Abstract
Histone deacetylases (HDACs) are negative regulators of transcription. Endochondral bone formation including chondrocyte and osteoblast maturation is regulated by HDACs. Very little is known about the role HDACs play in osteoclast differentiation. It has been previously reported that HDAC inhibitors, trichostatin A and sodium butyrate, suppress osteoclast differentiation through multiple mechanisms. In this study, we report that suppression of HDAC3 expression similar to HDAC inhibitors inhibits osteoclast differentiation, whereas osteoclasts suppressed for HDAC7 expression had accelerated differentiation when compared with control cells. Mitf, a transcription factor, is necessary for osteoclast differentiation. We demonstrate that Mitf and HDAC7 interact in RAW 264 cells and osteoclasts. The transcriptional activity of Mitf is repressed by HDAC7. Lastly, we show that either the amino or the carboxyl terminus of HDAC7 is sufficient for transcriptional repression and that the repression of HDAC7 is insensitive to trichostatin A, indicating that HDAC7 represses Mitf at least in part by deacetylation-independent mechanism.
Keywords: Bone, Cell Differentiation, Histone Deacetylase, shRNA, Transcription Regulation, HDAC, MITF, Osteoclast
Introduction
Formation of the skeleton is a complex process involving coordinated function of osteoclasts and osteoblasts. Subsequently, bone homeostasis is maintained by osteoblasts, which synthesize mineralized bone, and osteoclasts, highly specialized multinuclear cells that resorb bone. Many skeletal diseases such as osteoporosis, Paget disease of bone, rheumatoid arthritis, and bone metastases arise from an imbalance in the relative activities of osteoblasts and osteoclasts.
Histone deacetylases are best known for promoting transcriptional repression and silencing through the removal of acetyl groups from histone core proteins at target gene promoters, resulting in a less transcriptionally active state (1). They also catalyze deacetylation of non-histone substrates, thereby regulating the stability or activity of their substrates (2). The 18 human HDACs3 fall into four classes based on structural and biochemical characteristics. Class I HDACs include HDACs 1, 2, 3, and 8, whereas Class II HDACs include 4, 5, 6, 7, 9, and 10. Class II HDACs are further subdivided into two subclasses, IIa and IIb. Class IIa HDACS (HDACs 4, 5, 7, and 9) can also interact with additional non-deacetylase co-repressor proteins to bring about transcriptional repression (1). Broad spectrum HDAC inhibitors (HDIs) such as trichostatin A (TSA) and sodium butyrate (NaB) are thought to have an effect on cells by inhibition of HDACs. TSA is generally considered a nonspecific HDAC inhibitor as it has similar Ki for all isoforms examined (3), and NaB has been shown to inhibit all Class I and II HDACs.
A number of HDACs regulate chondrocyte and osteoblast differentiation and activity through interactions with transcription factors such as Runx2, Smads, Twist, and pRb (4–11). Interactions between HDACs and Runx2 inhibit the activity of Runx2, thereby suppressing osteoblast differentiation (11, 12), whereas HDIs accelerate osteoblastic differentiation (11).
Very little is known about how HDACs regulate osteoclast gene expression and differentiation. HDIs, TSA, and NaB have been shown to inhibit receptor activator of NF-κB ligand (RANKL)-mediated osteoclast differentiation due to inhibition of c-fos expression, NF-κB-dependent transcription, and p38 MAP kinase activity (13, 14). HDAC1 is recruited to the promoters of osteoclast genes by STAT3 and Eos-Mitf-Pu.1 complex (15, 16). Hu et al. demonstrated that co-repressors CtBP, HDAC1, and Sin3A were present on Ctsk and Acp5 promoters when osteoclast precursors were stimulated with macrophage colony-stimulating factor (M-CSF), but their levels were significantly reduced following 3 days of combined stimulation with M-CSF and RANKL (16). The cytokines RANKL and M-CSF are necessary and sufficient for osteoclast differentiation (17). The combination of these two factors activates transcription factors such as Nfatc1, Mitf, PU.1, and c-Fos (18–22), which are necessary for osteoclast differentiation. Mitf belongs to the MiT family of basic helix-loop-helix transcription factors that regulate gene expression in a variety of cell types including melanocytes, macrophages, and osteoclasts (23). The MiT family includes Mitf, Tfe3, Tfeb, and Tfec (24, 25). The importance of Mitf in osteoclast differentiation is confirmed by the lack of osteoclast differentiation and the resulting osteopetrotic phenotype observed in mice homozygous for the Mitf (mi) allele. A similar osteopetrotic phenotype is also noted in the double mutants homozygous for the Mitf (vga) hypomorphic allele and the Tfe3 null allele (24, 26, 27). Recent results indicate that the Mitf complex integrates signals necessary for the appropriate temporal regulation of osteoclast genes such as Ctsk and Acp5 during differentiation. M-CSF signaling alone can regulate Mitf nuclear localization and recruitment of Mitf to target promoters (28). However, Mitf does not activate gene expression with stimulation of M-CSF alone. Rather, combined stimulation with M-CSF and RANKL is required to induce expression of osteoclast differentiation genes (29).
In the current work, we demonstrate that suppression of HDAC3 by shRNA closely mirrors the inhibitory effect of HDIs on osteoclast formation. Unexpectedly, we show that suppression of HDAC7 has the opposite effect, enhancing osteoclast formation. Further experiments support a model in which HDAC7 inhibits osteoclast differentiation by repressing Mitf activity. Lastly, we show that repression of Mitf by HDAC7 is deacetylation-independent.
EXPERIMENTAL PROCEDURES
Cell Culture, Luciferase Assays, and Transfections
Osteoclasts were isolated from bone marrow of mice as described previously. Bone marrow was flushed from femurs, and the resulting cells were cultured for 3 days in the presence of 50 ng/ml M-CSF on non-tissue culture-coated dishes. The adherent cell population, containing the osteoclasts, was cultured for the indicated times and amounts of M-CSF and RANKL. RAW 264.7 c4 cells were grown in DMEM supplemented with 10% FBS, 25 units/ml penicillin, 25 mg/ml streptomycin. RAW 264.7 c4 is a cell clone derived in Dr. A. Ian Cassady's laboratory at the University of Queensland from commercially available RAW 264.7 cells (American Type Culture Collection, Manassas, VA) that requires both M-CSF and RANKL for efficient differentiation into osteoclast-like cells, but not for growth or survival. These cells were a gift obtained from Dr. A. Ian Cassady and Dr. David Hume. The conditions for differentiating RAW 264.7 c4 cells into osteoclasts-like cells were previously described (28, 30). Differentiation of RAW 264.7c4 and osteoclasts were optimized using 10 ng/ml M-CSF and 60 ng/ml RANKL (R&D Systems, Minneapolis, MN). NIH 3T3 and 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum, 2% l-glutamine, and 0.5% penicillin/streptomycin (Invitrogen). NIH 3T3 and 293T cells were transiently transfected by Lipofectamine Plus reagent (Invitrogen) according to the instructions of the manufacturer. The luciferase activities were measured using the Luciferase Assay System (Promega) according to the instructions of the manufacturer.
Antibodies and Chemicals
Polyclonal Mitf antibody was generated by 21st Century Biochemicals (Marlboro, MA) using a peptide containing mouse Mitf amino acids 85–96 as an immunogen. HDAC7 antibody (clone A7), Myc (clone 9E10), gal4 DNA binding domain (clone RK5C1), and actin (clone I-19) were purchased from Santa Cruz Biotechnology; HDAC3 (clone 7G6C5) and histone H3 (9715) were purchased from Cell Signaling; and acetylated histone H3 (06-599) was purchased from Upstate/Millipore. M-CSF and RANKL were purchased from R&D Systems and used at 10 ng/ml (M-CSF) or 60 ng/ml (RANKL). TSA was used at 20 nm (Sigma).
Immunoprecipitation and Western Blotting
24 h after transfection, the 293T cells were harvested in Nonidet P-40 lysis buffer (20 mm Tris, pH 8.0, 137 mm NaCl, 10% glycerol, 1% Nonidet P-40, and protease and phosphatase inhibitors). Extracts were incubated with target antibody and protein A/G beads (Pierce) overnight at 4 °C. On the next day, immunoprecipitates were washed three times in lysis buffer. The bound proteins were resuspended in sample buffer and resolved by SDS-PAGE. The resolved proteins were transferred to PVDF membrane, blocked, and blotted in primary antibody overnight at 4 °C. On the next day, the blot was incubated with horseradish peroxidase anti-rabbit or anti-mouse (GE Healthcare) for 1 h at room temperature. Antibody binding was detected using the ECL system (GE Healthcare).
Plasmids
FLAG Mitf was previously described (28). FLAG HDAC7 and FLAG HDAC7 1–478 and 472–912 were previously described (12). pMI-Mitf contains Mitf amino acid residues 1–185 cloned into an expression plasmid fusing the Gal4 DNA binding domain to the amino terminus of Mitf.
RNA Isolation and Real-time PCR
RNA was extracted using TRIzol (Invitrogen). 2 μg of purified total RNA was reverse-transcribed by the iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time RT-PCR was performed using the MyiQ single-color real-time PCR detection system (Bio-Rad) using 1 μl of the cDNA with 2× iQ SYBR Green supermix (Bio-Rad). Osteoclast genes were normalized to L4 RNA. Primer sequences used were NFATc1 (forward) 5′-TCA TCC TGT CCA ACA CCA AA-3′, (reverse) 5′-TCA CCC TGG TGT TCT TCC TC-3′; cathepsin K (forward) 5′-AGG GAA GCA AGC ACT GGA TA-3′, (reverse) 5′-GCT GGC TGG AAT CAC ATC TT-3′; DC-STAMP (forward) 5′-GGG CAC CAG TAT TTT CCT GA-3′, (reverse) 5′-TGG CAG GAT CCA GTA AAA GG-3′; HDAC3 (forward) 5′-CTG GCT TCT GCT ATG TCA AC-3′, (reverse) 5′-ACA TAT TCA ACG CAT TCC CCA-3′; and HDAC7 (forward) 5′-TGA AGA ATG GCT TTG CTG TG-3′, (reverse) 5′-CAC TGG GGT CCT GGT AGA AA-3′. All measurements were performed in triplicate, analyzed using the 2−ΔΔCt method, and amplified with equal efficiencies.
Lentiviral Infection of Osteoclasts
Lentiviral vectors encoding shRNAs against HDAC7 (Ref. 12 or Open Biosystems) or HDAC3 (Open Biosystems) or a control shRNA were used to produce replication-defective lentivirus according to the manufacturer's protocols. Viral stocks were titrated by infection in HeLa cells. Following infection, primary osteoclast cultures were stimulated with M-CSF and RANKL.
TRAP Staining
Cells were rinsed in PBS, fixed in 4% paraformaldehyde for 20 min, and stained using the leukocyte acid phosphatase kit (Sigma-Aldrich, 387-A). Cells were photographed and analyzed using Adobe Photoshop to measure the number and size of TRAP-positive osteoclasts.
Counting Nuclei
After cells were TRAP-stained and photographed, cells were stained with DAPI (Molecular Probes) for 5 min at room temperature. Cells were rinsed and stained with Vybrant cell-labeling dye (Molecular Probes) for 15 min at 37 °C. Cells were photographed at 10× magnification and counted using Adobe Photoshop.
Statistical Analysis
All experiments were run in triplicate, and results are expressed as mean ± S.D. Each experiment was done at least three times, and the error bars in Figs. 1–4, 6, and 7 represent independent experiments. Student's t tests or a one-way analysis of variance analysis followed by a Tukey's multiple comparison test were used to compare data; p < 0.05 indicates significance.
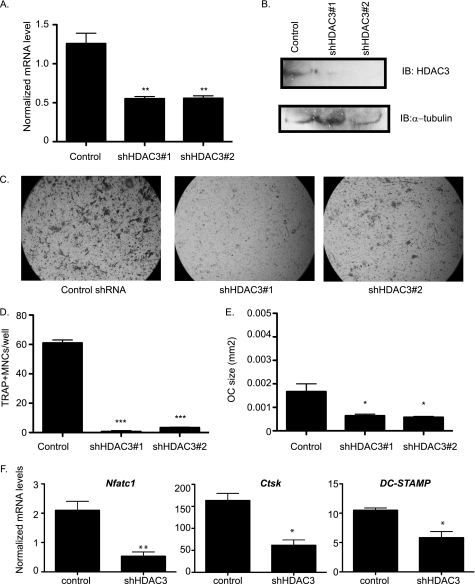
FIGURE 1.
Suppression of HDAC3 inhibits osteoclast differentiation. A, real-time RT-PCR of bone marrow. **, p ≤ 0.007 versus control shRNA. B, Western blot (IB) of RAW 264.7 lysates showing expression of HDAC3. C, TRAP staining of osteoclast cultures. D and E, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.0001 and *, p ≤ 0.05 versus control shRNA. F, expression profile of Nfat-c1, Ctsk, and DC-STAMP. *, p ≤ 0.05 and **, p ≤ 0.005 versus control shRNA following infection with control shRNA or HDAC3 shRNA lentiviral vectors.
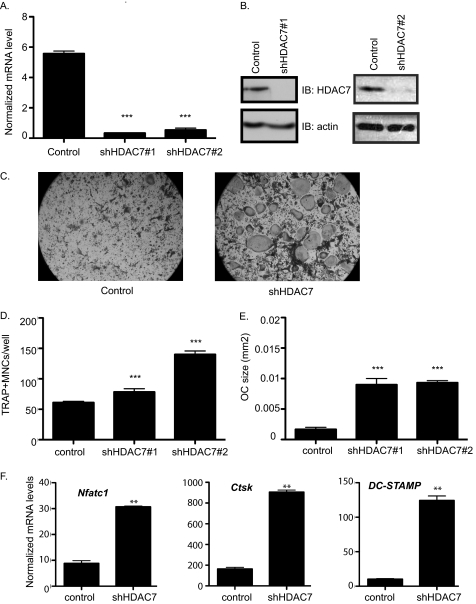
FIGURE 2.
Accelerated osteoclast differentiation in HDAC7-suppressed osteoclasts. A, real-time RT-PCR of bone marrow. ***, p ≤ 0.0001 versus control shRNA. B, Western blot (IB) of osteoclast lysates showing expression of HDAC7. C, TRAP staining of osteoclast cultures infected with control or HDAC7 shRNA-expressing lentiviruses. D and E, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.0001 versus control shRNA. F, expression profile of Nfat-c1, Ctsk, and DC-STAMP. **, p ≤ 0.001 versus control shRNA following infection with control shRNA or HDAC7 shRNA lentiviral vectors.
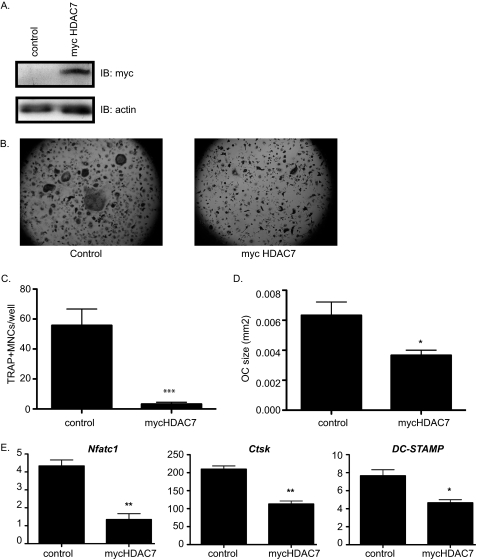
FIGURE 3.
Overexpression of Myc-HDAC7 inhibits osteoclast differentiation. A, Western blot (IB) of osteoclast lysates. B, TRAP-stained images. C and D, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.005 and *, p ≤ 0.05. E, expression profile of Nfat-c1, Ctsk, and DC-STAMP of osteoclast infected with control or Myc-HDAC7-expressing lentivirus. **, p ≤ 0.005 and *, p ≤ 0.05 versus control-infected cells.
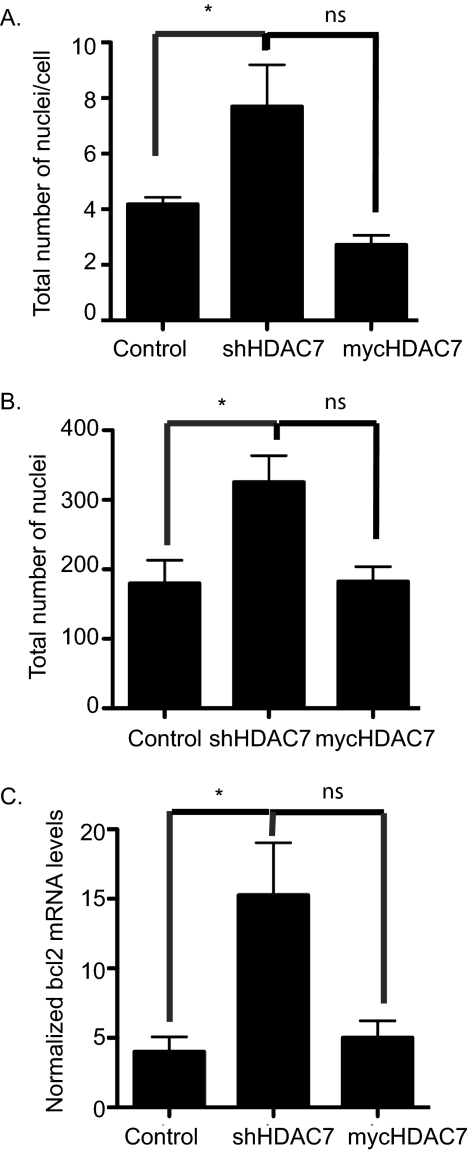
FIGURE 4.
HDAC7 expression inhibits cell-cell fusion. Day 5 osteoclast cultures were stained with DAPI and Vybrant cell-labeling dye, and histomorphometry was performed to assess the total number of nuclei per multinucleated cell (A) and the total number of nuclei (B). *, p < 0.01 versus control-infected cells, ns = not significant when compared with control-infected cells. C, expression profile of bcl2 in osteoclasts infected with control, shHDAC7-, or Myc-HDAC7-overexpressing lentivirus. *, p < 0.01 versus control-infected cells, ns = not significant when compared with control-infected cells.
FIGURE 6.
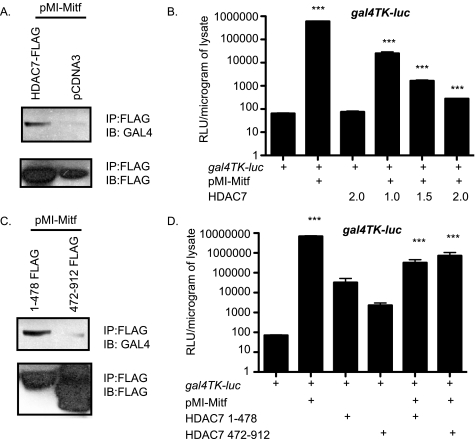
Amino terminus of Mitf is sufficient for HDAC7 repression. A, 293T cells were transfected with FLAG-HDAC7 and pMI-Mitf. Lysates were immunoprecipitated (IP) with FLAG antibody and immunoblotted (IB) with an antibody that recognizes the Gal4 DNA binding domain. B, NIH 3T3 cells were transiently transfected with 5× Gal4-TK-luciferase reporter construct and/or pMI-Mitf and full-length HDAC7. Reporter activity is presented as relative luciferase units (RLU), and the results of three experiments each performed in duplicate are presented. ***, p ≤ 0.0001 versus gal4-TK-luc or gal4-TK-luc+Mitf. C, 293T cells were transfected with FLAG-HDAC7 1–478 or 472–912 and pMI-Mitf. Lysates were immunoprecipitated with FLAG antibody and immunoblotted with an antibody that recognizes the Gal4 DNA binding domain. D, NIH 3T3 cells were transiently transfected with 5× Gal4-TK-luciferase reporter construct and/or pMI-Mitf and either HDAC7 1–478 or HDAC7 472–912. Reporter activity is presented as relative luciferase units, and the results of three experiments each performed in duplicate are presented. ***, p ≤ 0.0001 versus gal4-TK-luc or gal4-TK-luc+Mitf.
FIGURE 7.
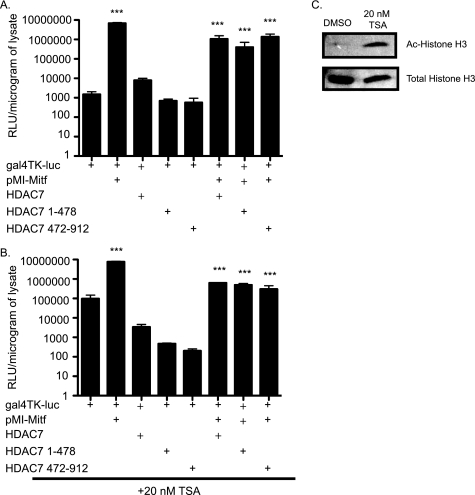
HDAC7 represses Mitf in a deacetylation-independent mechanism. A, NIH 3T3 cells were co-transfected with Gal4TK-luc, pMI-Mitf, and/or FLAG-HDAC7 1–428 or 422–912. Cells were treated with DMSO. Reporter activity is presented as relative luciferase units (RLU), and the results of three experiments each performed in duplicate are presented. B, repression of Mitf by HDAC7 is insensitive to TSA. Transfections were performed as described in A, and the cells were treated with 20 nm TSA for 16–20 h before harvest. ***, p ≤ 0.0001 versus gal4-TK-luc or gal4-TK-luc+Mitf. C, increased levels of acetylated histone H3 after TSA treatment. NIH 3T3 cells were treated with TSA in parallel to the experiment shown in B. Protein extracts were subjected to SDS-PAGE and immunoblotted against total and acetylated histone H3.
RESULTS
Suppression of HDAC3 Inhibits Osteoclast Differentiation
Previous reports indicated that the non-selective HDAC inhibitors TSA and NaB inhibit osteoclast formation (13). However, these data do not indicate the significance of individual HDACs to osteoclastogenesis. In light of its importance in regulating osteoblastic differentiation (11, 31), we asked how HDAC3 affects osteoclast formation. To this end, we made use of lentiviral vectors encoding shRNAs against HDAC3 or a control shRNA. To ensure that any effects were due to specific knockdown of HDAC3, we compared two distinct shRNAs against HDAC3. Infection of murine bone marrow cultures with either HDAC3 shRNA reduced HDAC3 mRNA expression by ∼50% when compared with the control shRNA (Fig. 1A). We observed a similar reduction in HDAC3 protein levels when we infected RAW 264.7 cells (Fig. 1B). Although control shRNA cultures readily formed TRAP-positive osteoclasts upon stimulation with RANKL, osteoclast formation was strongly reduced in cultures infected with either HDAC3 shRNA (Fig. 1C). Quantitative analysis of these cultures indicated that both the number and the size of TRAP-positive cells were significantly reduced in HDAC3 shRNA cultures when compared with the controls (Fig. 1, D and E). Similarly, quantitative RT-PCR analysis revealed that expression of osteoclast marker genes NFAT-c1, cathepsin K (Ctsk), and DC-STAMP was reduced in the HDAC3 shRNA cells (Fig. 1F). These data indicate that reduction of HDAC3 expression, like treatment with HDIs, impairs osteoclastogenesis and suggest that HDAC3 activity is necessary for osteoclast formation.
HDAC7 Activity Inhibits Osteoclast Differentiation
We next examined the effect of suppressing expression of the Class II deacetylase, HDAC7. Infection of bone marrow cultures with the previously described HDAC7 shRNA vector (12) or an HDAC7 shRNA purchased from Open Biosystems reduced HDAC7 mRNA levels by 16- or 10-fold, respectively, when compared with the control vector (Fig. 2A), and HDAC7 protein levels showed a clear reduction (Fig. 2B). In contrast to the HDAC3-suppressed cells, osteoclast differentiation in HDAC7-suppressed cultures was enhanced when compared with controls (Fig. 2C). The average size of TRAP-positive multinucleated osteoclasts in HDAC7-suppressed cells was increased 5.6- and 5.8-fold (Fig. 2E), and the mean number of TRAP-positive multinucleated cells per well showed a 1.3- and 2.3-fold increase (Fig. 2D). Real-time RT-PCR demonstrated that HDAC7-suppressed osteoclasts showed increased expression of osteoclast markers: Nfat-c1 (3.5-fold increase, p ≤ 0.009), Ctsk (5.5-fold increase, p ≤ 0.02), and DC-STAMP (12-fold increase, p ≤ 0.008, Fig. 2F). These results reveal that HDAC7 suppression enhances osteoclast formation, thus suggesting that HDAC7 activity inhibits osteoclastogenesis.
To further test the hypothesis that HDAC7 inhibits osteoclastogenesis, we made use of a lentiviral vector to overexpress Myc-tagged HDAC7. Immunoblotting detected strong expression of a Myc-tagged protein of 115 kDa, the predicted molecular mass of HDAC7 (Fig. 3A). When these cultures were stimulated with RANKL, their differentiation into TRAP-positive multinucleated osteoclasts was impaired when compared with control cells (Fig. 3B). Overexpression of Myc-HDAC7 reduced the number of TRAP-positive cells 17-fold (Fig. 3C) and reduced the size of the osteoclasts 1.75-fold (Fig. 3D). Expression of osteoclast marker genes NFAT-c1, Ctsk, and DC-STAMP was significantly reduced (Fig. 3E). Taken together, these data demonstrate that HDAC7 acts as an inhibitor of osteoclast differentiation.
Overexpression of HDAC7 Prevents Fusion of Osteoclast Precursors
We next wanted to determine whether the enhanced osteoclast differentiation we detected with suppressed HDAC7 expression was due to increased fusion of osteoclast precursors. When we compared nuclei per cell, we detected a 1.8-fold increase in nuclei per cell in the osteoclasts expressing the shRNA against HDAC7 (Fig. 4A), whereas the number of nuclei per cell was not statistically different in the mycHDAC7-overexpressing osteoclasts when compared with the control-infected cells (Fig. 4A). As shown in Fig. 4B, we detected significantly more total nuclei (1.7-fold increase, p ≤ 0.009) in osteoclasts expressing the shRNA against HDAC7 when compared with the control-infected osteoclasts. We detected no difference in the total nuclei in the mycHDAC7-overexpressing osteoclasts when compared with the control-infected cells (Fig. 4B). The increase in the total number of nuclei that we detect in shHDAC7-expressing osteoclasts suggests that HDAC7 functions to decrease proliferation or increase apoptosis in osteoclasts. Mitf has been shown to activate the bcl2 promoter in melanocytes and osteoclasts (39). We analyzed expression of bcl2 and detected a 3.75-fold increase in bcl2 expression in the HDAC7-suppressed cells when compared with control-infected cells (Fig. 4C). We detected no significant increase in bcl2 expression in the Myc-HDAC7-overexpressing cells when compared with the control-infected cells (Fig. 4C).
HDAC7 Binds to Mitf in Osteoclasts
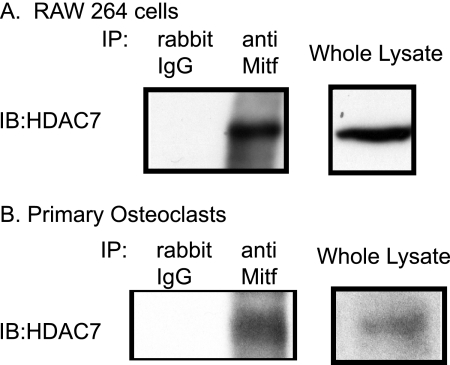
The Mitf transcription factor is one of the essential regulators of osteoclast formation. Although treatment with M-CSF recruits Mitf to target gene promoters, co-stimulation with M-CSF and RANKL is necessary for efficient activation of Mitf-dependent transcription. This suggests that corepressors prior to stimulation with RANKL might inhibit Mitf targets. Based on its effect as an inhibitor of osteoclast differentiation, we hypothesized that HDAC7 might interact with Mitf. We immunoprecipitated lysates from RAW 264.7 c4, a stable subclone of the commercially available RAW 264.7 cells, or primary osteoclasts with an antibody against Mitf or an IgG control and examined the lysates by immunoblotting against HDAC7. We detected strong interaction between Mitf and HDAC7 in RAW 264.7 c4 cells (Fig. 5A). A similar interaction between Mitf and HDAC7 was detected in primary osteoclasts, which were just stimulated with M-CSF (Fig. 5B). We did not detect any HDAC7 when cell lysates were immunoprecipitated with the control IgG antibody (Fig. 5, left lane). These data indicate that HDAC7 and Mitf can interact in RAW 264.7 c4 cells and osteoclasts.
FIGURE 5.
HDAC7 interacts with Mitf. A and B, representative Western blots (IB) of Mitf immunoprecipitates (IP) from RAW 264.7 c4 cells (A) and primary mouse osteoclasts (B) immunoblotted against HDAC7 (Abcam). Osteoclasts or RAW 264.7 c4 cells were stimulated with M-CSF (10 ng/ml) for 2 days.
HDAC7 Represses Mitf-dependent Transcription
We previously showed that the amino terminus of Mitf contains the domains necessary for activation of osteoclast genes (32). To further characterize the interaction between Mitf and HDAC7 and to determine whether the Mitf amino terminus is sufficient for functional interaction between HDAC7 and Mitf, we cloned Mitf amino acid residues 1–185, which encompass the transactivation domains of Mitf, into an expression plasmid fusing the Gal4 DNA binding domain to the amino terminus of Mitf (pMI-Mitf). To further map the interaction between Mitf and HDAC7, we overexpressed pMI-Mitf and FLAG-HDAC7 in 293T cells. Cell lysates were immunoprecipitated with an antibody that recognizes FLAG-HDAC7 and analyzed by Western blot for the presence of pMI-Mitf. We could only detect the amino terminus of Mitf in the presence of the FLAG-HDAC7 (Fig. 6A). In NIH 3T3 cells, the resulting pMI-Mitf activated the gal4-TK luciferase reporter nearly 100,000-fold, whereas expression of HDAC7 alone had no effect on the reporter (Fig. 6B). We decided to study the effect of HDAC7 on the activation of the gal-TK luciferase reporter by Mitf in NIH 3T3 cells because NIH 3T3 cells do not express endogenous Mitf like RAW 246.7 cells, and NIH 3T3 cells are easier to transfect than osteoclast precursors. Co-transfection of pMI-Mitf with increasing amounts of HDAC7 gave a dose-dependent significant repression of the Mitf activity (Fig. 6B, compare pMI-Mitf to pMI-Mitf+HDAC7).
To map the interaction between HDAC7 and Mitf, we overexpressed HDAC7 1–478 and HDAC7 472–912 and pMI-Mitf in 293T cells. Cell lysates were immunoprecipitated with antibody that recognizes either HDAC7 1–478 or HDAC7 472–912 and analyzed by Western blot for the presence of pMI-Mitf. We could only detect the amino terminus of Mitf in the presence of HDAC7 1–478 (Fig. 6C). To determine what region or regions of HDAC7 mediate Mitf repression, we transfected NIH 3T3 cells with gal4-TK luciferase reporter, pMI-Mitf, and either HDAC7 1–478 or HDAC 472–912, which encode the amino-terminal domain and the carboxyl-terminal deacetylase domain of HDAC7 (Fig. 6D). Both the amino-terminal and the carboxyl-terminal portions of HDAC7 gave strong and significant repression of Mitf activity.
Repression by HDAC7 Does Not Require Deacetylation
HDAC7 has been shown to repress transcription through deacetylation-dependent and -independent mechanisms. If HDAC7 is repressing Mitf activity through a deacetylation-independent mechanism, then HDAC7 should be able to repress the activation of Mitf in the presence of the broad spectrum HDAC inhibitor TSA. When we repeated the transfections shown in Fig. 7A in the presence of TSA, we found that HDAC7 represses the activation of the gal4-TK luciferase by Mitf similarly regardless of the presence of TSA (Fig. 7B). Moreover, the repressive activities of 1–478 and 472–912 HDAC7 fragments were also unaffected by TSA. To verify the activity of TSA in these experiments, parallel cultures were treated with TSA or DMSO vehicle and subjected to immunoblotting against total histone H3 and acetylated histone H3. Fig. 7C shows that overnight treatment with TSA increased the level of acetylated histone H3, consistent with reduced HDAC activity in the TSA-treated cells. These data indicate that multiple domains of HDAC7 are involved in repression of Mitf and that neither deacetylase catalytic activity nor the HDAC7 deacetylase domain is required for functional repression of Mitf.
DISCUSSION
Several studies have shown that HDIs, TSA, and NaB directly inhibit osteoclastogenesis (13, 14). The current study is among the first to identify the HDACs to be involved in this inhibitory effect on osteoclast formation. We show that inhibition of HDAC3 mirrors the effects of TSA and NaB. Thus, HDAC3 activity is necessary for osteoclast formation. Targets of HDAC3 in osteoclasts are not yet known, although HDIs inhibit RANKL induction of p38 activity and expression of c-fos and NFAT-c1 (13). Yi et al. found that HDIs lead to activation of apoptotic pathways in osteoclast precursors (33). HDIs are being investigated as potential therapeutic agents for a variety of cancers, HIV/AIDS, asthma, and central nervous system disorders (34–37). Thus, understanding their impact on skeletal physiology is important and potentially clinically relevant. HDIs accelerate osteoblastic differentiation both in vitro and in vivo (11, 12). This observation, together with the discovery that HDIs or suppression of HDAC3 inhibit osteoclast formation, might lead to the prediction that HDIs may lead to increased bone mass. However, studies have suggested that long term administration of the histone deacetylase inhibitor valproic acid for treatment of epilepsy may actually be associated with decreased bone mass (38). The cellular and physiological explanations for this effect remain unclear.
Our current studies reveal an unexpected function for HDAC7 in osteoclasts that is distinct from the function of HDAC3. Suppression of HDAC7 enhanced osteoclast formation, whereas overexpression of HDAC7 impairs osteoclast formation, indicating that HDAC7 represses osteoclast differentiation. Analyzing total nuclei of osteoclasts expressing the shRNA against HDAC7 indicates that suppression of HDAC7 expression leads to an increase in total nuclei when compared with control-infected cells. Furthermore, we were able to detect an increase in bcl2 mRNA expression in HDAC7-suppressed cells. Mitf has been shown to activate the bcl2 promoter in melanocytes and osteoclasts (39). We have hypothesized that loss of HDAC7 would lead to more Mitf activation of the bcl2 promoter, a prosurvival factor, and may account for the increase in the total number of nuclei that we detected in shHDAC7-expressing osteoclasts.
M-CSF stimulation recruits Mitf to the nucleus and localizes Mitf in a complex with PU.1 to osteoclast promoters (29). However, M-CSF stimulation alone is not sufficient to activate gene expression. We were able to detect an interaction between endogenous Mitf and HDAC7 when the osteoclasts were stimulated with M-CSF only. It is the combination of M-CSF and RANKL signaling that leads to phosphorylated, activated Mitf and recruitment of RNA polymerase II, and robust gene expression (29). Based on our ability to detect an interaction between Mitf and HDAC7 in osteoclasts, we hypothesize a model in which treatment with RANKL disrupts the interaction between Mitf and HDAC7, permitting Mitf-mediated transcriptional activation. Studies are currently underway to test this hypothesis by using ChIP analysis of Mitf target genes during osteoclast differentiation. This is similar to the scenario proposed for osteoblast differentiation where BMP2 stimulation transiently relocates HDAC7 from the nucleus to the cytoplasmic compartment, thereby permitting Runx2-dependent transcription (12). We have not detected a change in HDAC7 subcellular localization following RANKL treatment.4 RANKL stimulation leads to phosphorylation of Mitf on specific residues (20), suggesting possible mechanisms by which the Mitf-HDAC7 interaction might be regulated. However, we cannot rule out that HDAC7 also interacts with other transcription factors involved in osteoclast differentiation like PU.1 or NFAT-c1. It also remains unclear whether the altered expression of any particular gene in the HDAC7-suppressed cells reflects regulation by HDAC7 directly or indirectly.
The contrast between HDAC7 suppression and HDAC3 suppression or HDI treatments on osteoclast formation suggests that the effects of HDAC7 in osteoclasts are mediated through distinct mechanisms that do not require deacetylase activity. In further support of this, we found that repression of the gal4-TK promoter by HDAC7 is insensitive to treatment with TSA and showed that the HDAC7 deacetylase domain is dispensable for repression. The amino terminus of Class IIa HDACs represses transcription by a deacetylase-independent mechanism(s), which include interactions with CtBP and HP1 (40). The carboxyl-terminal portion of HDAC7 also possessed robust repressive activity, which was insensitive to TSA in our assays. This is similar to the physical and repressive interactions between HDAC7 and Runx2, which also appear to involve multiple regions of HDAC7 (12). As with Mitf, HDAC7 repression of Runx2 is insensitive to TSA and can be mediated by the amino-terminal domain (12). HDAC7 and other Class IIa HDACs can regulate transcription by recruiting large repressive complexes containing Class I HDACs, Sin3, and SMRT-N-CoR (41–45). The HDAC3 interaction domain of HDAC7 was mapped to amino acids 438–912 (42), which are included in the carboxyl-terminal construct we used. Hu et al. have demonstrated by ChIP assay that HDAC1, Sin3A, and CtBP are recruited to the Ctsk and Acp5 promoters during M-CSF stimulation, but their association is decreased with RANKL signaling (16). It is possible that one mechanism used by HDAC7 during M-CSF stimulation of osteoclast is based on recruitment of such repressive complexes. This recruitment of repressive complexes could explain why the carboxyl terminus of HDAC7 inhibits Mitf activation even when we detect no interaction between the C terminus of HDAC7 and Mitf by co-immunoprecipitation (Fig. 6).
This study establishes HDAC7 as a novel Mitf repressor. Understanding the mechanisms by which HDAC7 represses Mitf function, the significance of HDACs in the context of the larger transcription complex (c-Fos-Mitf-PU.1-Nfatc1) at osteoclast promoters, and the regulatory inputs that coordinate their activity will be significant areas for future studies to increase our understanding of skeletal development, maintenance, and pathological states.
Acknowledgment
We acknowledge A. Ian Cassady for the gift of the RAW 264.7 c4 cells.
This work was supported, in whole or in part, by National Institutes of Health Grants R03 DE020117 (to E. D. J.) and R01 AR056642 (to R. G.) and MinnCResT Grant T32 DE007288 from the NIDCR (to L. P.).
E. D. Jensen and K. C. Mansky, unpublished observation.
- HDAC
- histone deacetylase
- HDI
- HDAC inhibitor
- TSA
- trichostatin A
- RANKL
- receptor activator of NF-κB ligand
- TRAP
- tartrate-resistant acid phosphatase
- NaB
- sodium butyrate
- DMSO
- dimethyl sulfoxide
- CtBP
- C-terminal-binding protein.
REFERENCES
- 1. Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 [DOI] [PubMed] [Google Scholar]
- 2. Gregoretti I. V., Lee Y. M., Goodson H. V. (2004) J. Mol. Biol. 338, 17–31 [DOI] [PubMed] [Google Scholar]
- 3. Drummond D. C., Noble C. O., Kirpotin D. B., Guo Z., Scott G. K., Benz C. C. (2005) Annu. Rev. Pharmacol. Toxicol. 45, 495–528 [DOI] [PubMed] [Google Scholar]
- 4. Luo R. X., Postigo A. A., Dean D. C. (1998) Cell 92, 463–473 [DOI] [PubMed] [Google Scholar]
- 5. Westendorf J. J., Zaidi S. K., Cascino J. E., Kahler R., van Wijnen A. J., Lian J. B., Yoshida M., Stein G. S., Li X. (2002) Mol. Cell. Biol. 22, 7982–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang J. S., Alliston T., Delston R., Derynck R. (2005) EMBO J. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jeon E. J., Lee K. Y., Choi N. S., Lee M. H., Kim H. N., Jin Y. H., Ryoo H. M., Choi J. Y., Yoshida M., Nishino N., Oh B. C., Lee K. S., Lee Y. H., Bae S. C. (2006) J. Biol. Chem. 281, 16502–16511 [DOI] [PubMed] [Google Scholar]
- 8. Lee H. W., Suh J. H., Kim A. Y., Lee Y. S., Park S. Y., Kim J. B. (2006) Mol. Endocrinol. 20, 2432–2443 [DOI] [PubMed] [Google Scholar]
- 9. Westendorf J. J. (2006) J. Cell. Biochem. 98, 54–64 [DOI] [PubMed] [Google Scholar]
- 10. Hayashi M., Nimura K., Kashiwagi K., Harada T., Takaoka K., Kato H., Tamai K., Kaneda Y. (2007) J. Cell Sci. 120, 1350–1357 [DOI] [PubMed] [Google Scholar]
- 11. Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 12. Jensen E. D., Schroeder T. M., Bailey J., Gopalakrishnan R., Westendorf J. J. (2008) J. Bone Miner. Res. 23, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahman M. M., Kukita A., Kukita T., Shobuike T., Nakamura T., Kohashi O. (2003) Blood 101, 3451–3459 [DOI] [PubMed] [Google Scholar]
- 14. Kim H. N., Ha H., Lee J. H., Jung K., Yang D., Woo K. M., Lee Z. H. (2009) Eur. J. Pharmacol. 623, 22–29 [DOI] [PubMed] [Google Scholar]
- 15. Kim K., Lee J., Kim J. H., Jin H. M., Zhou B., Lee S. Y., Kim N. (2007) J. Immunol. 178, 5588–5594 [DOI] [PubMed] [Google Scholar]
- 16. Hu R., Sharma S. M., Bronisz A., Srinivasan R., Sankar U., Ostrowski M. C. (2007) Mol. Cell. Biol. 27, 4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wada T., Nakashima T., Hiroshi N., Penninger J. M. (2006) Trends Mol. Med. 12, 17–25 [DOI] [PubMed] [Google Scholar]
- 18. Matsuo K., Owens J. M., Tonko M., Elliott C., Chambers T. J., Wagner E. F. (2000) Nat. Genet. 24, 184–187 [DOI] [PubMed] [Google Scholar]
- 19. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 20. Mansky K. C., Sankar U., Han J., Ostrowski M. C. (2002) J. Biol. Chem. 277, 11077–11083 [DOI] [PubMed] [Google Scholar]
- 21. Weilbaecher K. N., Motyckova G., Huber W. E., Takemoto C. M., Hemesath T. J., Xu Y., Hershey C. L., Dowland N. R., Wells A. G., Fisher D. E. (2001) Mol. Cell 8, 749–758 [DOI] [PubMed] [Google Scholar]
- 22. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 23. Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A., Fisher D. E. (1994) Genes Dev. 8, 2770–2780 [DOI] [PubMed] [Google Scholar]
- 24. Hershey C. L., Fisher D. E. (2004) Bone 34, 689–696 [DOI] [PubMed] [Google Scholar]
- 25. Steingrímsson E., Copeland N. G., Jenkins N. A. (2004) Annu. Rev. Genet. 38, 365–411 [DOI] [PubMed] [Google Scholar]
- 26. Steingrímsson E., Moore K. J., Lamoreux M. L., Ferré-D'Amaré A. R., Burley S. K., Zimring D. C., Skow L. C., Hodgkinson C. A., Arnheiter H., Copeland N. G., et al. (1994) Nat. Genet. 8, 256–263 [DOI] [PubMed] [Google Scholar]
- 27. Steingrimsson E., Tessarollo L., Pathak B., Hou L., Arnheiter H., Copeland N. G., Jenkins N. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bronisz A., Sharma S. M., Hu R., Godlewski J., Tzivion G., Mansky K. C., Ostrowski M. C. (2006) Mol. Biol. Cell 17, 3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma S. M., Bronisz A., Hu R., Patel K., Mansky K. C., Sif S., Ostrowski M. C. (2007) J. Biol. Chem. 282, 15921–15929 [DOI] [PubMed] [Google Scholar]
- 30. Meadows N. A., Sharma S. M., Faulkner G. J., Ostrowski M. C., Hume D. A., Cassady A. I. (2007) J. Biol. Chem. 282, 1891–1904 [DOI] [PubMed] [Google Scholar]
- 31. Razidlo D. F., Whitney T. J., Casper M. E., McGee-Lawrence M. E., Stensgard B. A., Li X., Secreto F. J., Knutson S. K., Hiebert S. W., Westendorf J. J. (2010) PLoS One 5, e11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mansky K. C., Marfatia K., Purdom G. H., Luchin A., Hume D. A., Ostrowski M. C. (2002) J. Leukoc. Biol. 71, 295–303 [PubMed] [Google Scholar]
- 33. Yi T., Baek J. H., Kim H. J., Choi M. H., Seo S. B., Ryoo H. M., Kim G. S., Woo K. M. (2007) Exp. Mol. Med. 39, 213–221 [DOI] [PubMed] [Google Scholar]
- 34. Gottesfeld J. M., Pandolfo M. (2009) Future Neurol. 4, 775–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tan J., Cang S., Ma Y., Petrillo R. L., Liu D. (2010) J. Hematol. Oncol. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Routy J. P. (2005) Lancet 366, 523–524 [DOI] [PubMed] [Google Scholar]
- 37. Halili M. A., Andrews M. R., Sweet M. J., Fairlie D. P. (2009) Curr. Top. Med. Chem. 9, 309–319 [DOI] [PubMed] [Google Scholar]
- 38. Vestergaard P., Rejnmark L., Mosekilde L. (2004) Epilepsia 45, 1330–1337 [DOI] [PubMed] [Google Scholar]
- 39. McGill G. G., Horstmann M., Widlund H. R., Du J., Motyckova G., Nishimura E. K., Lin Y. L., Ramaswamy S., Avery W., Ding H. F., Jordan S. A., Jackson I. J., Korsmeyer S. J., Golub T. R., Fisher D. E. (2002) Cell 109, 707–718 [DOI] [PubMed] [Google Scholar]
- 40. Dressel U., Bailey P. J., Wang S. C., Downes M., Evans R. M., Muscat G. E. (2001) J. Biol. Chem. 276, 17007–17013 [DOI] [PubMed] [Google Scholar]
- 41. Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. (2002) Mol. Cell 9, 45–57 [DOI] [PubMed] [Google Scholar]
- 42. Fischle W., Dequiedt F., Fillion M., Hendzel M. J., Voelter W., Verdin E. (2001) J. Biol. Chem. 276, 35826–35835 [DOI] [PubMed] [Google Scholar]
- 43. Kao H. Y., Downes M., Ordentlich P., Evans R. M. (2000) Genes Dev. 14, 55–66 [PMC free article] [PubMed] [Google Scholar]
- 44. Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. (2000) EMBO J. 19, 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guenther M. G., Barak O., Lazar M. A. (2001) Mol. Cell. Biol. 21, 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]