Abstract
IL-32 was recently identified as a proinflammatory cytokine that is induced by IL-18 in natural killer (NK) cells and is highly correlated with inflammatory disorders. However, the relationship between IL-32 and tumor progression is still unknown. In this study, we investigated whether overexpression of IL-32 affects susceptibility of chronic myeloid leukemia (CML) cells to NK cells. Interestingly, IL-32α-overexpressing CML cell lines, K562, Kcl22, and BV173, showed higher NK cell-mediated killing. Flow cytometry analysis revealed that overexpression of IL-32α induced increased expression of Fas and UL16-binding protein 2 (ULBP2) in CML cells. The direct relationship between overexpression of surface molecules by IL-32α and increased NK cell-mediated killing was confirmed by Fas or ULBP2 siRNA transfection. IL-32α-induced Fas and ULBP2 expression are regulated p38 MAPK. In addition, the transcription factor Ets1 plays a key role in ULBP2 specific expression by IL-32α overexpression in ULBP family members. Taken together, these data show that IL-32α stimulates Fas and ULBP2 expression via activation of p38 MAPK, which increases NK susceptibility of CML cells. Enhanced NK cell susceptibility of CML cells by IL-32α overexpression may improve the efficiency of NK cell-based immunotherapy.
Keywords: Cancer Therapy, Cytokine, Interleukin, Leukemia, p38 MAPK, Ets-1, Fas, IL-32alpha, NK Cell, ULBP2
Introduction
IL-32 originally was identified as natural killer (NK)5 transcript 4 and is induced by IL-18 in NK cells (1). It has cytokine-like characteristics and plays a critical role in inflammation and was therefore renamed IL-32. It is enhanced by IFN-γ, IL-18, and IL-12 in various cell types such as NK cells and epithelial cells and stimulates production of various pro-inflammatory cytokines including TNF-α, IL-8, IL-6, IL-1β, and various chemokines through activation of NF-κB and p38 MAPK in monocytes and macrophages (1, 2). Kim et al. (1) found four isoforms of IL-32 by alternative splicing (IL-32α, IL-32β, IL-32γ, and IL-32δ). IL-32α is the most abundant transcript, whereas IL-32γ is the longest isoform and is the most powerful inducer of cytokine production (3). Two additional isoforms, IL-32ϵ and IL-32ζ, have been recently identified, although these cytokines are not frequently expressed in various cells except T cells (4). Because IL-32 is a proinflammatory cytokine, its effect in various inflammatory diseases such as rheumatoid arthritis and inflammatory bowel disease has been investigated. In most patients with rheumatoid arthritis or inflammatory bowel disease, expression of IL-32 is higher than in healthy controls. However, the function of IL-32 in tumor survival and progression is still unknown.
Fas and ULBP2 are known as critical molecules related with NK cytolytic activity. Fas is known as a death receptor. Triggering of Fas and its specific ligand, the Fas ligand, induces assembly of the death-inducing signaling complex and activates caspase cascades. These signaling cascades induce cellular apoptosis (5). High expression of Fas ligand is detected in activated T and NK cells, which have critical roles in elimination of Fas-expressing target cells (6). ULBPs (UL16 binding proteins) are the most typical ligands of NKG2D with MICA/B (MHC class I-chain-related protein A and B) (7, 8). As a general rule, NKG2D ligands, including ULBPs and MICA/B, are expressed on various tumor cells and are increased by stress and virus infection. Because NKG2D is a critical activating receptor on activated T and NK cells (9–11), increased NKG2D ligands on cells may be an important target of T and NK cells. In this study, we determined that IL-32α stimulates Fas and ULBP2 expression on the surface of cells through p38 MAPK activation, and increased Fas and ULBP2 affect enhancement of NK susceptibility of CML cells. These results suggest that IL-32α has anti-cancer characteristics in CML cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
Human chronic myeloid leukemia cell lines, K562, Kcl22, and BV173, and the human activated NK cell line, NK-92MI, were purchased from the ATCC (Manassas, VA). All CML cells were cultured in RPMI 1640 (Invitrogen) containing 2 mm l-glutamine, 100 unit/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated FBS (Invitrogen). NK-92MI cells were cultured in α modified minimum essential Eagle's medium (Invitrogen) supplemented with 2 mm l-gulutamine, 1.5 g/liter sodium bicarbonate, 1% MEM vitamin solution (Invitrogen), 0.1 mm 2-mercaptoethanol (Sigma), and 20% heat-inactivated FBS (Invitrogen). All cell lines were maintained in a 5% CO2 incubator at 37 °C in log phase growth.
Transfection
K562, Kcl22, and BV173 cells were transfected with IL-32α overexpressing vector (IL-32α/pcDNA3.1+) using the MicroPorator electroporation system (NanoEnTek, Inc., Seoul, Korea). Transfection conditions were 1000 V/50 ms/1 pulse for K562 cells, 1450 V/20 ms/2 pulse for Kcl22 cells, and 1300 V/30 ms/1 pulse for BV173 cells. pcDNA3.1+ was also transfected into each cell line as a vector control. The transfected cells were cultured in RPMI 1640 growth medium containing 500 μg/ml G418 (Clontech, Palo Alto, CA) for selection. Ten different G418-resistant clones were isolated and screened for effects of IL-32α overexpression. More than 90% of transfectants showed similar responses. The representative data are shown.
NK Cytotoxicity Assay
Target cells were stained with 0.5 μm carboxyfluorescein succinimidyl ester (Molecular Probes, Inc., Eugene, OR) in media containing 10% FBS at 37 °C and then washed with PBS three times. Stained target cells were incubated with NK-92MI cells at an E:T ratio of 0.25:1, 0.5:1, or 1:1 for K562 cells and of 2:1 for Kcl22 and BV173 cells for 1 h at 37 °C. After incubation, cells were stained with 3 μl of 7-AAD (BD Pharmingen). Target cells killed by NK-92MI cells were measured by determining the percentage of CFSE (FL-1) and 7-Aminoactinomycin D (FL-3) double-positive regions using flow cytometry.
Flow Cytometry Analysis
Direct FACS analysis was performed to detect the expression of surface molecules in cells. The cells (5 × 105) were washed twice with PBS and then stained with fluorochrome-conjugated antibodies for 30 min on ice. After incubation, stained cells were washed twice with PBS, and the expression level was measured using FACS caliber (Becton Dickinson, Franklin Lakes, NJ). The antibodies were phycoerythrin-conjugated anti-human Fas (CD95), phycoerythrin-conjugated anti-ICAM-1 (CD54), PE-conjugated anti-human ULBP2, and PE-conjugated anti-human MICA/B. Fas and ICAM-1 antibodies were purchased from BD Pharmingen (San Diego, CA), and ULBP2 and MICA/B were purchased from R&D Systems (Minneapolis, MN).
RT-PCR
Total RNA was isolated using TRIzol (Invitrogen), and cDNA was made using 2 μg of total RNA. The cDNAs were used as a template for PCR amplification with primers purchased from Bioneer (Daejeon, Korea). The primer sequences were as follows: β-actin, 5′-TCA CCC ACA CTG TGC CCA TCT ACG-3′ (forward) and 5′-CAG CGG AAC CGC TCA TTG CCA ATG-3′ (reverse); Fas, 5′-CAG AAC TTG GAA GGC CTG CA-3′ (forward) and 5′-TCT GTT CTG CTG TGT CTT GG- 3′ (reverse); ULBP1, 5′-GCA GAG GAT CTT GGC AGT TC-3′ (forward) and 5′-ATG AGA AGG CTC CAG GGA CT-3′ (reverse); ULBP2, 5′-CAG AGC AAC TGC GTG ACA TT-3′ (forward) and 5′-CAT GCC CAT CAA GAA GTC CT-3′ (reverse); ULBP3, 5′-TGG AAC TGG CTG ACA CTG AG-3′ (forward) and 5′-GCC TCT TCT TCC TGT GCA TC-3′ (reverse); and Ets-1, 5′-AGC ATA GTT GTG ATC GCC TCA-3′ (forward) and 5′-CCA TAG CTG GAT TGG TCC ACT-3′ (reverse). PCR cycle conditions were 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. The results were quantified using densitometry analysis (TotalLabTM program).
siRNA Transfection
IL-32α-overexpressing and vector control K562 cells (1 × 106) were transfected transiently with 100 nm Fas and ULBP2 siRNA or 40 pm Ets-1 siRNA (Santa Cruz Biotechnology) using a MicroPorator electroporation system (NanoEnTek, Inc., Seoul, Korea). Transfected cells were cultured in serum-containing media without antibiotics. After 48 h, the cells were used for flow cytometry analysis and cytotoxicity assay. The siRNA sequences used were as follows: nonrelated control siRNA (GL3), 5′-CUU ACG CUG AGU UCG A-dTdT-3′ (sense) and 5′-UCG AAG UAC UCA GCG UAA G-dTdT-3′ (antisense); Fas siRNA, 5′-GCC AAG AAG GGA AGG AGU A-dTdT-3′ (sense) and 5′-UAC UCC UCC CCU UCU UGG C-dTdT-3′ (antisense); and ULBP2 siRNA, 5′-GGA CAU ACU UAC AGA GCA A-dTdT-3′ (sense) and 5′-UUG CUC UGU AAG UAU GUC C-dTdT-3′ (antisense).
Western Blot Analysis
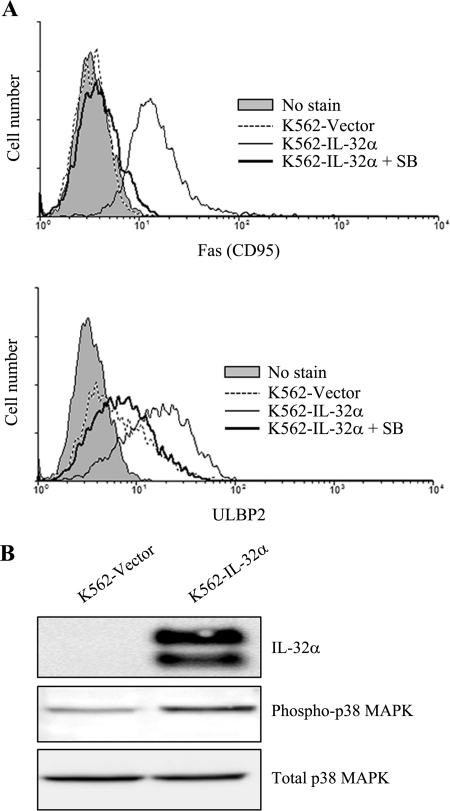
Cells were washed with ice-cold PBS and lysed with ice-cold cell lysis buffer (50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% deoxycholic acid sodium salt, 150 mm NaCl, 1 mm EDTA, and protein inhibitor mixture) containing a phosphatase inhibitor mixture (Sigma). After collecting cell lysates, protein quantity was determined using a Bradford protein assay (Bio-Rad). An equal volume of protein from each group was separated by SDS-PAGE under reducing conditions, and separated proteins were transferred to PVDF membranes (Bio-Rad). After transfer, the membrane was blocked with 5% nonfat dried skim milk for 1 h at room temperature and then incubated with monoclonal antibodies against IL-32 (mouse anti-human IL-32, KU32–52) (12), mouse anti-α-tubulin antibodies (Sigma), rabbit anti-total p38 MAPK antibodies (Cell Signaling Technology, Danvers, MA), or rabbit anti-phospho-p38 MAPK antibodies overnight at 4 °C. After incubation with the primary antibody, the membrane was washed three times with PBS containing 0.1% Tween 20 (PBST) and then incubated with HRP-conjugated secondary antibody for 1 h at room temperature. After washing, each protein was detected using an ECL system (Amersham Biosciences).
RESULTS
IL-32α Stimulates NK Susceptibility of CML Cells
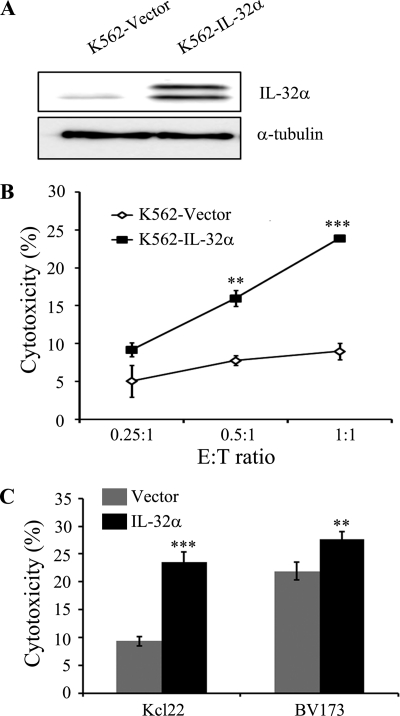
It has been reported that marrow stromal cells from CML patients have low expression of IL-32, and NK cell activity in CML patients is lower than in healthy controls. In this study, we hypothesized that the expression of IL-32 may correlate with NK cell activation in CML. To confirm this, we constructed IL-32α-overexpressing K562 cells. Fig. 1A shows high expression of IL-32α is detected in IL-32α-overexpressing K562 cells. To study the effect of IL-32α on the susceptibility of CML cells to NK cells, we performed an NK cytotoxicity assay. NK-92MI, an IL-2 transfected human NK cell line, was used as effector cells, and K562 cells were used as target cells. Fig. 1B shows that NK cytolytic activity increased with higher E:T ratio in all groups of target cells. Surprisingly, IL-32α-overexpressing K562 cells were more sensitive to NK-92MI cells than K562 vector control cells. As shown in Fig. 1C, overexpression of IL-32α in other CML cell lines, Kcl22 and BV173, significantly increased susceptibility to NK cells.
FIGURE 1.
Elevated NK susceptibility by IL-32α in CML cells. A, IL-32α expression in IL-32α-overexpressing K562 cells. K562 transfectants (vector or IL-32α) were lysed in cell lysis buffer as described under “Experimental Procedures,” the cell lysates were resolved on SDS-PAGE, and IL-32 expression was assessed by Western blotting with KU32–52 (mouse anti-human IL-32). The blot is representative of three independent experiments. B, shown is the NK susceptibility of K562 stable transfectants (vector and IL-32α). K562-stable transfectants (vector or IL-32α) were used as target cells, and IL-2-transfected human NK cells, NK-92MI, were used as effector cells. Each group of target cells was stained using CFSE for distinction from effector cells during flow cytometry analysis. CFSE-stained target cells were incubated with effector cells at a E:T ratio (0.25:1, 0.5:1, and 1:1) for 1 h. After incubation, co-incubated effector cells and target cells were stained with 7-AAD. The killing effect was analyzed by measurement of percentage of the CFSE and 7-AAD double-positive region. C, shown is the NK susceptibility of IL-32α-overexpressing CML Kcl22 and BV173 cell lines. Each transfectant was assayed for NK cytotoxicity. NK-92MI cells were used as effector cells at an E:T ratio of 2:1. Data presented are representative of three independent data sets for each condition (**, p < 0.05; ***, p < 0.001 versus vector control).
Overexpression of IL-32α Enhances Fas and ULBP2 Expression at both mRNA and Protein Levels in CML Cells
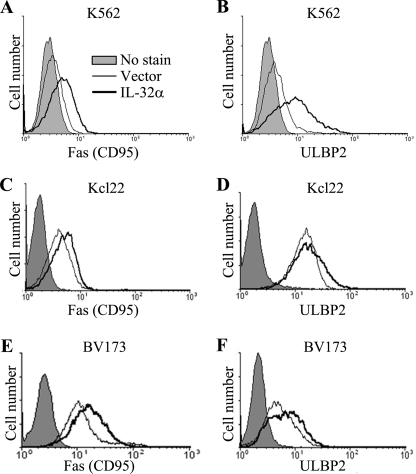
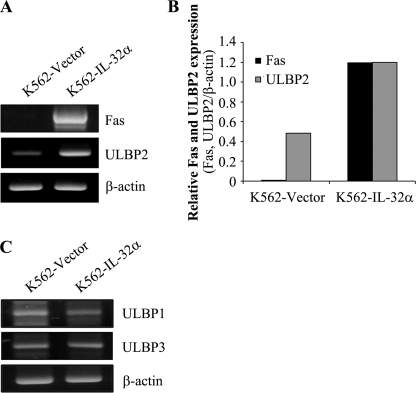
To identify the mechanism involved in IL-32α-enhanced NK susceptibility of CML cells, vector- and IL-32α-stable transfectants were used to test the expression of various surface molecules related to NK susceptibility. It has been reported that Fas transfers apoptotic signaling after engagement of Fas ligand on immune cells, including NK cells and T cells (13). ULBP2 and MICA/B are known ligands of NKG2D, which is a typical NK-activating receptor expressed on NK cell surface (7, 8). Additionally, ICAM-1 plays a critical role in conjugate formation between target cells and NK cells (14). Interestingly, cell surface expression of Fas and ULBP2 was dramatically increased in IL-32α-overexpressing K562 cells (Fig. 2, A and B), Kcl22 cells (Fig. 2, C and D), and BV173 cells (Fig. 2, E and F). However, IL-32α did not affect the expression of MICA/B and ICAM-1 (data not shown). In addition, to confirm whether IL-32α also stimulates Fas and ULBP2 mRNA expression, we performed RT-PCR. In Fig. 3, A and B, the expression of Fas and ULBP2 was increased in IL-32α-overexpressing K562 cells. In addition to ULBP2, ULBP1 and ULBP3 also are critical NKG2D ligands that interact with NKG2D and stimulate NK cytolytic activity and also are enhanced during tumor progression (15). To examine whether IL-32α stimulates ULBP2 specifically or also affects ULBP1 and ULBP3 expression, we tested ULBP1 and ULBP3 expression in IL-32α-overexpressing K562 cells using RT-PCR. Interestingly, ULBP1 and ULBP3 mRNA expression was not affected by IL-32α (Fig. 3C). These data demonstrate that IL-32α induced Fas and ULBP2 expression at both mRNA and protein levels, and the effect of IL-32α on ULBPs is limited to ULBP2.
FIGURE 2.
Induction of Fas and ULBP2 protein expression by IL-32α in CML cells. Surface expression of Fas and ULBP2 after IL-32α overexpression was measured in CML cell lines. CML transfectants were stained with PE-conjugated anti-human Fas and PE-conjugated anti-human ULBP2. FAS and ULBP2 expression in K562 (A and B), Kcl22 (C and D), and BV173 (E and F) was analyzed using flow cytometry. Data presented are representative of three independent data sets for each condition.
FIGURE 3.
Induction of Fas and ULBP2 expression by IL-32α at mRNA level in K562 cells. Total RNAs were purified from K562 stable transfectants (vector or IL-32α) using TRIzol, and cDNAs were made using 2 μg of total RNA. cDNAs were used as templates for PCR amplification with Fas, ULBP1, ULBP2, ULBP3, and β-actin primers. A, Fas and ULBP2 expression by RT-PCR. B, Fas and ULBP2 expression by densitometry analysis using the TotalLabTM program. C, ULBP1 and ULBP3 expression by RT-PCR. Data presented are representative of three independent data sets for each condition.
Increased NK Susceptibility by IL-32α Occurred through Up-regulation of Fas and ULBP2
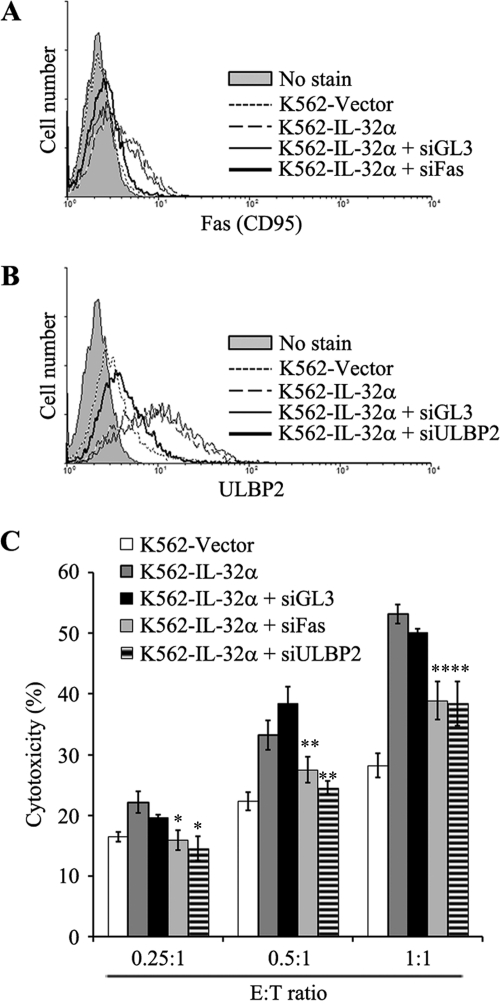
To test whether IL-32α-induced Fas and ULBP2 expression is functional in elevated NK susceptibility by IL-32α, we performed an NK cytotoxicity assay after blocking Fas and ULBP2 expression in IL-32α-overexpressing K562 cells using siRNA. Fig. 4, A and B, show that siRNAs of Fas and ULBP2 effectively suppressed Fas and ULBP2 expression in IL-32α-overexpressing K562 cells. Using these cells as target cells, we tested NK susceptibility. In Fig. 4C, IL-32α-induced cell death of K562 cells were effectively inhibited by Fas siRNA or ULBP2 siRNA. This indicates that IL-32α-induced NK susceptibility of K562 cells is due to up-regulation of Fas and ULBP2 by IL-32α.
FIGURE 4.
Blocking effect of siRNA against Fas and ULBP2 in IL-32α-induced NK susceptibility in K562 cells. IL-32α-induced Fas and ULBP2 were blocked with specific siRNA. IL-32α-overexpressing K562 cells were transiently transfected with nonrelated control siRNA (GL3 siRNA), Fas siRNA, and ULBP2 siRNA using MicoPorator electroporation. After 48 h, K562 vector, K562-IL-32α, and siRNA-transfected K562-IL-32α (siGL3, siFas, and siULBP2) cells were analyzed. A, Fas expression in K562-vector, K562-IL-32α, and siRNA-transfected K562-IL-32α (siGL3 and siFas) cells. B, ULBP2 expression in K562 vector, K562-IL-32α, and siRNA-transfected K562-IL-32α (siGL3 and siULBP2) cells. C, NK susceptibility test. K562 vector, K562-IL-32α, and siRNA-transfected K562-IL-32α (siGL3, siFas, and siULBP2) cells were used as target cells, and NK-92MI cells were used as effector cells. Target cells were stained with CFSE as described under “Experimental Procedures,” and then the cells were incubated with NK-92MI cells at an optimal E:T ratio (E:T ratio: 0.25:1, 0.5:1, and 1:1). After a 1-h incubation, co-incubated effector cells and target cells were stained with 7-AAD. The killing effect was analyzed by measurement of percentage of CFSE and 7-AAD double-positive region. Data presented are representative of three independent data sets for each condition (*, p < 0.01; **, p < 0.05 versus K562-IL-32α).
IL-32α Stimulates Fas and ULBP2 Expression through p38 MAPK Activation
It has been demonstrated that IL-32 stimulates inflammatory response through phosphorylation of p38 MAPK (1), indicating that p38 MAPK plays an important role in the IL-32 signaling pathway. Additionally, it is known that activation of p38 MAPK is also involved in Fas expression (16). We tested whether activation of p38 MAPK is required for IL-32α-induced Fas and ULBP2 expression. IL-32α-overexpressing K562 cells were incubated with various concentrations of p38 MAPK specific inhibitor, SB203580 (5, 10, 20, and 40 μm), for 48 h, and Fas and ULBP2 expression was analyzed in K562 vector transfectants, IL-32α-overexpressing K562 cells, and SB203580-treated IL-32α-overexpressing K562 cells. Interestingly, IL-32α-induced Fas and ULBP2 expression were reduced by SB203580 treatment in a dose-dependent manner (data not shown), and maximal effect was shown with 40 μm of SB203580 treatment (Fig. 5A). This indicates that activation of p38 MAPK may be involved in IL-32α-induced Fas and ULBP2 expression. To test whether IL-32α stimulates activation of p38 MAPK in K562 cells, K562 cells were transiently transfected with IL-32α, and phosphorylation of p38 MAPK was analyzed. We confirmed overexpression of IL-32α and elevated phosphorylation of p38 MAPK (Fig. 5B). These data suggest that IL-32α stimulates Fas and ULBP2 expression through phosphorylation of p38 MAPK.
FIGURE 5.
p38 MAPK activation in IL-32α-induced Fas and ULBP2 expression. IL-32α-overexpressing K562 cells were incubated with 40 μm SB203580 (SB) for 48 h. After incubation time, the cells were stained with PE-conjugated anti-human Fas (CD95) Ab and PE-conjugated anti human ULBP2 Ab. A, Fas and ULBP2 expression by histogram analysis. B, K562 cells were transiently transfected with vector (pcDNA3.1+) or IL-32α cDNA (IL-32α/pcDNA3.1+). Αfter 24 h, K562 vector transfectants and IL-32α transfectants were lysed with cell lysis buffer as described under “Experimental Procedures.” Whole protein lysates were assessed with KU32–52 (mouse anti-human IL-32), rabbit anti-phosphor-p38 MAPK, and rabbit anti-total p38 MAPK. Data presented are representative of three independent data sets for each condition.
Transcriptional Regulation by Ets-1 Induces ULBP2-specific Up-regulation
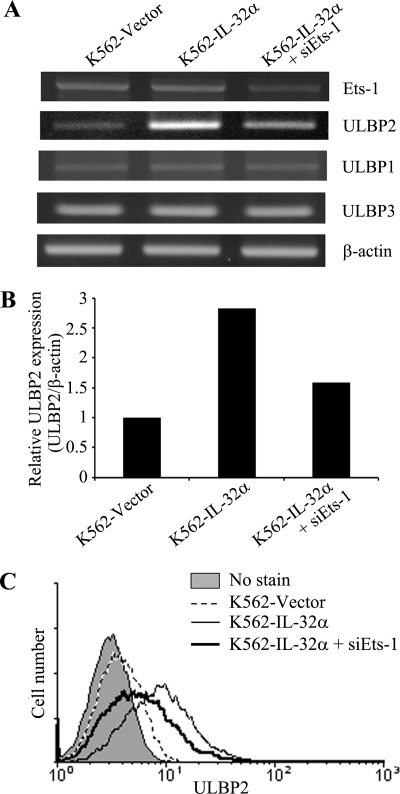
Activated p38 MAPK has been shown to phosphorylate several transcription factors (17). Moreover, screening of transcription factor binding sites using MATCHTM (version 1.0) revealed that the Ets-1 binding site in the promoter region of ULBP2 was differentially located from ULBP1 and ULBP3. To test whether activation of Ets-1 plays a key role in the expression of ULBP2, we analyzed the mRNA level of ULBP family members after Ets-1 siRNA transfection in IL-32α-overexpressing K562 cells. Interestingly, up-regulated ULBP2 mRNA in IL-32α-overexpressing K562 cells was decreased by suppression of Ets-1 (Fig. 6, A and B). However, the expression level of ULBP1 and ULBP3 was not affected by Ets-1 knockdown (Fig. 6A). In addition, down-regulation of Ets-1 also decreased surface expression of ULBP2 in IL-32α-overexpressing K562 cells (Fig. 6B). These data suggest that Ets-1 is an influential transcription factor for ULBP2-specific expression by IL-32α overexpression in CML cells.
FIGURE 6.
Blocking effect of Ets-1 in IL-32α-induced ULBP2 expression. K562 stable transfectants (vector or IL-32α) were transfected with Ets-1 siRNA. After incubation for 24 h, the cells were used for mRNA isolation, and the expression level of Ets-1 and ULBP family members was assessed. A, Ets-1 and ULBP family member expression by RT-PCR. B, ULBP2 expression by densitometry analysis using TotalLabTM program. C, protein level of ULBP2 on the Ets-1 down-regulated K562 stable transfectants (vector or IL-32α) was analyzed using flow cytometry. Data presented are representative of three independent data sets for each condition.
DISCUSSION
IL-32 is a newly discovered proinflammatory cytokine that is induced by TNF-α, IL-8, IL-6, and IL-1β. Additionally, it stimulates production of various inflammatory mediators such as TNF-α, IL-8, and IL-6 (1, 2) and has an important role in progression of various inflammatory disorders such as rheumatoid arthritis and inflammatory bowel disease. Although the expression and role of IL-32 in inflammatory disorders are well known, the effect of IL-32 in other diseases, including tumors, is still unclear. It has been reported that various proinflammatory mediators, including cyclooxygenase-2 and prostaglandin E2, affect tumor progression (18). Additionally, proinflammatory cytokine IL-18 also affects tumor processes, including proliferation, migration, invasion, and immune escape of tumor cells (19). Because IL-32 is a known IL-18-induced gene in NK cells, it is possible that IL-18 may also stimulate IL-32 expression in tumor cells and subsequently affect tumor progression.
This study demonstrated that IL-32α enhances NK susceptibility of CML cells through up-regulation of Fas and ULBP2. Because it has been demonstrated that many proinflammatory cytokines, including IL-1β, ΤΝF-α, IFN-γ, and IL-18, stimulate Fas and ULBP2 expression (20–22), induction of Fas and ULBP2 by IL-32α may be a common function of proinflammatory cytokine. Fas is a typical death receptor that binds to the Fas ligand on immune cells such as NK and T cells. Additionally, ULBP2 is a good target against NKG2D, an activating receptor of NK cells and T cells. Therefore, our data indicates that IL-32α has an anti-cancerous effect in CML cells. Marcondes et al. (23) also investigated the anti-cancerous effect of IL-32. They demonstrated that IL-32 is induced by TNF-α and stimulates apoptosis of cell in CML, which corroborates with our data. However, some studies have suggested a procancerous effect of IL-32. Nishida et al. (24) demonstrated that IL-32 is highly expressed in pancreatic cancer cells in comparison with normal pancreatic cells, has pro-cancerous effects that inhibit apoptosis through enhancement of antiapoptotic proteins, and stimulates DNA synthesis in proliferation of pancreatic cancer cells. In stomach cancer, IL-32 also is highly expressed in cancerous tissue and serum of patients (25). These findings indicate that IL-32 has dual effects in tumor progression.
The Standard proinflammatory cytokine IL-18 also has both anticancerous and procancerous effects. It stimulates invasion and migration of cancer cells and regulates immune escape, although it also enhances immune response of T cells and NK cells. Therefore, IL-18-inducible IL-32 may also have dual effects in tumorigenesis. As reports have demonstrated, the anticancerous effect of IL-32 is limited to leukemia, but the procancerous effect of IL-32 is found in many types of solid tumors. Therefore, there is the possibility that the dual effect of IL-32 is due to differences between leukemia and solid tumors. Another possible explanation for the duel effects is the difference among IL-32 isoforms. IL-32 has six isoforms, IL-32α, IL-32β, IL-32γ, IL-32δ, IL-32ϵ, and IL-32ζ. However, the effect of each isoform is still unclear. Although this study demonstrates that IL-32α has anticancerous effects by stimulating expression of Fas and ULBP2, the function of other isoforms exclusive of IL-32α is still unknown. Therefore, more studies are needed to determine the function of each IL-32 isoform in tumor progression.
It has been reported that p38 MAPK is involved in IL-32-induced inflammatory response, and many transcription factors such as ATF1 and -2, MEF2A, Sap-1, Elk-1, NF-κB, Ets-1, and p53 are activated by p38 MAPK (17, 26). Even though one report has shown that transcription factor Sp3 is the essential activator of ULBP1 transcription (27), the exact regulating mechanism of p38 MAPK in the expression of ULBP family members is not well known. Because the environment of transcription binding sites influences the regulation of target gene expression (28), variation in the transcription factor binding site might be one of the causes of differential transcription of target gene. Among many transcription factors, Ets-1 is regulated by p38 MAPK and has various biological functions, including inducing cellular growth and differentiation and organ development (29). Ets-1 also enhances target gene transcription synergistically with other transcription factors (30). In this study, we found that ULBP2 contains four Ets-1 binding sites in the promoter region, which were within 94 base pairs of each other (−211 to −202, −196 to −187, −130 to −121 and −126 to −117), whereas ULBP3 has only one Ets-1 binding site on the promoter region (−52 to −43). ULBP1 does not enclose an Ets-1 binding site. On the other hand, there is no Ets-1 binding site in the Fas promoter region, and the expression level of Fas was not regulated by Ets-1 siRNA transfection (data not shown). Even though these findings suggest that differential transcriptional regulation by Ets-1 is a possible candidate for ULBP2 specific overexpression, the direct effect of Ets-1 in ULBP2-specific regulation should be elucidated by promoter assay.
In summary, this study investigated the effect of IL-32α on chronic myeloid leukemia. IL-32α stimulates Fas and ULBP2 expression in CML cells through p38 MAPK activation, and IL-32α-induced Fas and ULBP2 play an important role in enhancement of NK susceptibility of CML cells. This indicates that IL-32α has an anticancerous effect and may be a good candidate for therapy of CML.
This work was supported by Grant 10033778 (2009) from the Ministry of Knowledge Economy and National Research Foundation of Korea Grant R11-2005-017-05003-0 funded by the Korean government (Ministry of Education & Science Technology).
- NK
- natural killer
- CML
- chronic myeloid leukemia
- ULBP
- UL16 binding protein
- PE
- phycoerythrin
- 7-AAD
- 7-aminoactinomycin D.
REFERENCES
- 1. Kim S. H., Han S. Y., Azam T., Yoon D. Y., Dinarello C. A. (2005) Immunity 22, 131–142 [DOI] [PubMed] [Google Scholar]
- 2. Joosten L. A., Netea M. G., Kim S. H., Yoon D. Y., Oppers-Walgreen B., Radstake T. R., Barrera P., van de Loo F. A., Dinarello C. A., van den Berg W. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi J. D., Bae S. Y., Hong J. W., Azam T., Dinarello C. A., Her E., Choi W. S., Kim B. K., Lee C. K., Yoon D. Y., Kim S. J., Kim S. H. (2009) Immunology 126, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goda C., Kanaji T., Kanaji S., Tanaka G., Arima K., Ohno S., Izuhara K. (2006) Int. Immunol. 18, 233–240 [DOI] [PubMed] [Google Scholar]
- 5. Gloire G., Charlier E., Piette J. (2008) Biochem. Pharmacol. 76, 1451–1458 [DOI] [PubMed] [Google Scholar]
- 6. Liang S., Zhang J., Wei H., Sun R., Tian Z. (2005) Int. Immunopharmacol. 5, 839–848 [DOI] [PubMed] [Google Scholar]
- 7. Bauer S., Groh V., Wu J., Steinle A., Phillips J. H., Lanier L. L., Spies T. (1999) Science 285, 727–729 [DOI] [PubMed] [Google Scholar]
- 8. Bahram S., Bresnahan M., Geraghty D. E., Spies T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6259–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jamieson A. M., Diefenbach A., McMahon C. W., Xiong N., Carlyle J. R., Raulet D. H. (2002) Immunity 17, 19–29 [DOI] [PubMed] [Google Scholar]
- 10. Raulet D. H. (2003) Nat. Rev. Immunol. 3, 781–790 [DOI] [PubMed] [Google Scholar]
- 11. Trinchieri G. (2003) Nat. Immunol. 4, 509–510 [DOI] [PubMed] [Google Scholar]
- 12. Kim K. H., Shim J. H., Seo E. H., Cho M. C., Kang J. W., Kim S. H., Yu D. Y., Song E. Y., Lee H. G., Sohn J. H., Kim J., Dinarello C. A., Yoon D. Y. (2008) J. Immunol. Methods 333, 38–50 [DOI] [PubMed] [Google Scholar]
- 13. Wajant H. (2006) Cancer Treat. Res. 130, 141–165 [DOI] [PubMed] [Google Scholar]
- 14. Lee H. R., Yoon S. Y., Kang H. B., Park S., Kim K. E., Cho Y. H., Kim S., Kim C. W., Cho B. J., Lee W. J., Bang S. I., Park H., Cho D. (2009) Immunol. Lett. 123, 72–76 [DOI] [PubMed] [Google Scholar]
- 15. Sutherland C. L., Rabinovich B., Chalupny N. J., Brawand P., Miller R., Cosman D. (2006) Blood 108, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 16. Chen K. C., Kao P. H., Lin S. R., Chang L. S. (2009) J. Cell Biochem. 106, 93–102 [DOI] [PubMed] [Google Scholar]
- 17. Kyriakis J. M., Avruch J. (2001) Physiol. Rev. 81, 807–869 [DOI] [PubMed] [Google Scholar]
- 18. Greenhough A., Smartt H. J., Moore A. E., Roberts H. R., Williams A. C., Paraskeva C., Kaidi A. (2009) Carcinogenesis 30, 377–386 [DOI] [PubMed] [Google Scholar]
- 19. Kim K. E., Song H., Kim T. S., Yoon D., Kim C. W., Bang S. I., Hur D. Y., Park H., Cho D. H. (2007) Oncogene 26, 1468–1476 [DOI] [PubMed] [Google Scholar]
- 20. Song H., Kim K. E., Hur D., Lim J. S., Yang Y., Cho B. J., Kim C. H., Kim T., Bang S., Lee W. J., Park H., Cho D. (2008) Immunol. Lett. 120, 103–107 [DOI] [PubMed] [Google Scholar]
- 21. Kondo M., Murakawa Y., Harashima N., Kobayashi S., Yamaguchi S., Harada M. (2009) Immunology 128, e589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gannagé M., Buzyn A., Bogiatzi S. I., Lambert M., Soumelis V., Dal Cortivo L., Cavazzana-Calvo M., Brousse N., Caillat-Zucman S. (2008) Transplantation 85, 911–915 [DOI] [PubMed] [Google Scholar]
- 23. Marcondes A. M., Mhyre A. J., Stirewalt D. L., Kim S. H., Dinarello C. A., Deeg H. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2865–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishida A., Andoh A., Inatomi O., Fujiyama Y. (2009) J. Biol. Chem. 284, 17868–17876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seo E. H., Kang J., Kim K. H., Cho M. C., Lee S., Kim H. J., Kim J. H., Kim E. J., Park D. K., Kim S. H., Choi Y. K., Kim J. M., Hong J. T., Yoon D. Y. (2008) J. Microbiol. Biotechnol. 18, 1606–1612 [PubMed] [Google Scholar]
- 26. Netea M. G., Lewis E. C., Azam T., Joosten L. A., Jaekal J., Bae S. Y., Dinarello C. A., Kim S. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3515–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. López-Soto A., Folgueras A. R., Seto E., Gonzalez S. (2009) Oncogene 28, 2370–2382 [DOI] [PubMed] [Google Scholar]
- 28. Dai Z., Dai X., Xiang Q., Feng J. (2009) Genomics Proteomics Bioinformatics 7, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe D., Takagi H., Suzuma K., Suzuma I., Oh H., Ohashi H., Kemmochi S., Uemura A., Ojima T., Suganami E., Miyamoto N., Sato Y., Honda Y. (2004) Am. J. Pathol. 164, 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Logan S. K., Garabedian M. J., Campbell C. E., Werb Z. (1996) J. Biol. Chem. 271, 774–782 [DOI] [PubMed] [Google Scholar]