profiles are indistinguishable, and the overlap analysis indicates that no major peaks were lost due to deletion of both the N and C termini of E2F1 (Table 2).
Having ruled out the involvement of the N and C terminus in recruiting E2F1 to its genomic targets, we next evaluated the contribution of the E2F1 marked box domain to genomic recruitment. The marked box (MB) domain has been implicated previously in several protein-protein interactions with other transcription factors. Specifically, this domain has been shown to be involved in recruiting E2F2 and E2F3 to promoter regions (31–33) and has been suggested to be important for the interaction of E2F1 with DP1 (34). To investigate the role of the MB domain in genomic recruitment of E2F1, we created an E2F1 expression construct spanning from amino acids 1–244, thereby deleting the marked box domain and the transactivation domain. We made a stable MCF7 cell line containing E2F1ΔMB, treated the cells with 4-OHT, performed ChIP assays, and tested a series of known E2F1 binding sites using PCR. Surprisingly, our ChIP-PCR experiments using this mutant did not detect binding of the mutant E2F1 protein at any E2F1 target sites (data not shown). Further investigation revealed that this E2F1 construct failed to translocate into the nucleus upon treatment with 4-OHT (supplemental Fig. S2A), suggesting a role for the marked box domain in nuclear translocation. The failure of this E2F1 mutant protein to translocate into the nucleus prevented our evaluation of the role of the marked box in binding specificity. Therefore, we created a second E2F1ΔMB mutant that contained the SV-40 large T antigen nuclear localization signal immediately upstream of the E2F1 coding sequence (downstream of the HA and ER domains). This alternative cloning strategy proved to be successful in moving the fusion protein into nucleus upon treatment with 4-OHT (supplemental Fig. S2B). Therefore, we created an MCF7 cell line stably expressing this new E2F1ΔMB, induced expression with 4-OHT, and performed ChIP-seq analysis to assess which genomic sites are bound by an E2F1 protein lacking both the marked box and the transactivation domain. Again, we found that the pattern of peaks called using Sole-Search for the E2F1ΔMB is very similar to the pattern of peaks for WT E2F1 (Fig. 2, top panel), the actual binding profiles are indistinguishable, and the overlap analysis indicates that no major peaks were lost due to deletion of both the marked box and transactivation domains of E2F1 (Table 2). As an additional test of the requirement for the various E2F1 protein interaction domains, we used the Sole-Search overlap program to identify the few regions that appeared to be specifically bound by WT E2F1 but not by the N- or C-terminal deletion mutants, and reanalyzed a set of these sites by ChIP PCR. We found that either these sites were false positives in the E2F1 data set or false negatives in the mutant ChIP-seq datasets (supplemental Fig. S3).
The results presented above suggest that the most critical domain for recruiting E2F1 to the human genome must be the DNA binding domain. To provide support for this hypothesis, we created a stable cell line that inducibly expresses a tagged version of an E2F1 protein that harbors a two amino acid change in the DNA binding domain. This mutation has been shown previously to abolish E2F1 binding to DNA in vitro (25) but not to affect other functions of E2F1 such as binding to pocket proteins (59–61). Using ChIP-seq, we have now shown that essentially all binding of E2F1 to the genome is abolished in the DNA binding mutant. Although a small number of peaks (508) were called by Sole-Search (Table 1), closer inspection revealed that these were false positives. Unlike true peaks, the peaks in the DNA binding domain mutant ChIP-seq data set did not resemble a bell-shaped curve, and/or they were located in telomeric or centromeric repeat regions (see supplemental Fig. S4).
Most of our analyses have focused on the promoters of protein-coding genes. However, it was possible that E2F1 genomic recruitment might be different at different types of promoters. Therefore, we analyzed the binding of E2F1 to the promoters of miRNA genes. A previous study used the genomic coordinates of H3K4me3-enriched loci derived from multiple cell types to identify putative promoters for 578 human miRNAs (62). With the caveat that we do not know how many of these miRNA promoters are correctly localized or which are in open chromatin in MCF7 cells, we determined which of the 578 putative miRNA promoters were bound by E2F1. To do so, the top 10,000 peaks identified by ChIP-seq for the WT E2F1 and the E2F1 deletion mutants were overlapped with the miRNA promoters, allowing a 200-bp gap between the E2F1 peak and the putative core miRNA promoter region. We identified 96 miRNA promoters that were bound by E2F1, 128 promoters that were bound by the C-terminal deletion mutant, 90 promoters that were bound by the mutant deleted for both the N and C termini, and 85 promoters that were bound by the marked box mutant. Thus, 17–22% of the miRNA promoters were bound by E2F1. Visual inspection indicated that the miRNA promoters bound by E2F1 were also bound by the E2F1 deletion mutants. Thus, E2F1 is recruited to miRNA promoters in a similar manner as it is recruited to the promoters of coding genes. In summary, E2F1 protein derivatives lacking characterized protein interaction domains behaved identically to WT E2F1 in respect to their recruitment to the human genome, whereas the DNA binding domain mutant was completely unable to stably bind to the genome.
E2F1 in Vivo Consensus Motif Differs from in Vitro Consensus Motif
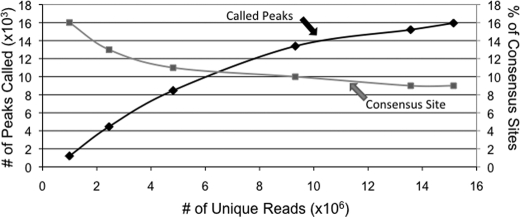
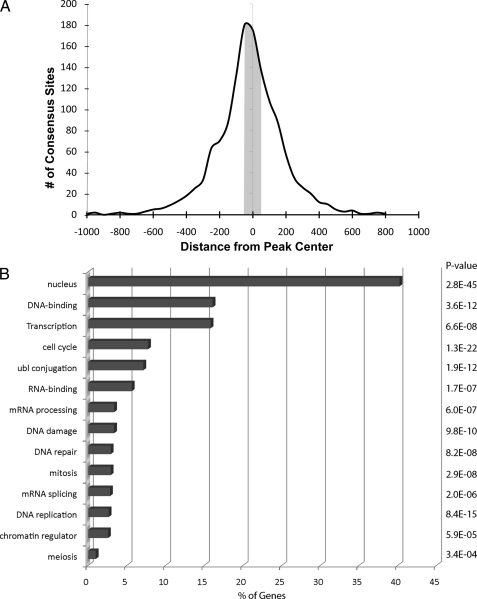
The results described above indicate that the DNA binding domain of E2F1 is the major (if not only) mechanism by which E2F1 is directed to the genome. The fact that protein-protein interactions do not appear to influence E2F1 recruitment suggests that E2F1 DNA binding studies performed in vitro should provide relevant information for the in vivo binding studies. Previous work using in vitro protein-DNA binding analyses has derived an E2F1 consensus motif of TTTSSCGC, where S can be either C or G (4, 24). To determine whether the in vitro derived motif is used in vivo, we analyzed the sequences under the E2F1 ChIP-seq peaks. For these analyses, we only used the top 10,000 peaks (supplemental Table S2) so that we could eliminate the very small peaks at the bottom of the ranked list. We found that only 12% of the top 10,000 ranked WT E2F1 peaks contained a match to the in vitro consensus motif (see supplemental Table S3 for a list of these sites). To further investigate the relationship between the number of consensus sites, the number of sequenced tags, and the number of called peaks, we performed the following analysis. We combined both replicate WT E2F1 ChIP-seq lanes, producing more than 22 million mapped reads. We then randomly selected increasingly larger subsets of the reads and called peaks on the different sets of reads. As expected, the number of called peaks increased as the number of reads increased, until a plateau was reached at about 15,000–16,000 peaks (Fig. 3). We then determined the percentage of the peaks that contain a match to the E2F1 consensus motif. As shown in Fig. 3, the percentage of peaks containing a match to the consensus motif was similar, no matter how many peaks were called; the percentage of consensus-containing peaks was highest (16%) when only 1,000 peaks was called, but did not drop below 9% even when 16,000 peaks were called. Thus, the low percentage of consensus-containing peaks is not a consequence of calling too few or too many peaks. In fact, many of the well characterized E2F target promoters that contain a consensus motif are near the middle or bottom of the ranked list. For example, the very first mammalian E2F binding site characterized is a consensus site located in between the bidirectionally transcribed DHFR and MSH3 genes (4, 63–68). This particular E2F binding site (TTTCGCGC) is one of the strongest sites in vitro (as determined by gel shift competition studies) but is ranked number 8408 in the set of in vivo E2F1 binding sites. Thus, the presence of a consensus motif within the E2F1 binding site does not necessarily determine the height of the peak in E2F1 ChIP-seq experiments. Importantly, analysis of a set of 1000 randomly selected promoters revealed that 12% contain a match to the E2F consensus motif. Thus, the set of E2F1 in vivo binding sites has the same percentage of E2F consensus motifs as does the set of all human promoters. Other studies have shown that a consensus motif can be found within 50 nt ± the center of the peak for many human transcription factors analyzed by ChIP-seq (1). However, if we limit the search to the 50 nucleotides on either side of the center of the in vivo E2F1 peaks, we find that only 5% of the peaks contain an E2F1 in vitro consensus motif (Fig. 4). Therefore, the in vitro consensus motif is not a primary determinant of E2F1 binding.
FIGURE 3.
Percentage of consensus sites does not change with increasing reads. Different numbers of uniquely mapped reads were isolated randomly from the total number of uniquely mapped reads in the HA ER E2F1 merged WT ChIP-seq data sets. The number of significant peaks in each set of these randomly selected mapped reads is shown by the line labeled “called peaks”; the corresponding scale for this plot is shown on the left y axis. The number of consensus sites within each set of called peaks is plotted as a percentage of all the peaks in that set (and labeled as “consensus site”); the corresponding scale for the “consensus site” plot is shown on the right y axis.
FIGURE 4.
The consensus motif is not near the center of the binding site for most E2F1 consensus-containing peaks. A, 1337 E2F consensus motif-containing peaks were identified from the top 10,000 ranked peaks of the E2F1 WT merged data set. The distance of the E2F consensus motif relative to the center of the corresponding peak is plotted along the x axis (using a bin size of 50 nt). The y axis indicates the number of consensus motifs found in each bin. The E2F consensus motifs identified within ± 50 nt from the center of a peak is highlighted in gray. B, the functional annotations of the 1337 E2F consensus motif-containing targets were determined using the program Database for Annotation, Visualization, and Integrated Discovery (74). The percentage of the E2F consensus targets represented by the different functional categories is indicated on the x axis, and the p value for each identified category is shown on the right side of the graph.
We next performed a de novo motif analysis using our W-ChIPmotifs program (69). First, we eliminated the consensus-containing peaks from the top 10,000 peak set, producing a set of ∼8,800 peaks for the de novo motif analysis. Then, we selected the top 1000 promoters, the middle 1000 promoters, and the bottom 1000 promoters from this ranked list of nonconsensus-containing E2F1 binding sites for our de novo motif analyses. We chose only the 50 nucleotides on either side of the center of the ChIP-seq peak to eliminate common core promoter elements that are not directly responsible for E2F1 recruitment. The motif CGCGC was identified as a predominant motif in all three peak sets (supplemental Files S2–S4). In fact, ∼70% of the peaks in the top half of the ranked list contained at least one CGCGC motif (Table 3). However, the percentage of sites containing CGCGC does decline as the position of the peaks falls lower in the ranked set of E2F1 binding sites, with only 46% of the bottom 1000 ranked E2F1 peaks containing CGCGC. Analysis of a set of 1000 randomly selected promoters revealed that 70% contained a match to the CGCGC motif.
TABLE 3.
De novo motif analysis of E2F1 peaks
The top 10,000 ranked peaks for the merged E2F1 wild type ChIP-seq replicates were searched for the E2F1 consensus motif (TTTSSCGC, where S can be a C or a G). The E2F1 peaks were further subdivided into a set of the top 1,000 peaks, a set of peaks ranked 5,000–6,000, and a set of peaks ranked 9,000–10,000. Next, the peaks containing a match to the E2F consensus motif were removed from each of the sets of 1,000 peaks, and the sequence +/− 50 bp from the center of the remaining peaks was then analyzed for the presence of a de novo motif using ChIPMotifs. The most prevalent motif identified in each set was CGCGC (see supplemental Figs. S2–S4). The percentage of peaks containing the de novo motif CGCGC is indicated for each set of 1,000 peaks and for a set of 1,000 randomly chosen promoters. The percentage of peaks containing the E2F consensus motif within each of the sets is also indicated. N.A., not analyzed.
| % with a match to TTTSSCGC | % with a match to CGCGC | |
|---|---|---|
| Top 10,000 | 12 | N.A. |
| 1–1,000 | 27 | 73 |
| 5,000–6,000 | 12 | 69 |
| 9,000–10,000 | 6 | 59 |
| 14,000–15,000 | 4 | 46 |
| Random 1,000 promoters | 12 | 70 |
Comparison of E2F1 Target Promoters versus Promoters Not Bound by E2F1
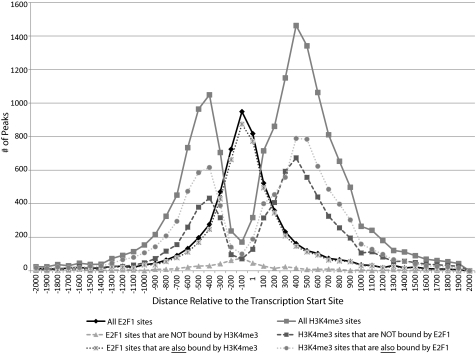
As indicated in Table 1, E2F1 binds to a large number of places in the genome and most of the binding sites are at core promoters (supplemental Fig. S1). Therefore, E2F1 binds to many, but not all, core promoters in the human genome. Although it is difficult to directly correlate E2F1 binding with gene expression (due to the fact that the repressive E2F proteins also bind to the same genomic sites), we have previously shown that most, but not all, promoters bound by E2F1 are also bound by RNAPII and are transcriptionally active (48, 49). We were interested in whether the promoters that were bound by E2F1 had different characteristics than promoters that were not bound by E2F1. However, we did not simply want to analyze all promoters that were not bound by E2F1 because many of these promoters may be unavailable for E2F1 binding due to being located in large repressive chromatin domains. In other cell types, these promoters might in fact be E2F1 targets. Therefore, we analyzed only those promoters that are located in active chromatin in MCF7 cells. To do so, we used ChIP-seq data from the ENCODE Consortium corresponding to the H3K4me3 mark in MCF7 cells. As expected, the H3K4me3 sites had a bimodal pattern with a peak upstream and a peak downstream of the transcription start site (see Fig. 5, all H3K4me3 sites). However, it was possible that the promoters bound by E2F1 had a different H3K4me3 pattern that the promoters not bound by E2F1. Therefore, we divided the H3K4me3 peaks into two sets, those that overlapped with E2F1 peaks (10,937 peaks) and those that did not (9,063 peaks). We found that there is essentially no difference in the patterns of H3K4me3 at the promoters also bound by E2F1 versus those bound only by H3K4me3 (Fig. 5). Therefore, the trimethylation of lysine 4 on histone H3 is not the determinant of E2F1 binding specificity because many promoters not bound by E2F1 have the same bimodal H3K4me3 pattern as the promoters that do bind E2F1. Interestingly, E2F1 binds in between the two nucleosomes in the shared target promoters. Therefore, it was possible that the sequences in between the two nucleosomes on either side of the start site of transcription may be different in the set of promoters bound by E2F1 versus the set of promoters not bound by E2F1. Because we have identified a GC-rich motif in the set of promoters that are bound by E2F1, we compared the GC content of the internucleosomal space of the two sets of promoters. We found that a 200-nt region centered between the two H3K4me3 peaks was 68% GC-rich in the promoters that are bound by both H3K4me3 and E2F1, whereas the 200-nt regions centered between the two H3K4me3 peaks was 67% GC-rich in the promoters that were bound by H3K4me3 but not by E2F1. For both sets of promoters, the area under the H3K4me3 peaks was 54% GC-rich. Therefore, the GC content of the internucleosomal space is not different in promoters bound by versus not bound by E2F1. As a final analysis of E2F1 bound promoters, we determined the enrichment of the CGCGC motif in the promoters bound by both H3K4me3 and E2F1 versus the promoters bound by H3K4me3 but not by E2F1. Because we have shown that E2F1 binds between the two H3K4me3 peaks, we examined the 200 nt on either side of the transcription start site in the promoters bound by both H3K4me3 and E2F1 and in the promoters bound only by H3K4me3. We found that 49% of the promoters bound by both H3K4me3 and E2F1 contained the CGCGC motif, whereas 39% of the promoters bound only by H3K4me3 contained the CGCGC motif. Thus, there is modest enrichment of the CGCGC motif in the set of promoters bound by E2F1.
FIGURE 5.
The relationship of E2F1 binding and the H3K4me3 pattern. An overlap analysis between the top 10,000 ranked peaks for E2F1 and the top 20,000 H3K4me3 peaks was performed. The pattern for all 20,000 H3K4me3 sites is shown, as is the pattern for the H3K4me3 sites bound by E2F1 and the H3K4me3 sites not bound by E2F1. Also shown is the binding pattern for all E2F1 sites, for E2F1 sites also bound by H3K4me3, and for E2F1 sites not also bound by H3K4me3. The number of peaks (indicated on the y axis) found at each given distance relative to the transcription start site (indicated on the x axis in base pairs) is plotted for all of the above-mentioned categories.
DISCUSSION
In this study, we have investigated the role of protein interaction domains and the DNA binding domain in recruiting E2F1 to the human genome. The in vitro DNA binding motif that has been shown to correspond to high affinity E2F1 binding is an 8-nt motif (TTTSSCGC) found throughout the human genome. For example, in the 1% of the genome that was analyzed by the ENCODE pilot project, there are 511 perfect matches to this motif. Upon extrapolation, this suggests that there are ∼51,000 perfect matches in the entire genome, with the number expanding dramatically if even one mismatch is allowed. Clearly, there are more motifs than E2F1 binding sites, so there must be a mechanism by which E2F1 is localized only to the “correct” sites in the genome. One possible mechanism is that interaction of E2F1 with other site-specific transcription factors could anchor E2F1 at the correct genomic location. We have tested this hypothesis using MCF7 cells containing stably integrated, inducibly regulated, HA-tagged E2F1 constructs. Surprisingly, we found that we could delete all of the characterized domains (other than the DNA binding domain) in the E2F1 protein without affecting its genomic binding pattern. Also, although our findings suggest that E2F1 genomic binding specificity may be conferred by a high affinity interaction between E2F1 and a very specific binding motif, analysis of the ChIP-seq data revealed that the in vitro consensus site is not present under most E2F1 peaks but rather E2F1 binds to a GC-rich sequence present in the majority of human promoters. The only structural study focusing on E2F family members bound to DNA is an analysis of a heterodimer of the E2F4 DNA binding domain (equivalent to amino acids 112–195 of E2F1) and the DP2 DNA binding domain; a 15-nt DNA duplex containing an E2F consensus motif (TTTTCGCGCGGTTTT) was used as the binding site (18). This study revealed that both the E2F4 and the DP2 DNA binding domains are related to the winged-helix DNA binding motif, which consists of three α helices and a β sheet. They found that E2F4 and DP2 each contact half of the central GC-rich motif using a conserved Arg-Arg-Xaa-Tyr-Asp in their α 3 helices (corresponding to amino acids 157–161 of E2F1); E2F4 contacts CGC on one strand and DP2 contacts CGC on the other strand. The T-rich portion of the consensus is contacted by residues 16–19 of E2F4 (corresponding to amino acids 117–120 of E2F1). Thus, the structural studies support the identification of a CGCGC motif in the E2F1 ChIP-seq peaks. However, the absence of a T-rich extension in most E2F1 binding sites suggests that the interaction of amino acids 117–120 of E2F1 with DNA might not be critical in vivo.
The fact that E2F1 binds almost directly over the transcription start site and that the binding pattern of E2F1 is very similar to that of RNA polymerase II and TAF1 (48–50), raised the interesting possibility that E2F1 might be recruited to transcription start sites via an interaction of its C-terminal transactivation domain along with components of the general transcriptional machinery (38–46). For example, previous studies have shown that E2F1 can interact with TBP and TFIIH, and one can imagine that a “tethered” recruitment of E2F1 to start sites via these components of the general transcriptional machinery might be the mechanism that accounts for essentially all of the E2F1 peaks being located over the transcription start site. However, we showed that an E2F1 mutant protein having a deletion of the entire transactivation domain still bound to all the same genomic locations as did WT E2F1. Therefore, we have ruled out the possibility that tethering via its transactivation domain is a commonly used recruitment mechanism for E2F1 in MCF7 cells.
Another protein domain that we deleted was the marked box domain. Previous studies have shown that this domain can mediate interactions with site-specific binding factors such as TFE-3 and RYBP (31–33), suggesting that this domain may help to direct E2F1 to subsets of promoters that are bound by these other factors. However, our data suggests that if this mechanism is utilized, the interactions must be important only at a small number of promoters. We have shown that the strongest sites bound by WT E2F1 are also bound by the marked box domain mutant (97% of the top 40% of WT E2F1 sites are in the ΔMB peak set and vice versa). The great majority of the sites that are not bound by the ΔMB mutant protein are very small and likely to be false positives. We tested this possibility by performing ChIP-PCR on a subset of the small peaks that were identified in the WT peak set but not in the ChIP-seq data from the E2F1 mutants lacking the C terminus. We could not confirm any of these sites as being bound by WT E2F1. It remains possible that there are a few places in the genome to which E2F1 is recruited using a marked box domain-dependent mechanism, but it would require multiple ChIP-seq experiments to identify these sites. We also note that previous studies have shown that the marked box domain is critical for the interaction of E2F1 with its heterodimerization partner DP1 (34). Because we find that the E2F1 marked box mutant binds to over 28,000 sites in the genome, this raises the possibility that DP1 might not be an obligate interaction partner of E2F1 in vivo. To test this hypothesis, we have performed ChIP analysis for DP1 using all commercially available antibodies but have not been able to successfully demonstrate reproducible binding of DP1 to any E2F1 target sites. However, negative DP1 ChIP results (even when performed alongside E2F1 ChIP experiments) are hard to interpret because it is possible that none of the DP1 antibodies are functional in ChIP assays. We have also transiently expressed an HA-tagged DP1 and performed ChIP experiments using the HA antibody. Again, we were not able to demonstrate binding of the tagged DP1 to any E2F1 target. We are left with the conclusion that the DP1 protein is either masked in the E2F1-DP1 complex that binds to the genome or that DP1 is not bound with E2F1 to target sites.
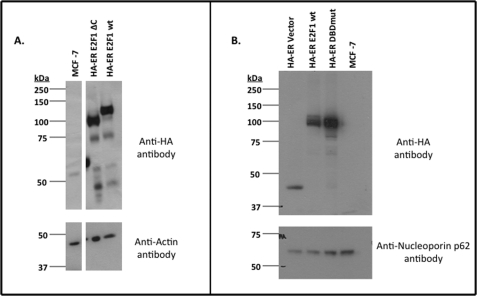
It is also interesting to note that there are more ChIP-seq peaks detected for the E2F1 deletion mutants than for WT E2F1 and, moreover, some of the peaks common to both the WT E2F1 and deletion mutants are actually larger in the E2F1 deletion mutant ChIP-seq data sets. There are several possibilities that may account for these observations. First, it has already been established that the C terminus of WT E2F1 contains the binding site for p14ARF, which normally flags E2F1 for ubiquitination via the proteasome pathway (70–73). Thus, all of the E2F1 deletion mutant proteins missing their C terminus domain may be more stable in the cell than the WT E2F1 protein and consequently may be more readily available to bind to their genomic targets. However, Western blot analysis indicates that the WT E2F1 and the C-terminal deletion mutant protein are expressed at similar levels (Fig. 6). Second, the C terminus of E2F1 interacts with numerous nuclear proteins, which may result in the sequestering of some WT E2F1 protein into complexes that are nonproductive for DNA binding. Finally, it is also possible that the smaller sizes of the E2F1 deletion mutants simply allows them to access the core promoter regions much more readily than the full-length E2F1 protein.
FIGURE 6.
Expression levels of E2F1 wild type and mutant proteins. A, shown are the protein levels of the HA-ER-E2F1ΔC (lane 2) and HA-ER-E2F1 wild type (lane 3) fusion proteins that were detected in whole cell extracts from the corresponding MCF7 stable cell lines using an anti-HA antibody (top). The parental MCF7 cells (lane 1) were used as a negative control and an antibody to actin was used as a loading control (bottom). An unrelated lane was removed between lanes 1 and 2, but all of the lanes shown here are from the same blot. B, shown are the protein levels of the HA-ER (lane 1), HA-ER-E2F1 wild type (lane 2), and HA-ER-E2F1 DNA binding domain mutant (lane 3) fusion proteins that were detected in whole cell extracts from the corresponding MCF7 stable cell lines using an anti-HA antibody (top); adapted from supplemental Fig. S5 of Rabinovich et al. (50). The parental MCF7 cells (lane 4) were used as a negative control and an antibody to nucleoporin p62 (NUP62) was used as a loading control (bottom).
In summary, we have tested the hypothesis that recruitment of E2F1 to the human genome can be mediated by either a direct interaction between E2F1 and a consensus motif and/or selective recruitment of E2F1 to some sites due to protein-protein interactions. We conclude that the in vitro E2F consensus motif is not present at most in vivo E2F1 binding sites, but instead, a shorter GC-rich motif that is common to most human promoters is enriched in the sequences corresponding to E2F1 ChIP-seq peaks. Also, we can find no evidence that protein-protein interactions are required for recruitment of E2F1 to its genomic target sites.
Acknowledgments
We thank the members of the Farnham laboratory and Dave Segal for helpful discussions Cheryl Serchen for analytical assistance, and Kristian Helin for the wild type HA-tagged E2F1 construct. The MCF7 H3K4me3 ChIP-seq data was generated at the University of Washington by the ENCODE group led by John Stamatoyannopoulos. These data was collected as part of the ENCODE Project Consortium.
This work was supported in part by United States Public Health Service Grants CA45240 and HG004558.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Files S1–S4, Table S1, and Figs. S1–S4.
- DBD
- DNA binding domain
- ER
- estrogen receptor
- MB
- marked box
- miRNA
- micro RNA
- nt
- nucleotide(s)
- 4-OHT
- 4-hydroxytamoxifen.
REFERENCES
- 1. Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. (2008) Nucleic. Acids. Res. 36, 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cooper S. J., Trinklein N. D., Nguyen L., Myers R. M. (2007) Genome Res. 17, 136–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stender J. D., Kim K., Charn T. H., Komm B., Chang K. C., Kraus W. L., Benner C., Glass C. K., Katzenellenbogen B. S. (2010) Mol. Cell. Biol. 30, 3943–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slansky J. E., Farnham P. J. (1996) in Transcriptional Control of Cell Growth: The E2F Gene Family (Farnham P. J. ed.) pp. 1–30, Springer-Verlag, New York [Google Scholar]
- 5. Dimova D. K., Dyson N. J. (2005) Oncogene 24, 2810–2826 [DOI] [PubMed] [Google Scholar]
- 6. Kong L. J., Chang J. T., Bild A. H., Nevins J. R. (2007) Oncogene 26, 321–327 [DOI] [PubMed] [Google Scholar]
- 7. DeGregori J., Johnson D. G. (2006) Curr. Mol. Med. 6, 739–748 [DOI] [PubMed] [Google Scholar]
- 8. Krek W., Livingston D. M., Shirodkar S. (1993) Science 262, 1557–1560 [DOI] [PubMed] [Google Scholar]
- 9. Helin K., Wu C. L., Fattaey A. R., Lees J. A., Dynlacht B. D., Ngwu C., Harlow E. (1993) Genes Dev. 7, 1850–1861 [DOI] [PubMed] [Google Scholar]
- 10. Bandara L. R., Buck V. M., Zamanian M., Johnston L. H., La Thangue N. B. (1993) EMBO. J. 12, 4317–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J., Ran C., Li E., Gordon F., Comstock G., Siddiqui H., Cleghorn W., Chen H. Z., Kornacker K., Liu C. G., Pandit S. K., Khanizadeh M., Weinstein M., Leone G., de Bruin A. (2008) Dev. Cell. 14, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christensen J., Cloos P., Toftegaard U., Klinkenberg D., Bracken A. P., Trinh E., Heeran M., Di Stefano L., Helin K. (2005) Nucleic Acids. Res. 33, 5458–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Logan N., Graham A., Zhao X., Fisher R., Maiti B., Leone G., La Thangue N. B. (2005) Oncogene 24, 5000–5004 [DOI] [PubMed] [Google Scholar]
- 14. Maiti B., Li J., de Bruin A., Gordon F., Timmers C., Opavsky R., Patil K., Tuttle J., Cleghorn W., Leone G. (2005) J. Biol. Chem. 280, 18211–18220 [DOI] [PubMed] [Google Scholar]
- 15. Logan N., Delavaine L., Graham A., Reilly C., Wilson J., Brummelkamp T. R., Hijmans E. M., Bernards R., La Thangue N. B. (2004) Oncogene 23, 5138–5150 [DOI] [PubMed] [Google Scholar]
- 16. Di Stefano L., Jensen M. R., Helin K. (2003) EMBO J. 22, 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Bruin A., Maiti B., Jakoi L., Timmers C., Buerki R., Leone G. (2003) J. Biol. Chem. 278, 42041–42049 [DOI] [PubMed] [Google Scholar]
- 18. Zheng N., Fraenkel E., Pabo C. O., Pavletich N. P. (1999) Genes Dev. 13, 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Milton A., Luoto K., Ingram L., Munro S., Logan N., Graham A. L., Brummelkamp T. R., Hijmans E. M., Bernards R., La Thangue N. B. (2006) Oncogene 25, 3212–3218 [DOI] [PubMed] [Google Scholar]
- 20. Rogers K. T., Higgins P. D., Milla M. M., Phillips R. S., Horowitz J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7594–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu C. L., Zukerberg L. R., Ngwu C., Harlow E., Lees J. A. (1995) Mol. Cell. Biol. 15, 2536–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Chellappan S. P. (1995) Oncogene 10, 2085–2093 [PubMed] [Google Scholar]
- 23. Ormondroyd E., de la Luna S., La Thangue N. B. (1995) Oncogene. 1437–1446 [PubMed] [Google Scholar]
- 24. Tao Y., Kassatly R. F., Cress W. D., Horowitz J. M. (1997) Mol. Cell. Biol. 17, 6994–7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cress W. D., Johnson D. G., Nevins J. R. (1993) Mol. Cell. Biol. 13, 6314–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karlseder J., Rotheneder H., Wintersberger E. (1996) Mol. Cell. Biol. 16, 1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shin E. K., Tevosian S. G., Yee A. S. (1996) J. Biol. Chem. 271, 12261–12268 [DOI] [PubMed] [Google Scholar]
- 28. Majello B., De Luca P., Suske G., Lania L. (1995) Oncogene 10, 1841–1848 [PubMed] [Google Scholar]
- 29. Lin S. Y., Black A. R., Kostic D., Pajovic S., Hoover C. N., Azizkhan J. C. (1996) Mol. Cell. Biol. 16, 1668–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin J. M., Collins P. J., Trinklein N. D., Fu Y., Xi H., Myers R. M., Weng Z. (2007) Genome Res. 17, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giangrande P. H., Hallstrom T. C., Tunyaplin C., Calame K., Nevins J. R. (2003) Mol. Cell. Biol. 23, 3707–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giangrande P. H., Zhu W., Rempel R. E., Laakso N., Nevins J. R. (2004) EMBO J. 23, 1336–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schlisio S., Halperin T., Vidal M., Nevins J. R. (2002) EMBO J. 21, 5775–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vidal M., Braun P., Chen E., Boeke J. D., Harlow E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10321–10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cress W. D., Nevins J. R. (1996) Mol. Cell. Biol. 16, 2119–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu W., Giangrande P. H., Nevins J. R. (2004) EMBO. J. 23, 4615–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen C. R., Kang Y., Siegel P. M., Massagué J. (2002) Cell 110, 19–32 [DOI] [PubMed] [Google Scholar]
- 38. Ross J. F., Liu X., Dynlacht B. D. (1999) Mol. Cell. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 39. Pearson A., Greenblatt J. (1997) Oncogene 15, 2643–2658 [DOI] [PubMed] [Google Scholar]
- 40. Fry C. J., Pearson A., Malinowski E., Bartley S. M., Greenblatt J., Farnham P. J. (1999) J. Biol. Chem. 274, 15883–15891 [DOI] [PubMed] [Google Scholar]
- 41. Hagemeier C., Cook A., Kouzarides T. (1993) Nucleic Acids. Res. 21, 4998–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emili A., Ingles C. J. (1995) J. Biol. Chem. 270, 13674–13680 [DOI] [PubMed] [Google Scholar]
- 43. Trouche D., Cook A., Kouzarides T. (1996) Nucleic Acids. Res. 24, 4139–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D. (1998) Cell 94, 363–374 [DOI] [PubMed] [Google Scholar]
- 45. Lang S. E., McMahon S. B., Cole M. D., Hearing P. (2001) J. Biol. Chem. 276, 32627–32634 [DOI] [PubMed] [Google Scholar]
- 46. Taubert S., Gorrini C., Frank S. R., Parisi T., Fuchs M., Chan H. M., Livingston D. M., Amati B. (2004) Mol. Cell. Biol. 24, 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Farnham P. J. (2009) Nat. Rev. Genet. 10, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bieda M., Xu X., Singer M. A., Green R., Farnham P. J. (2006) Genome Res. 16, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu X., Bieda M., Jin V. X., Rabinovich A., Oberley M. J., Green R., Farnham P. J. (2007) Genome Res. 17, 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rabinovich A., Jin V. X., Rabinovich R., Xu X., Farnham P. J. (2008) Genome Res. 18, 1763–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Batsché E., Muchardt C., Behrens J., Hurst H. C., Crémisi C. (1998) Mol. Cell. Biol. 18, 3647–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Decary S., Decesse J. T., Ogryzko V., Reed J. C., Naguibneva I., Harel-Bellan A., Cremisi C. E. (2002) Mol. Cell. Biol. 22, 7877–7888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Revenko A. S., Kalashnikova E. V., Gemo A. T., Zou J. X., Chen H. W. (2010) Mol. Cell. Biol. 30, 5260–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krek W., Ewen M. E., Shirodkar S., Arany Z., Kaelin W. G., Jr., Livingston D. M. (1994) Cell 78, 161–172 [DOI] [PubMed] [Google Scholar]
- 55. Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. (2007) Nat. Methods 4, 1–2 [DOI] [PubMed] [Google Scholar]
- 56. Lan X., Bonneville R., Apostolos J., Wu W., Jin V. X. (2010) Bioinformatics 27, 428–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee T. A., Farnham P. J. (2000) Oncogene 19, 2257–2268 [DOI] [PubMed] [Google Scholar]
- 58. Blahnik K. R., Dou L., O'Geen H., McPhillips T., Xu X., Cao A. R., Iyengar S., Nicolet C. M., Ludäscher B., Korf I., Farnham P. J. (2010) Nucleic Acids Res. 38, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Helin K., Harlow E., Fattaey A. (1993) Mol. Cell. Biol. 13, 6501–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Helin K., Harlow E. (1994) J. Virol. 68, 5027–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Halaban R., Cheng E., Zhang Y., Mandigo C. E., Miglarese M. R. (1998) Oncogene 16, 2489–2501 [DOI] [PubMed] [Google Scholar]
- 62. Marson A., Levine S. S., Cole M. F., Frampton G. M., Brambrink T., Johnstone S., Guenther M. G., Johnston W. K., Wernig M., Newman J., Calabrese J. M., Dennis L. M., Volkert T. L., Gupta S., Love J., Hannett N., Sharp P. A., Bartel D. P., Jaenisch R., Young R. A. (2008) Cell 134, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Slansky J. E., Farnham P. J. (1996) BioEssays 18, 55–62 [DOI] [PubMed] [Google Scholar]
- 64. Means A. L., Slansky J. E., McMahon S. L., Knuth M. W., Farnham P. J. (1992) Mol. Cell. Biol. 12, 1054–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schilling L. J., Farnham P. J. (1994) Crit. Rev. Euk. Gene Exp. 4, 19–53 [DOI] [PubMed] [Google Scholar]
- 66. Schilling L. J., Farnham P. J. (1995) Cell Growth Differ. 6, 541–548 [PubMed] [Google Scholar]
- 67. Wade M., Blake M. C., Jambou R. C., Helin K., Harlow E., Azizkhan J. C. (1995) J. Biol. Chem. 270, 9783–9791 [DOI] [PubMed] [Google Scholar]
- 68. Wade M., Kowalik T. F., Mudryj M., Huang E. S., Azizkhan J. C. (1992) Mol. Cell. Biol. 12, 4364–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jin V. X., Apostolos J., Nagisetty N. S., Farnham P. J. (2009) Bioinformatics 25, 3191–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., Vousden K. H. (1998) Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]
- 71. Marti A., Wirbelauer C., Scheffner M., Krek W. (1999) Nat. Cell. Biol. 1, 14–19 [DOI] [PubMed] [Google Scholar]
- 72. Martelli F., Hamilton T., Silver D. P., Sharpless N. E., Bardeesy N., Rokas M., DePinho R. A., Livingston D. M., Grossman S. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Campanero M. R., Flemington E. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3. [PubMed] [Google Scholar]